PAX9 Is Involved in Periodontal Ligament Stem Cell-like Differentiation of Human-Induced Pluripotent Stem Cells by Regulating Extracellular Matrix
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Reverse Transcription Polymerase Chain Reaction
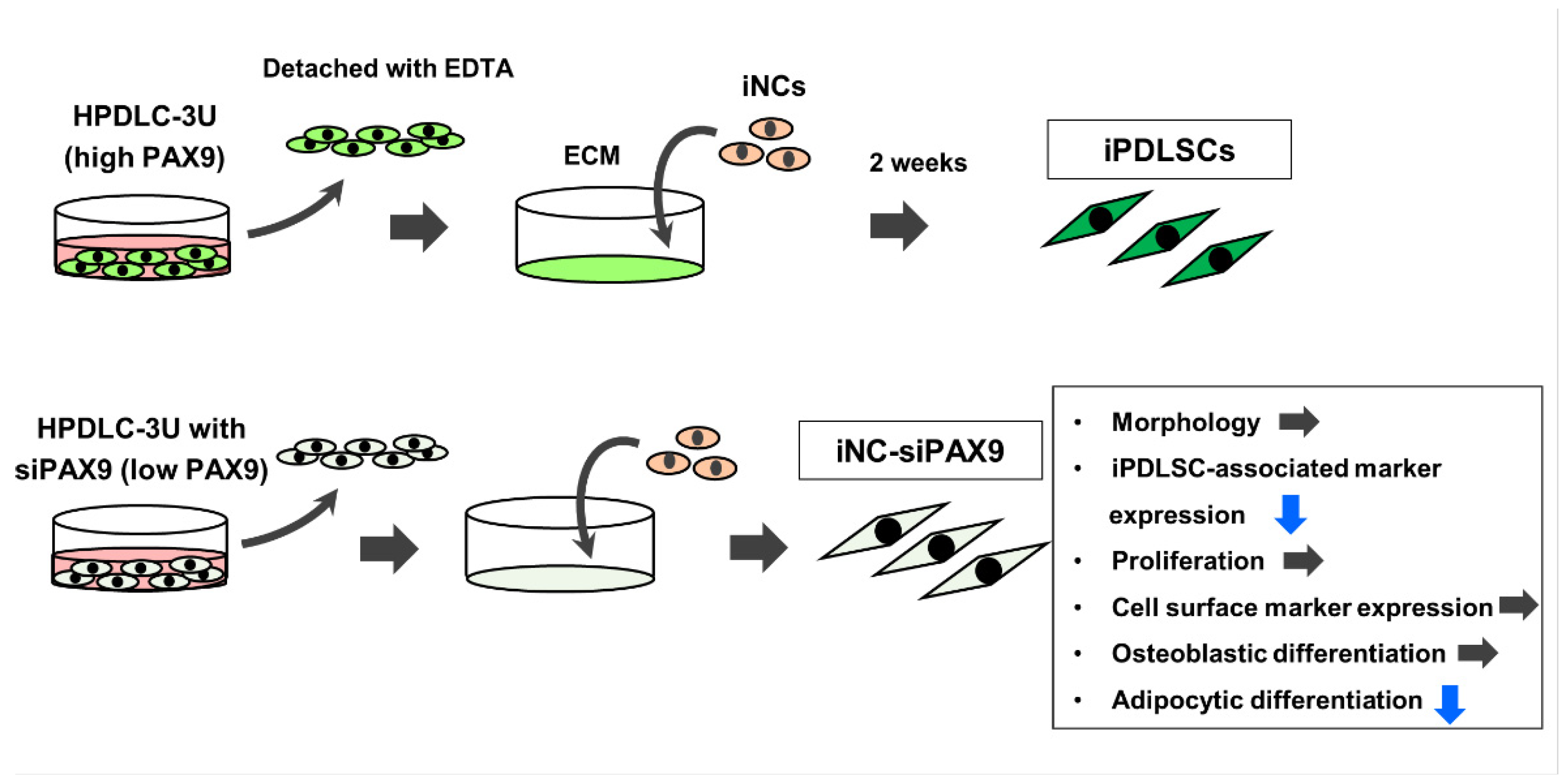
2.3. Differentiation of iPSC-Derived Neural Crest Cells (iNCs) Cultured on ECM Derived from HDF, HPDLC-3S, or HPDLC-3U
2.4. Cap Analysis Gene Expression and Gene Ontology Enrichment Analysis
2.5. Small Interfering RNA Transfection
2.6. Differentiation of iPSC-Derived Neural Crest Cells (iNCs) Cultured on ECM Derived from Unt-3U, 3U + siCont, and 3U + siPAX9
2.7. Immunofluorescence Staining
2.8. Flow Cytometric Analysis
2.9. Proliferation Assay
2.10. Osteoblastic Differentiation
2.11. Adipocytic Differentiation
2.12. Statistical Analysis
3. Results
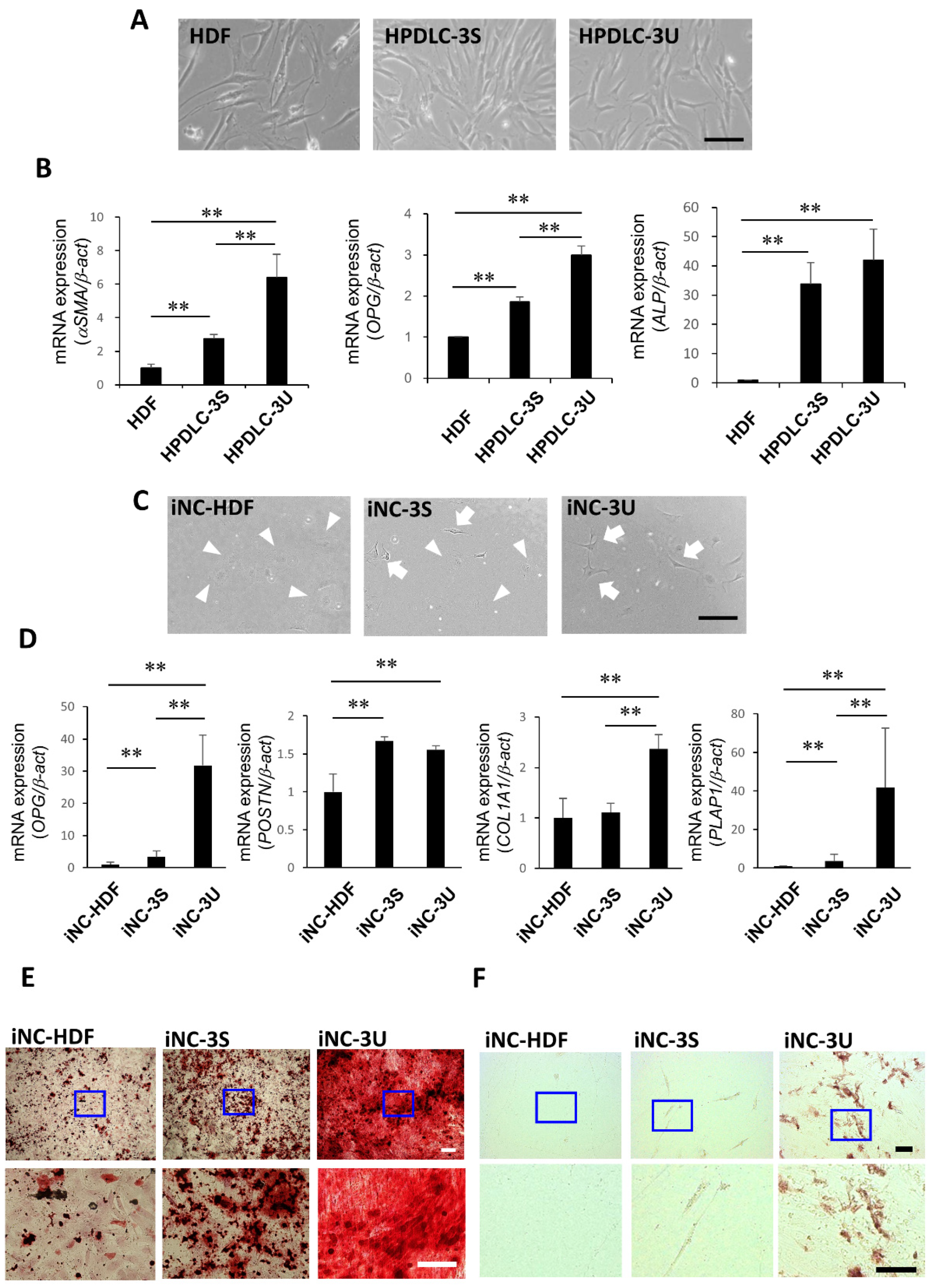
3.1. Periodontal Ligament Stem Cell Characteristics of iNC-HDF, iNC-3S, and iNC-3U
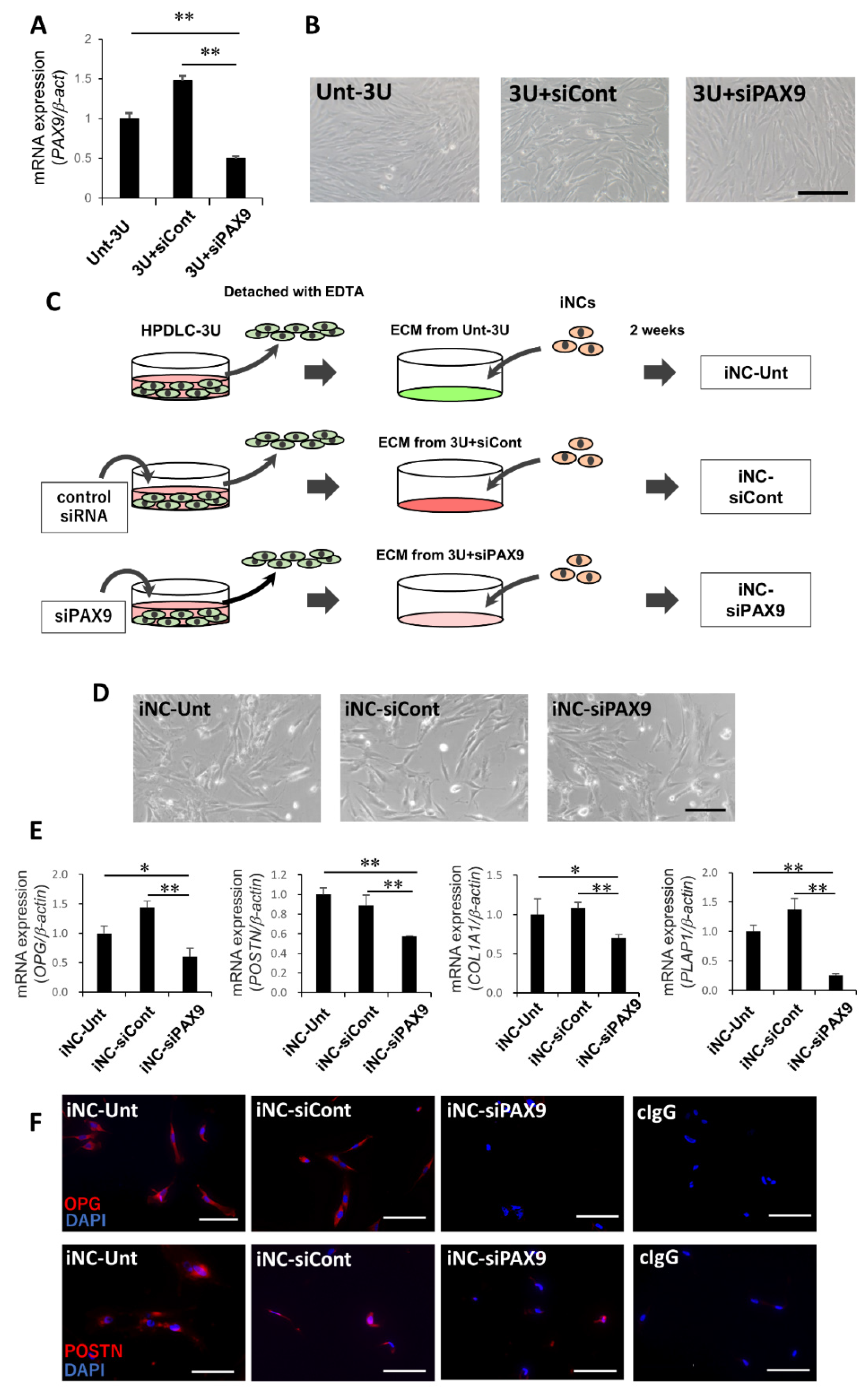
3.2. Identification of Transcription Factor PAX9 Responsible for the Induction of iNCs to iPDLSCs
3.3. PAX9 Gene Expression in Unt-3U, 3U + siCont, and 3U + siPAX9
3.4. iPDLSC-Associated Marker Expression in iNC-Unt, iNC-siCont, and iNC-siPAX9
3.5. Proliferation and Cell Surface Marker Expression in iNC-Unt, iNC-siCont, and iNC-siPAX9
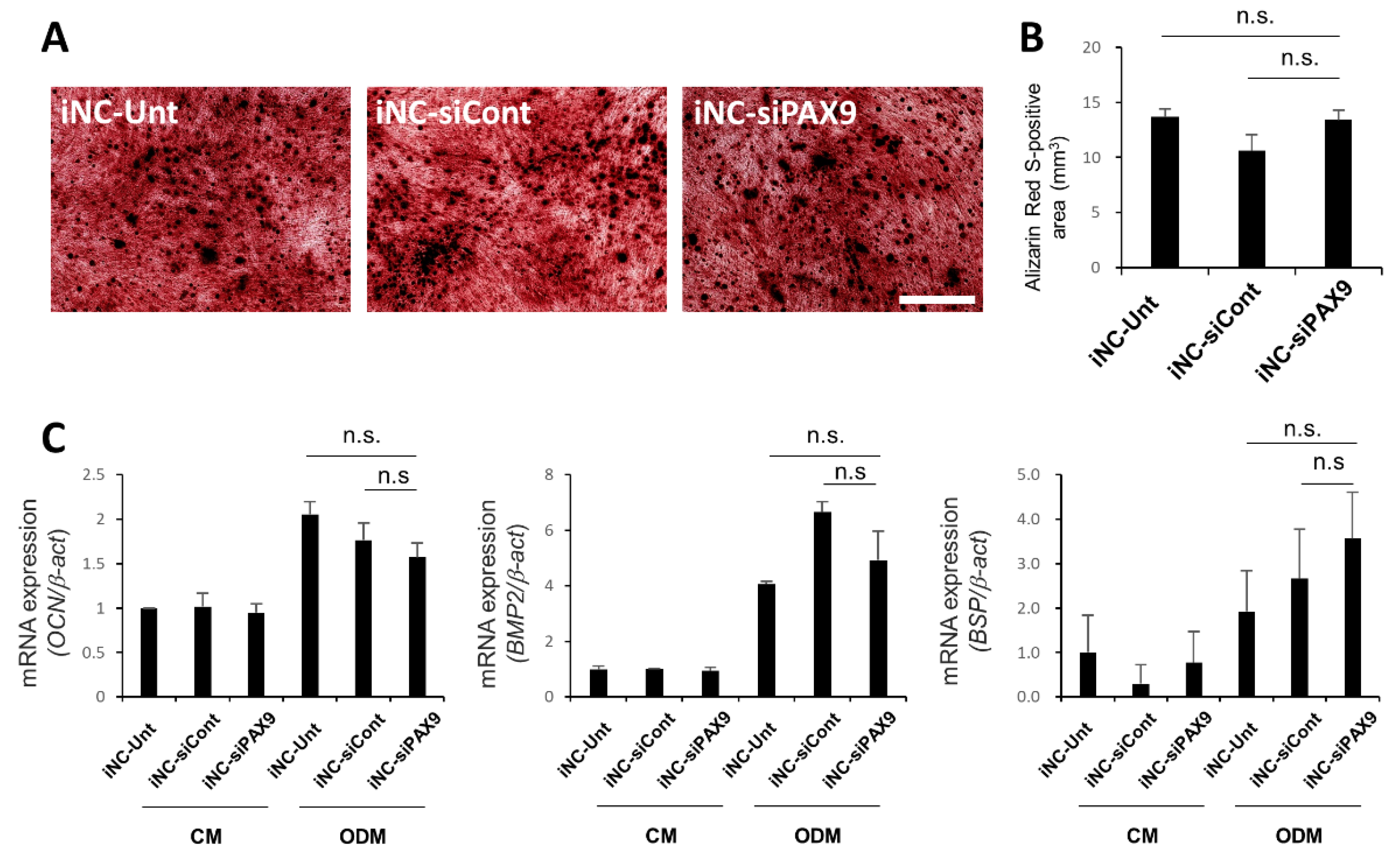
3.6. Osteoblastic Differentiation of iNC-Unt, iNC-siCont, and iNC-siPAX9
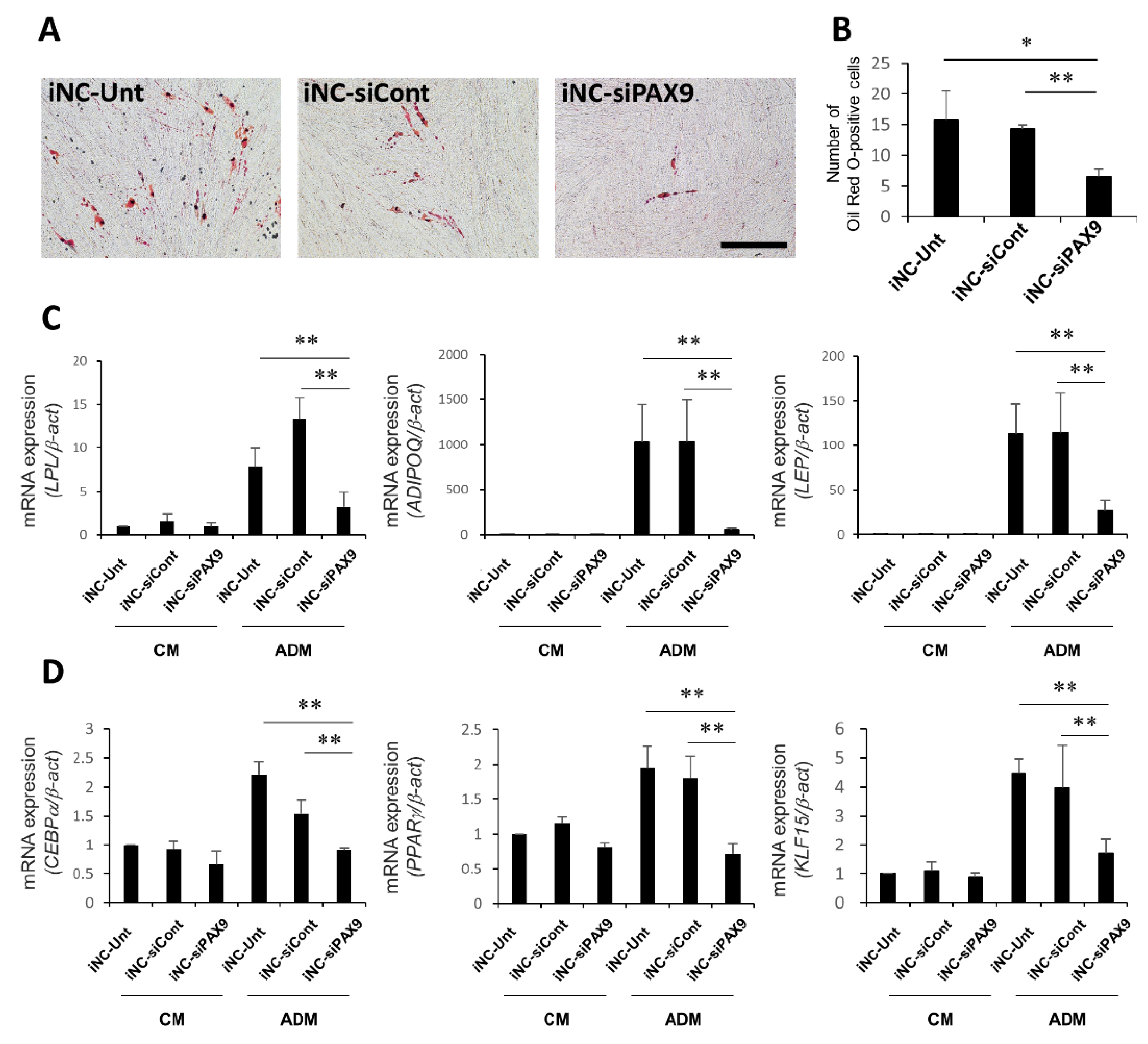
3.7. Adipocytic Differentiation of iNC-Unt, iNC-siCont, and iNC-siPAX9
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carnes, D.L.; Maeder, C.L.; Graves, D.T. Cells with osteoblastic phenotypes can be explanted from human gingiva and periodontal ligament. J. Periodontol. 1997, 68, 701–707. [Google Scholar] [CrossRef]
- Tomokiyo, A.; Maeda, H.; Fujii, S.; Wada, N.; Shima, K.; Akamine, A. Development of a multipotent clonal human periodontal ligament cell line. Differentiation 2008, 76, 337–347. [Google Scholar] [CrossRef]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Wada, N.; Menicanin, D.; Shi, S.; Bartold, P.M.; Gronthos, S. Immunomodulatory properties of human periodontal ligament stem cells. J. Cell Physiol. 2009, 219, 667–676. [Google Scholar] [CrossRef]
- Liu, D.; Xu, J.; Liu, O.; Fan, Z.; Liu, Y.; Wang, F.; Ding, G.; Wei, F.; Zhang, C.; Wang, S. Mesenchymal stem cells derived from inflamed periodontal ligaments exhibit impaired immunomodulation. J. Clin. Periodontol. 2012, 39, 1174–1182. [Google Scholar] [CrossRef]
