Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fibroblasts Isolation and Culture
2.2. Fibroblast Populated Collagen Lattice (FPCL) Contraction Assay
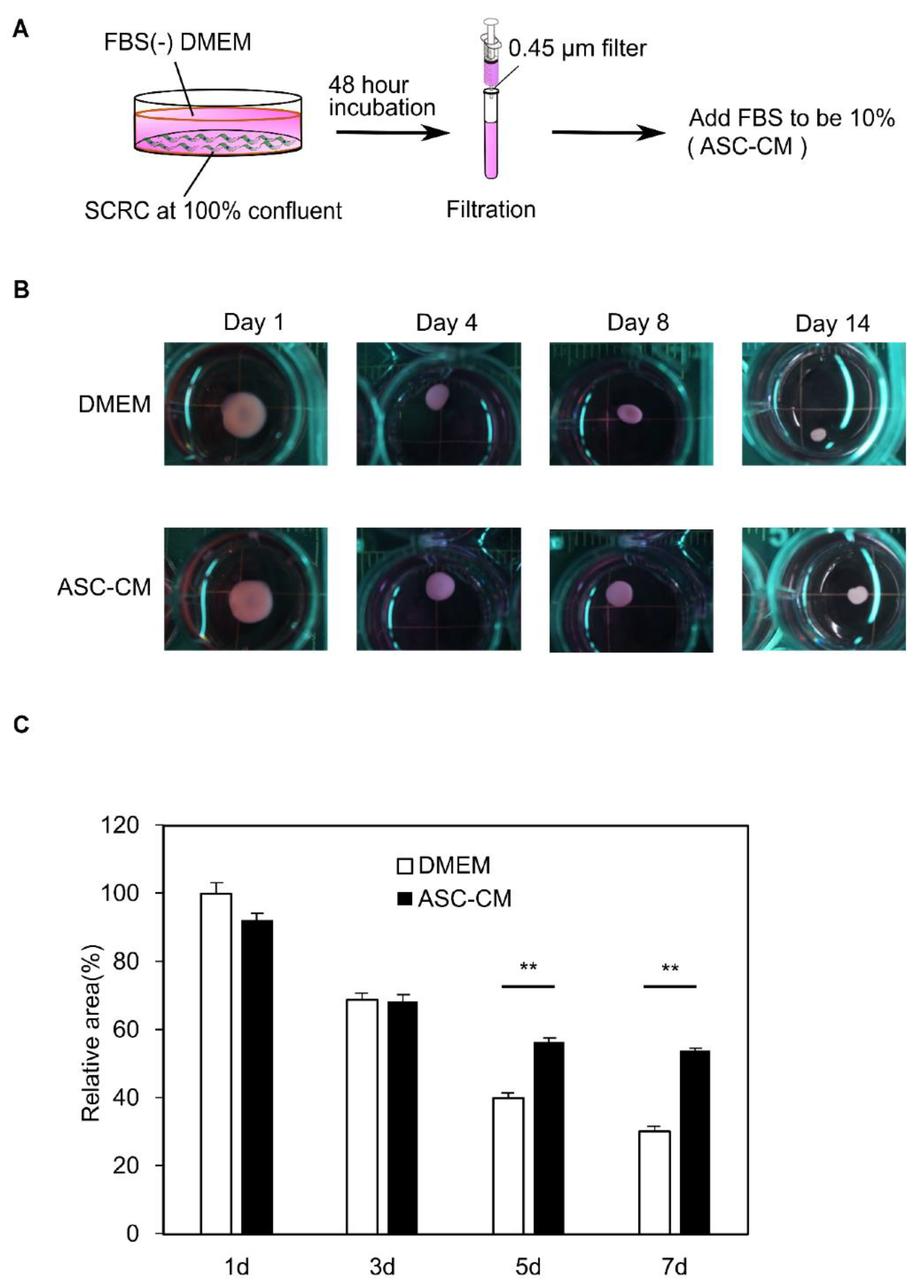
2.3. The Preparation of Adipose Derived Mesenchymal Stem Cell-Conditioned Medium (ASC-CM)
2.4. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.5. Gene Expression Analysis (RNA-Sequencing)