- Liu, O.; Xu, J.; Ding, G.; Liu, D.; Fan, Z.; Zhang, C.; Chen, W.; Ding, Y.; Tang, Z.; Wang, S. Periodontal ligament stem cells regulate B lymphocyte function via programmed cell death protein 1. Stem Cells 2013, 31, 1371–1382. [Google Scholar] [CrossRef]
- Shin, C.; Kim, M.; Han, J.A.; Choi, B.; Hwang, D.; Do, Y.; Yun, J.H. Human periodontal ligament stem cells suppress T-cell proliferation via down-regulation of non-classical major histocompatibility complex-like glycoprotein CD1b on dendritic cells. J. Periodontal. Res. 2017, 52, 135–146. [Google Scholar] [CrossRef]
- Shalini, H.S.; Vandana, K.L. Direct application of autologous periodontal ligament stem cell niche in treatment of periodontal osseous defects: A randomized controlled trial. J. Indian Soc. Periodontol. 2018, 22, 503–512. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Shrestha, R. Induced pluripotent stem cells are Japanese brand sources for therapeutic cells to pretrial clinical research. Prog. Stem Cell 2020, 7, 296–303. [Google Scholar] [CrossRef]
- Sinenko, S.A.; Ponomartsev, S.V.; Tomilin, A.N. Pluripotent stem cell-based gene therapy approach: Human de novo synthesized chromosomes. Cell. Mol. Life Sci. 2021, 78, 1207–1220. [Google Scholar] [CrossRef]
- Skuratovskaia, D.; Litvinova, L.; Vulf, M.; Zatolokin, P.; Popadin, K.; Mazunin, I. From Normal to Obesity and Back: The Associations between Mitochondrial DNA Copy Number, Gender, and Body Mass Index. Cells 2019, 8, 430. [Google Scholar] [CrossRef]
- Attwood, S.W.; Edel, M.J. iPS-Cell Technology and the Problem of Genetic Instability-Can It Ever Be Safe for Clinical Use? J. Clin. Med. 2019, 8, 288. [Google Scholar] [CrossRef]
- Denham, M.; Dottori, M. Neural differentiation of induced pluripotent stem cells. Methods Mol. Biol. 2011, 793, 99–110. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Kasahara, T.; Sueta, S.I.; Araoka, T.; Sakamoto, S.; Okada, C.; Mae, S.I.; Nakajima, T.; Okamoto, N.; Taura, D.; et al. A Modular Differentiation System Maps Multiple Human Kidney Lineages from Pluripotent Stem Cells. Cell Rep. 2020, 31, 107476. [Google Scholar] [CrossRef]
- Karakikes, I.; Ameen, M.; Termglinchan, V.; Wu, J.C. Human induced pluripotent stem cell-derived cardiomyocytes: Insights into molecular, cellular, and functional phenotypes. Circ. Res. 2015, 117, 80–88. [Google Scholar] [CrossRef]
- Sun, J.; Mandai, M.; Kamao, H.; Hashiguchi, T.; Shikamura, M.; Kawamata, S.; Sugita, S.; Takahashi, M. Protective Effects of Human iPS-Derived Retinal Pigmented Epithelial Cells in Comparison with Human Mesenchymal Stromal Cells and Human Neural Stem Cells on the Degenerating Retina in rd1 mice. Stem Cells 2015, 33, 1543–1553. [Google Scholar] [CrossRef]
- Takayama, K.; Negoro, R.; Yamashita, T.; Kawai, K.; Ichikawa, M.; Mori, T.; Nakatsu, N.; Harada, K.; Ito, S.; Yamada, H.; et al. Generation of Human iPSC-Derived Intestinal Epithelial Cell Monolayers by CDX2 Transduction. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 513–526. [Google Scholar] [CrossRef]
- El Hokayem, J.; Cukier, H.N.; Dykxhoorn, D.M. Blood Derived Induced Pluripotent Stem Cells (iPSCs): Benefits, Challenges and the Road Ahead. J. Alzheimers Dis. Parkinsonism 2016, 6, 275. [Google Scholar] [CrossRef]
- Hamano, S.; Tomokiyo, A.; Hasegawa, D.; Yoshida, S.; Sugii, H.; Mitarai, H.; Fujino, S.; Wada, N.; Maeda, H. Extracellular Matrix from Periodontal Ligament Cells Could Induce the Differentiation of Induced Pluripotent Stem Cells to Periodontal Ligament Stem Cell-Like Cells. Stem Cells Dev. 2018, 27, 100–111. [Google Scholar] [CrossRef]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Ramakrishnan, P.R.; Lin, W.L.; Sodek, J.; Cho, M.I. Synthesis of noncollagenous extracellular matrix proteins during development of mineralized nodules by rat periodontal ligament cells in vitro. Calcif. Tissue Int. 1995, 57, 52–59. [Google Scholar] [CrossRef]
- Worapamorn, W.; Li, H.; Haas, H.R.; Pujic, Z.; Girjes, A.A.; Bartold, P.M. Cell surface proteoglycan expression by human periodontal cells. Connect. Tissue Res. 2000, 41, 57–68. [Google Scholar] [CrossRef]
- Tomokiyo, A.; Yoshida, S.; Hamano, S.; Hasegawa, D.; Sugii, H.; Maeda, H. Detection, Characterization, and Clinical Application of Mesenchymal Stem Cells in Periodontal Ligament Tissue. Stem Cells Int. 2018, 2018, 5450768. [Google Scholar] [CrossRef]
- Tomokiyo, A.; Wada, N.; Maeda, H. Periodontal Ligament Stem Cells: Regenerative Potency in Periodontium. Stem Cells Dev. 2019, 28, 974–985. [Google Scholar] [CrossRef]