2.6. Histological Analysis
2.7. Statistical Analysis
3. Results
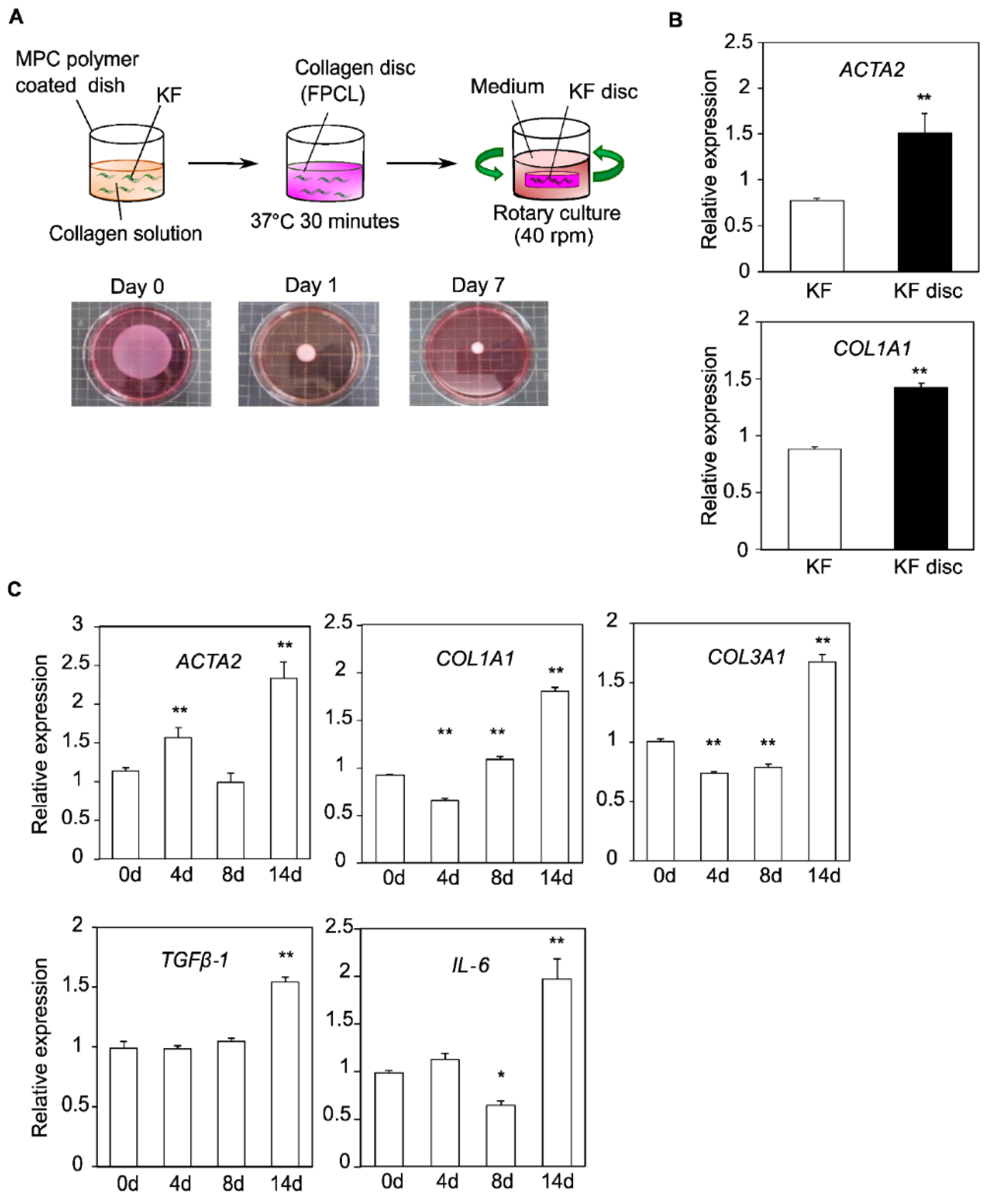
3.1. Construction of the FPCL Model
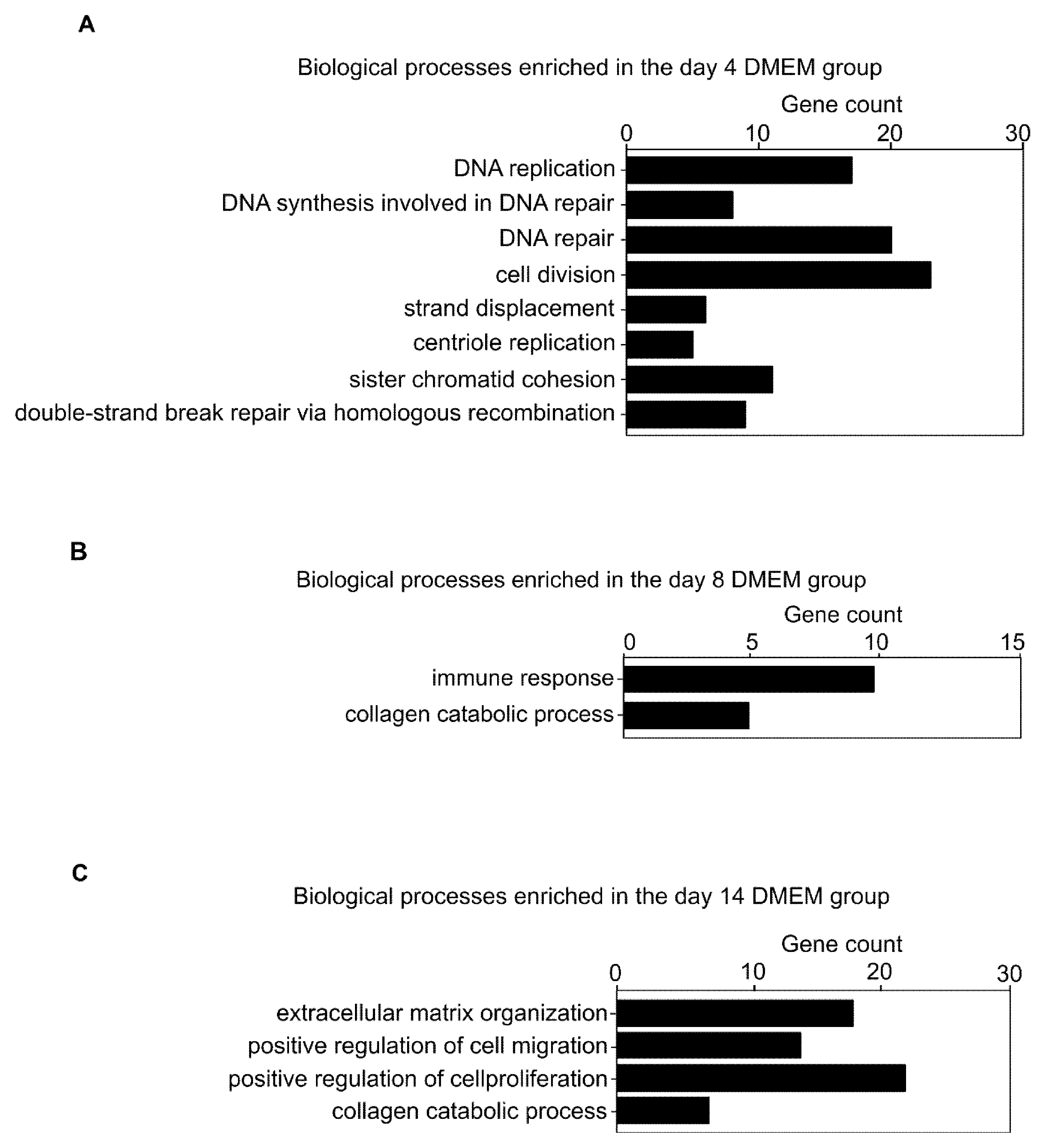
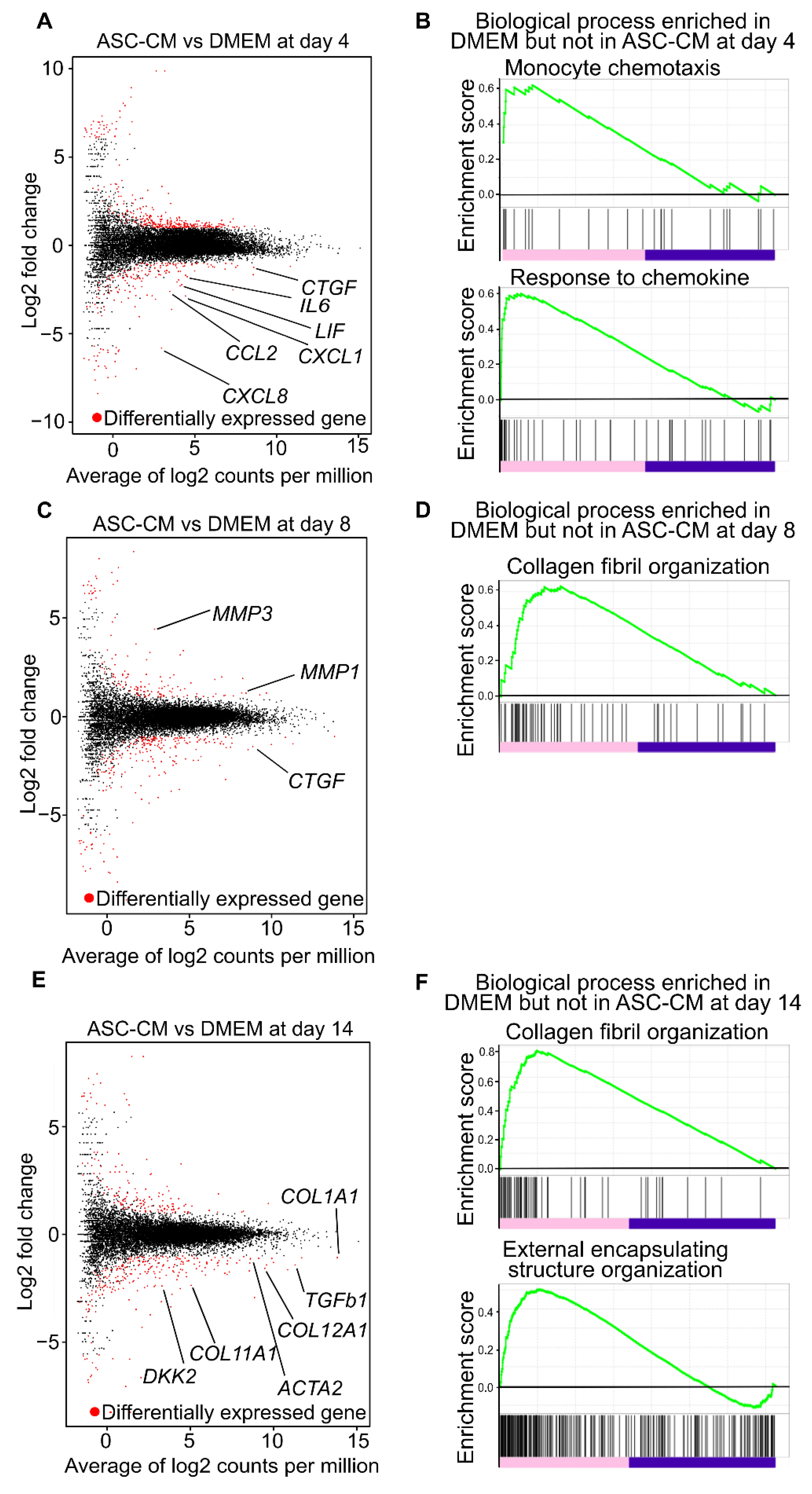
3.2. Comprehensive Gene Expression Analysis in the Collagen Discs
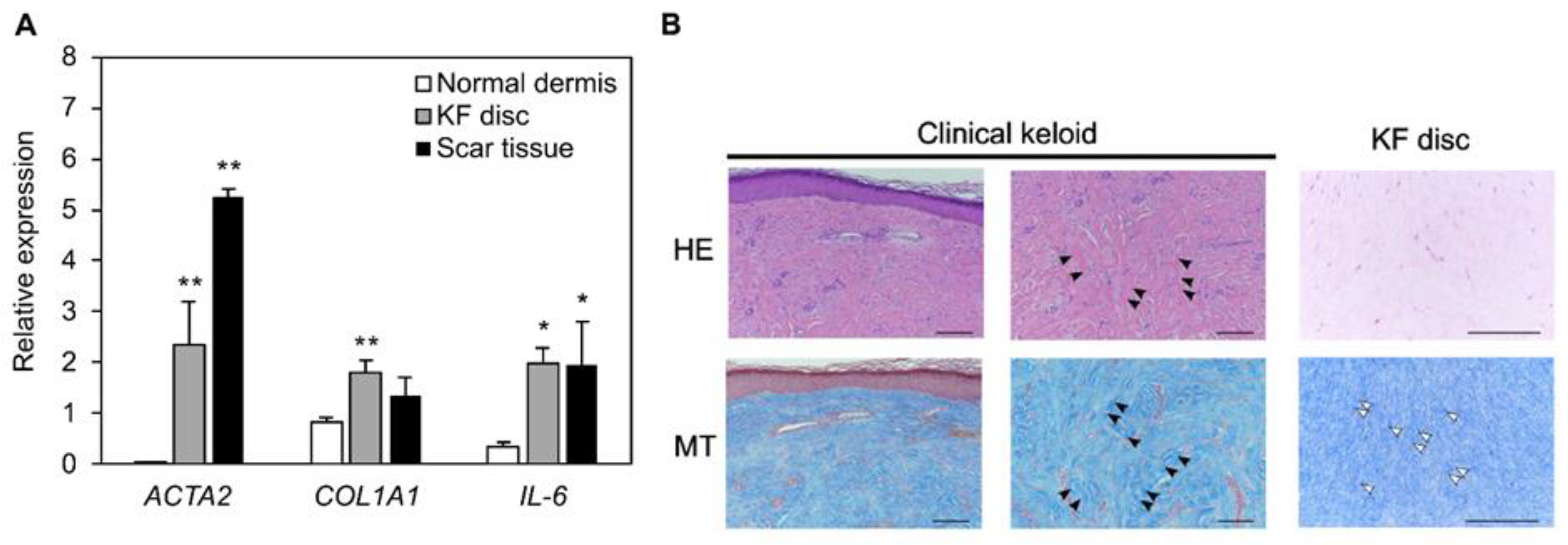