- McKee, T.J.; Perlman, G.; Morris, M.; Komarova, S.V. Extracellular matrix composition of connective tissues: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 10542. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Chen, S.J.; Yuan, W.; Lo, S.; Trojanowska, M.; Varga, J. Interaction of smad3 with a proximal smad-binding element of the human alpha2(I) procollagen gene promoter required for transcriptional activation by TGF-beta. J. Cell. Physiol. 2000, 183, 381–392. [Google Scholar] [CrossRef]
- Larouche, K.; Leclerc, S.; Salesse, C.; Guérin, S.L. Expression of the alpha 5 integrin subunit gene promoter is positively regulated by the extracellular matrix component fibronectin through the transcription factor Sp1 in corneal epithelial cells in vitro. J. Biol. Chem. 2000, 275, 39182–39192. [Google Scholar] [CrossRef] [Green Version]
- Rockel, J.S.; Bernier, S.M.; Leask, A. Egr-1 inhibits the expression of extracellular matrix genes in chondrocytes by TNFalpha-induced MEK/ERK signalling. Arthritis Res. Ther. 2009, 11, R8. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, P. The Nrf2 transcription factor: A multifaceted regulator of the extracellular matrix. Matrix Biol. Plus 2021, 10, 100057. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Spencer, V.A.; Bissell, M.J. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J. Biol. Chem. 2007, 282, 14992–14999. [Google Scholar] [CrossRef]
- Kook, S.H.; Hwang, J.M.; Park, J.S.; Kim, E.M.; Heo, J.S.; Jeon, Y.M.; Lee, J.C. Mechanical force induces type I collagen expression in human periodontal ligament fibroblasts through activation of ERK/JNK and AP-1. J. Cell. Biochem. 2009, 106, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Chiba, T.; Miyazaki, T.; Yagasaki, L.; Nakamichi, R.; Iwata, T.; Moriyama, K.; Harada, H.; Asahara, H. Single Cell RNA Sequencing Reveals Critical Functions of Mkx in Periodontal Ligament Homeostasis. Front. Cell Dev. Biol. 2022, 10, 795441. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Maeda, H.; Tanabe, K.; Tsuda, E.; Yano, K.; Nakamuta, H.; Akamine, A. Periodontal ligament cells secrete the factor that inhibits osteoclastic differentiation and function: The factor is osteoprotegerin/osteoclastogenesis inhibitory factor. J. Periodontal. Res. 2001, 36, 56–63. [Google Scholar] [CrossRef]
- Lee, G.; Chambers, S.M.; Tomishima, M.J.; Studer, L. Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 2010, 5, 688–701. [Google Scholar] [CrossRef]