3.3. The Similarity between Collagen Discs and Patient-Derived Scar Tissue in Histology and Gene Expression
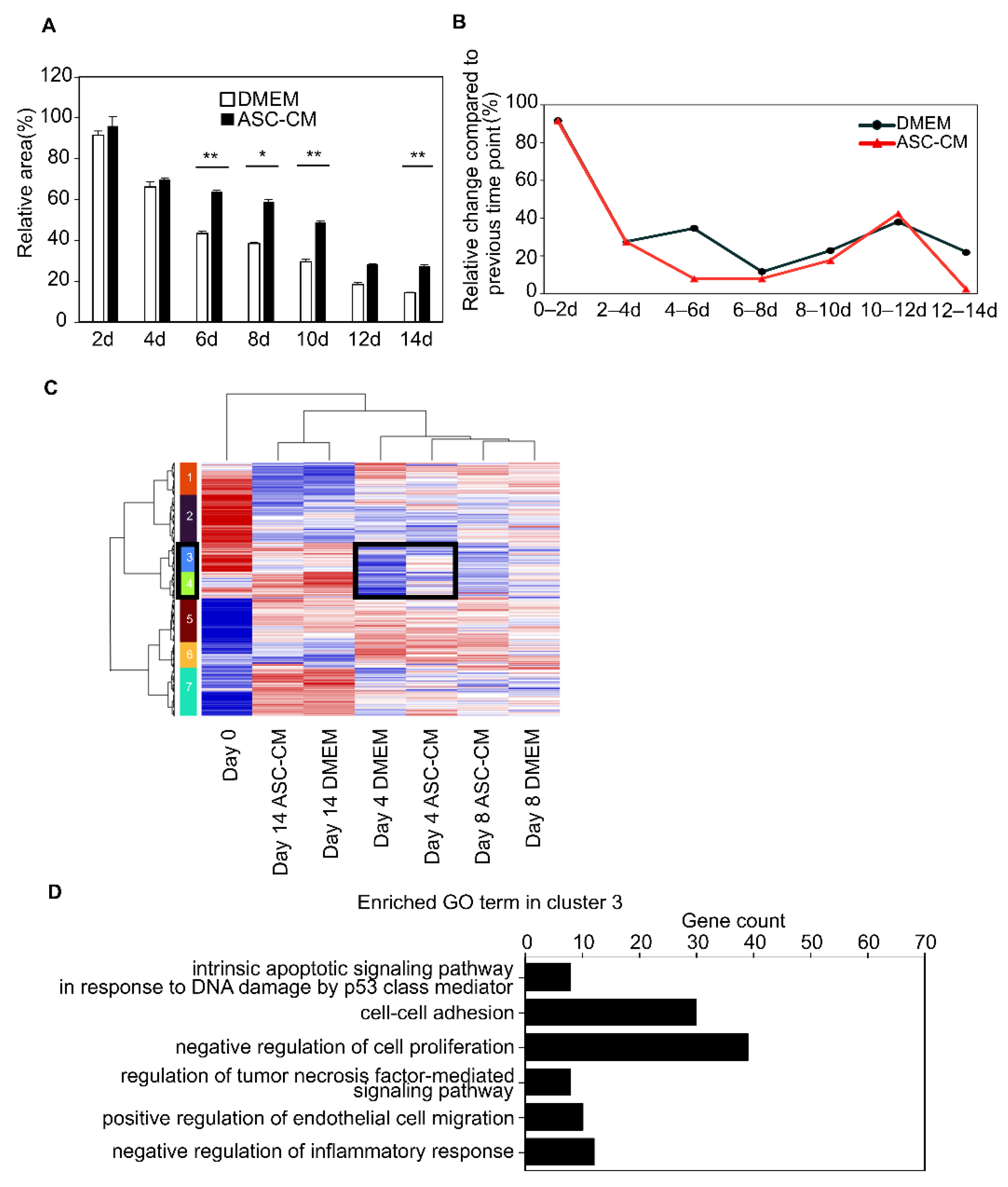
3.4. Suppression of the Collagen Disc Contraction by the ASC-CM
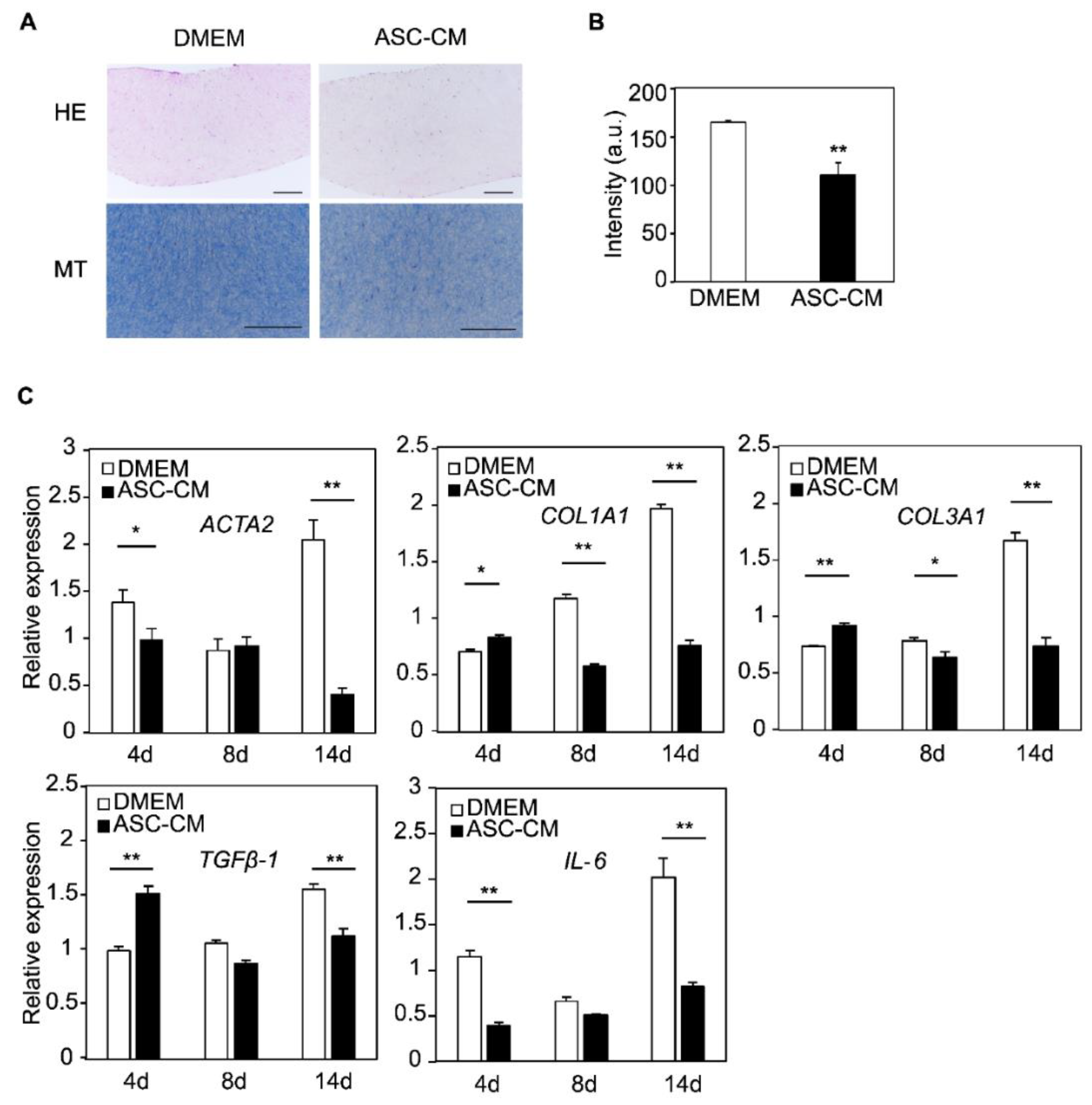
3.5. Changes in Histology and Gene Expression by the ASC-CM
3.6. Alteration of Global Trend of Gene Expression by the Addition of ASC-CM