- Haberle, V.; Forrest, A.R.; Hayashizaki, Y.; Carninci, P.; Lenhard, B. CAGEr: Precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015, 43, e51. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Ipposhi, K.; Tomokiyo, A.; Ono, T.; Yamashita, K.; Alhasan, M.A.; Hasegawa, D.; Hamano, S.; Yoshida, S.; Sugii, H.; Itoyama, T.; et al. Secreted Frizzled-Related Protein 1 Promotes Odontoblastic Differentiation and Reparative Dentin Formation in Dental Pulp Cells. Cells 2021, 10, 2491. [Google Scholar] [CrossRef] [PubMed]
- Morgner, J.; Ghatak, S.; Jakobi, T.; Dieterich, C.; Aumailley, M.; Wickström, S.A. Integrin-linked kinase regulates the niche of quiescent epidermal stem cells. Nat. Commun. 2015, 6, 8198. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, S.; Kang, M.S.; Georgiou, K.; Kumar, S.; Schaffer, D.V. Dynamics of Mechanosensitive Neural Stem Cell Differentiation. Stem Cells 2017, 35, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Z.; Zhou, B.; Duan, X.; Weng, W.; Cheng, K.; Wang, H.; Lin, J. Cell-Sheet-Derived ECM Coatings and Their Effects on BMSCs Responses. ACS Appl. Mater. Interfaces 2018, 10, 11508–11518. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, X.; Zhang, X.; Duan, X.; Yang, P.; Zhao, F.; Ao, Y. Effects of mechanical stress on chondrocyte phenotype and chondrocyte extracellular matrix expression. Sci. Rep. 2016, 6, 37268. [Google Scholar] [CrossRef]
- He, Q.; Lin, Y.; Liao, B.; Zhou, L.; Ai, J.; Jin, X.; Li, H.; Wang, K. The role of interleukin-6/interleukin-6 receptor signaling in the mechanical stress-induced extracellular matrix remodeling of bladder smooth muscle. Arch. Biochem. Biophys. 2021, 702, 108674. [Google Scholar] [CrossRef]
- Tewksbury, C.D.; Callaghan, K.X.; Fulks, B.A.; Gerstner, G.E. Individuality of masticatory performance and of masticatory muscle temporal parameters. Arch. Oral Biol. 2018, 90, 113–124. [Google Scholar] [CrossRef]
- Xiong, X.; Yang, X.; Dai, H.; Feng, G.; Zhang, Y.; Zhou, J.; Zhou, W. Extracellular matrix derived from human urine-derived stem cells enhances the expansion, adhesion, spreading, and differentiation of human periodontal ligament stem cells. Stem Cell Res. Ther. 2019, 10, 396. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Paiboonrungruang, C.; Li, Y.; Peters, H.; Kist, R.; Xiong, Z. PAX9 in Cancer Development. Int. J. Mol. Sci. 2022, 23, 5589. [Google Scholar] [CrossRef]
- Peters, H.; Schuster, G.; Neubüser, A.; Richter, T.; Höfler, H.; Balling, R. Isolation of the Pax9 cDNA from adult human esophagus. Mamm. Genome 1997, 8, 62–64. [Google Scholar] [CrossRef]
- Bannykh, S.I.; Emery, S.C.; Gerber, J.K.; Jones, K.L.; Benirschke, K.; Masliah, E. Aberrant Pax1 and Pax9 expression in Jarcho-Levin syndrome: Report of two Caucasian siblings and literature review. Am. J. Med. Genet. A 2003, 120a, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Nakatomi, M.; Wang, X.P.; Key, D.; Lund, J.J.; Turbe-Doan, A.; Kist, R.; Aw, A.; Chen, Y.; Maas, R.L.; Peters, H. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 2010, 340, 438–449. [Google Scholar] [CrossRef]
- Seki, D.; Takeshita, N.; Oyanagi, T.; Sasaki, S.; Takano, I.; Hasegawa, M.; Takano-Yamamoto, T. Differentiation of Odontoblast-Like Cells From Mouse Induced Pluripotent Stem Cells by Pax9 and Bmp4 Transfection. Stem Cells Transl. Med. 2015, 4, 993–997. [Google Scholar] [CrossRef]
- Sivakamasundari, V.; Kraus, P.; Sun, W.; Hu, X.; Lim, S.L.; Prabhakar, S.; Lufkin, T. A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol. Open 2017, 6, 187–199. [Google Scholar] [CrossRef]
- Kearns, S.M.; Laywell, E.D.; Kukekov, V.K.; Steindler, D.A. Extracellular matrix effects on neurosphere cell motility. Exp. Neurol. 2003, 182, 240–244. [Google Scholar] [CrossRef]
- Antoon, R.; Yeger, H.; Loai, Y.; Islam, S.; Farhat, W.A. Impact of bladder-derived acellular matrix, growth factors, and extracellular matrix constituents on the survival and multipotency of marrow-derived mesenchymal stem cells. J. Biomed. Mater. Res. A 2012, 100, 72–83. [Google Scholar] [CrossRef]
- Bi, Y.; Ehirchiou, D.; Kilts, T.M.; Inkson, C.A.; Embree, M.C.; Sonoyama, W.; Li, L.; Leet, A.I.; Seo, B.M.; Zhang, L.; et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 2007, 13, 1219–1227. [Google Scholar] [CrossRef]
- Hupe, M.; Li, M.X.; Kneitz, S.; Davydova, D.; Yokota, C.; Kele, J.; Hot, B.; Stenman, J.M.; Gessler, M. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci. Signal. 2017, 10, eaag2476. [Google Scholar] [CrossRef]
- Ormestad, M.; Astorga, J.; Landgren, H.; Wang, T.; Johansson, B.R.; Miura, N.; Carlsson, P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 2006, 133, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Tamakoshi, T.; Uezato, T.; Shu, F.; Kanzaki-Kato, N.; Fu, Y.; Koseki, H.; Yoshida, N.; Sugiyama, T.; Miura, N. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev. Biol. 2003, 259, 83–94. [Google Scholar] [CrossRef]
- Yu, L.; Wynn, J.; Ma, L.; Guha, S.; Mychaliska, G.B.; Crombleholme, T.M.; Azarow, K.S.; Lim, F.Y.; Chung, D.H.; Potoka, D.; et al. De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet. 2012, 49, 650–659. [Google Scholar] [CrossRef]