3.7. Effect of ASC-CM on the Early Stage of KF Disc Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gabbiani, G.; Ryan, G.B.; Majno, G. Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction. Experientia 1971, 27, 549–550. [Google Scholar] [CrossRef]
- Shibuya, Y.; Hokugo, A.; Okawa, H.; Kondo, T.; Khalil, D.; Wang, L.; Roca, Y.; Clements, A.; Sasaki, H.; Berry, E.; et al. Therapeutic downregulation of neuronal PAS domain 2 (Npas2) promotes surgical skin wound healing. elife 2022, 11, 71074. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Minn, K.W. The effect of myofibroblast on contracture of hypertrophic scar. Plast. Reconstr. Surg. 2004, 113, 633–640. [Google Scholar] [CrossRef]
- Desmouliere, A.; Redard, M.; Darby, I.; Gabbiani, G. Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar. Am. J. Pathol. 1995, 146, 56. [Google Scholar]
- Tissue Repair, Contraction, and the Myofibroblast: EBSCOhost. Available online: https://web-s-ebscohost-com.ezproxy.tulips.tsukuba.ac.jp/ehost/pdfviewer/pdfviewer?vid=2&sid=b62068ff-8af4-4bdb-9020-47fd8333f5ac%40redis (accessed on 9 March 2022).
- Ogawa, R. Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis. Int. J. Mol. Sci. 2017, 18, 606. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; McCauley, R.L.; Zhang, W. Elevated interleukin-6 expression in keloid fibroblasts. J. Surg. Res. 2000, 89, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, X.F.; Wang, Z.C.; Lou, D.; Fang, Q.Q.; Hu, Y.Y.; Zhao, W.Y.; Zhang, L.Y.; Wu, L.H.; Tan, W.Q. Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed. Pharmacother. 2020, 129, 110287. [Google Scholar] [CrossRef] [PubMed]
- Wulandari, E.; Jusman, S.W.A.; Moenadjat, Y.; Jusuf, A.A.; Sadikin, M. Expressions of collagen I and III in hypoxic keloid tissue. Kobe J. Med. Sci. 2016, 62, E58–E69. [Google Scholar]
- Bell, E.; Ivarsson, B.; Merrill, C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc. Natl. Acad. Sci. USA 1979, 76, 1274. [Google Scholar] [CrossRef] [PubMed]
- Dallon, J.C.; Ehrlich, H.P. A review of fibroblast-populated collagen lattices. Wound Repair Regen. 2008, 16, 472–479. [Google Scholar] [CrossRef]
- Guidry, C.; Grinnell, F. Studies on the mechanism of hydrated collagen gel reorganization by human skin fibroblasts. J. Cell Sci. 1985, 79, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Maeda, T.; Setoguchi, Y.; Yamazaki, M.; Matsumura, H.; Tsuboi, R. Pirfenidone suppresses keloid fibroblast-embedded collagen gel contraction. Arch. Dermatol. Res. 2012, 304, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Kamamoto, F.; Oliveira Paggiaro, A.; Rodas, A.; Roma Herson, M.; Beatriz Mathor, M.; Castro Ferreira, M. A wound contraction experimental model for studying keloids and wound-healing modulators. Artif. Organs 2003, 27, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, R.; Akita, S.; Akaishi, S.; Aramaki-Hattori, N.; Dohi, T.; Hayashi, T.; Kishi, K.; Kono, T.; Matsumura, H.; Muneuchi, G.; et al. Diagnosis and Treatment of Keloids and Hypertrophic Scars—Japan Scar Workshop Consensus Document 2018. Burn. Trauma 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, H.; Long, Y.; You, Z.; Ogawa, R.; Du, Y.; Huang, C. Asporin inhibits collagen matrix-mediated intercellular mechanocommunications between fibroblasts during keloid progression. FASEB J. 2021, 35, e21705. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, T.; Zhan, X. Silencing circular RNAPTPN12 promoted the growth of keloid fibroblasts by activating Wnt signaling pathway via targeting microRNA-21-5p. Bioengineered 2022, 13, 3503–3515. [Google Scholar] [CrossRef]
- Chen, H.; Hou, K.; Wu, Y.; Liu, Z. Use of Adipose Stem Cells Against Hypertrophic Scarring or Keloid. Front. Cell Dev. Biol. 2022, 9, 823694. [Google Scholar] [CrossRef]
- Frese, L.; Dijkman, P.E.; Hoerstrup, S.P. Adipose Tissue-Derived Stem Cells in Regenerative Medicine. Transfus. Med. Hemotherapy 2016, 43, 268. [Google Scholar] [CrossRef]
- Tucci-Viegas, V.M.; Hochman, B.; Frana, J.P.; Ferreira, L.M. Keloid explant culture: A model for keloid fibroblasts isolation and cultivation based on the biological differences of its specific regions. Int. Wound J. 2010, 7, 339–348. [Google Scholar] [CrossRef]
- Hsieh, S.C.; Wu, C.C.; Hsu, S.L.; Feng, C.H.; Yen, J.H. Gallic acid attenuates TGF-β1-stimulated collagen gel contraction via suppression of RhoA/Rho-kinase pathway in hypertrophic scar fibroblasts. Life Sci. 2016, 161, 19–26. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 18 March 2020).