- Neubüser, A.; Peters, H.; Balling, R.; Martin, G.R. Antagonistic interactions between FGF and BMP signaling pathways: A mechanism for positioning the sites of tooth formation. Cell 1997, 90, 247–255. [Google Scholar] [CrossRef]
- Vieira, A.R.; Meira, R.; Modesto, A.; Murray, J.C. MSX1, PAX9, and TGFA contribute to tooth agenesis in humans. J. Dent. Res. 2004, 83, 723–727. [Google Scholar] [CrossRef]
- Ogawa, T.; Kapadia, H.; Feng, J.Q.; Raghow, R.; Peters, H.; D’Souza, R.N. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J. Biol. Chem. 2006, 281, 18363–18369. [Google Scholar] [CrossRef]


| Target Gene (Abbreviation) | GenBank ID | Forward (Top) and Reverse (Bottom) Primer Sequences | Size of Amplified Products (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| β-actin | NM_001101.5 | 5′-ATTGCCGACAGGATGCAGA-3′/ 5′-GAGTACTTGCGCTCAGGAGGA-3′ | 89 | 60 |
| Alpha smooth muscle actin (aSMA) | NM_001613.4 | 5′-GACAATGGCTCTGGGCTCTGTA-3′/ 5′-CTGTGCTTCGTCACCCACGTA-3′ | 147 | 60 |
| Osteoprotegerin (OPG) | NM_002546.4 | 5′-CTCGAAGGTGAGGTTAGCATGTC-3′/ 5′-TGGCACCAAAGTAAACGCAGAG-3′ | 196 | 60 |
| Alkaline phosphatase (ALP) | NM_001177520.3 | 5′-GGACCATTCCCACGTCTTCAC-3′/ 5′-CCTTGTAGCCAGGCCCATTG-3′ | 137 | 60 |
| Periostin (POSTN) | NM_006475.2 | 5′-CATTGATGGAGTGCCTGTGGA-3′/ 5′-CAATGAATTTGGTGACCTTGGTG-3′ | 167 | 60 |
| Type1 collagen Alpha 1 (COL1A1) | NM_000088.3 | 5′-CCCGGGTTTCAGAGACAACTTC-3′/ 5′-TCCACATGCTTTATTCCAGCAATC-3′ | 148 | 60 |
| Periodontal ligament-associated protein 1 (PLAP1) | NM_017680.5 | 5′-ATGGGAGTCTTGCTAACATACCAC-3′/ 5′-CAGAAGTCATTTACTCCCACTCTTG-3′ | 154 | 60 |
| Forkhead box F2 (FOXF2) | NM_001452.2 | 5′-TCGCCTTACCTCAAGCAGC-3′/ 5′-AGAGTGATGCTGGTAACGGG-3′ | 165 | 60 |
| SIX homeobox 2 (SIX2) | NM_016932.5 | 5′-GGCCGAGGCCAAGGAA-3′/ 5′-GGGCTGGATGATGAGTGGTC-3′ | 144 | 60 |
| Distal-less homeobox 5 (DLX5) | NM_005221.6 | 5′-CAGAAGACTCAGTACCTCGCC-3′/ 5′-GTTACACGCCATTGGGTCG-3′ | 180 | 60 |
| Paired box 9 (PAX9) | NM_001372076.1 | 5′-GCAGGAAGCCAAGTACGGT-3′/ 5′-TGTCACAGTTGTGGGGAGAC-3′ | 200 | 60 |
| Transcription factor AP-4 (TFAP4) | NM_003223.3 | 5′-CACATCCCGGCAAAATCTGG-3′/ 5′-CCATGGCGTCACTGTCTGAG-3′ | 185 | 60 |
| Osteocalcin (OCN) | NM_199173.4 | 5′-CCCAGGCGCTACCTGTATCAA-3′/ 5′-GGTCAGCCAACTCGTCACCAGTC-3′ | 112 | 60 |
| Bone morphogenetic protein 2 (BMP2) | NM_001200.2 | 5′-TCCACTAATCATGCCATTGTTCAGA-3′/ 5′-GGGACACAGCATGCCTTAGGA-3′ | 73 | 60 |
| Bone sialoprotein (BSP) | MN_004967.3 | 5′-CTGGCACAGGGTATACAGGGTTAG-3′/ 5′-ACTGGTGCCGTTTATGCCTTG-3′ | 181 | 60 |
| Lipoprotein lipase (LPL) | NM_000237.2 | 5′-GACTCGTTCTCAGATGCCCT-3′/ 5′-ACTTCAGGCAGAGTGAATGGG-3′ | 145 | 60 |
| Adiponectin (ADIPOQ) | NM_001177800.2 | 5′-CAGGAAACCACGACTCAAGGG-3′/ 5′-CCGGTTTCACCGATGTCTCC-3′ | 200 | 60 |
| Leptin (LEP) | NM_000230.3 | 5′-GCTGTGCCCATCCAAAAAGTC-3′/ 5′-CCAGTGTCTGGTCCATCTTGG-3′ | 178 | 60 |
| CCAAT enhancer binding protein alpha (CEBPa) | NM_004364.4 | 5′-GGTGGACAAGAACAGCAACGA-3′/ 5′-GTCATTGTCACTGGTCAGCTC-3′ | 136 | 60 |
| Proliferator-activated receptor gamma (PPARg) | NM_138712.5 | 5′-TATTCTCAGTGGAGACCGCC-3′/ 5′-TGAGGACTCAGGGTGGTTCA-3′ | 115 | 60 |
| Kruppel like factor 15 (KLF15) | NM_014079.4 | 5′-TACACCAAAAGCAGCCACCTC-3′/ 5′-CTGGTACGGCTTCACACCTG-3′ | 153 | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugiura, R.; Hamano, S.; Tomokiyo, A.; Hasegawa, D.; Yoshida, S.; Sugii, H.; Fujino, S.; Adachi, O.; Kadowaki, M.; Yamashita, D.; et al. PAX9 Is Involved in Periodontal Ligament Stem Cell-like Differentiation of Human-Induced Pluripotent Stem Cells by Regulating Extracellular Matrix. Biomedicines 2022, 10, 2366. https://doi.org/10.3390/biomedicines10102366
Sugiura R, Hamano S, Tomokiyo A, Hasegawa D, Yoshida S, Sugii H, Fujino S, Adachi O, Kadowaki M, Yamashita D, et al. PAX9 Is Involved in Periodontal Ligament Stem Cell-like Differentiation of Human-Induced Pluripotent Stem Cells by Regulating Extracellular Matrix. Biomedicines. 2022; 10(10):2366. https://doi.org/10.3390/biomedicines10102366
Chicago/Turabian StyleSugiura, Risa, Sayuri Hamano, Atsushi Tomokiyo, Daigaku Hasegawa, Shinichiro Yoshida, Hideki Sugii, Shoko Fujino, Orie Adachi, Masataka Kadowaki, Daiki Yamashita, and et al. 2022. "PAX9 Is Involved in Periodontal Ligament Stem Cell-like Differentiation of Human-Induced Pluripotent Stem Cells by Regulating Extracellular Matrix" Biomedicines 10, no. 10: 2366. https://doi.org/10.3390/biomedicines10102366
APA StyleSugiura, R., Hamano, S., Tomokiyo, A., Hasegawa, D., Yoshida, S., Sugii, H., Fujino, S., Adachi, O., Kadowaki, M., Yamashita, D., & Maeda, H. (2022). PAX9 Is Involved in Periodontal Ligament Stem Cell-like Differentiation of Human-Induced Pluripotent Stem Cells by Regulating Extracellular Matrix. Biomedicines, 10(10), 2366. https://doi.org/10.3390/biomedicines10102366