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Fresno, C.; Fernández, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T.; Kojima, K.; Tamada, Y. Higher gene expression related to wound healing by fibroblasts on silk fibroin biomaterial than on collagen. Molecules 2020, 25, 1939. [Google Scholar] [CrossRef]
- Nirodi, C.S.; Devalaraja, R.; Nanney, L.B.; Arrindell, S.; Russell, S.; Trupin, J.; Richmond, A. Chemokine and chemokine receptor expression in keloid and normal fibroblasts. Wound Repair Regen. 2000, 8, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liao, N.; Lu, F.; Peng, H.; Gao, J. The role of Duffy antigen receptor for chemokines in keloids. Gene 2015, 570, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.M.; Takagawa, S.; Fresco, R.; Zhu, X.; Varga, J.; DiPietro, L.A. Diminished induction of skin fibrosis in mice with MCP-1 deficiency. J. Investig. Dermatol. 2006, 126, 1900–1908. [Google Scholar] [CrossRef] [PubMed]
- Albrengues, J.; Bourget, I.; Pons, C.; Butet, V.; Hofman, P.; Tartare-Deckert, S.; Feral, C.C.; Meneguzzi, G.; Gaggioli, C. LIF Mediates Proinvasive Activation of Stromal Fibroblasts in Cancer. Cell Rep. 2014, 7, 1664–1678. [Google Scholar] [CrossRef]
- Sato, C.; Yamamoto, Y.; Funayama, E.; Furukawa, H.; Oyama, A.; Murao, N.; Hosono, H.; Kawakubo, K.; Sakamoto, N.; Ohnishi, S. Conditioned medium obtained from amnion-derived mesenchymal stem cell culture prevents activation of keloid fibroblasts. Plast. Reconstr. Surg. 2018, 141, 390–398. [Google Scholar] [CrossRef]
- Bojanic, C.; To, K.; Hatoum, A.; Shea, J.; Seah, K.T.M.; Khan, W.; Malata, C.M. Mesenchymal stem cell therapy in hypertrophic and keloid scars. Cell Tissue Res. 2020, 383, 915–930. [Google Scholar] [CrossRef]
- Monika, P.; Waiker, P.V.; Chandraprabha, M.N.; Rangarajan, A.; Murthy, K.N.C. Myofibroblast progeny in wound biology and wound healing studies. Wound Repair Regen. 2021, 29, 531–547. [Google Scholar] [CrossRef]
- Bell, E.; Ehrlich, H.P.; Buttle, D.J.; Nakatsuji, T. Living tissue formed in vitro and accepted as skin-equivalent tissue of full thickness. Science 1981, 211, 1052–1054. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Akagi, Y.; Imai, Y.; Takayama, Y.; Kida, Y.S. Fabrication of Perfusable Vascular Channels and Capillaries in 3D Liver-like Tissue. Sci. Rep. 2020, 10, 5646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gir, P.; Oni, G.; Brown, S.A.; Mojallal, A.; Rohrich, R.J. Human adipose stem cells: Current clinical applications. Plast. Reconstr. Surg. 2012, 129, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- O’Halloran, N.; Courtney, D.; Kerin, M.J.; Lowery, A.J. Adipose-Derived Stem Cells in Novel Approaches to Breast Reconstruction: Their Suitability for Tissue Engineering and Oncological Safety. Breast Cancer 2017, 11, 1178223417726777. [Google Scholar] [CrossRef] [PubMed]
- Gardin, C.; Ferroni, L.; Bellin, G.; Rubini, G.; Barosio, S.; Zavan, B. Therapeutic Potential of Autologous Adipose-Derived Stem Cells for the Treatment of Liver Disease. Int. J. Mol. Sci. 2018, 19, 4064. [Google Scholar] [CrossRef]
- Suarez, E.; Syed, F.; Rasgado, T.A.; Walmsley, A.; Mandal, P.; Bayat, A. Skin equivalent tensional force alters keloid fibroblast behavior and phenotype. Wound Repair Regen. 2014, 22, 557–568. [Google Scholar] [CrossRef]
- Derderian, C.A.; Bastidas, N.; Lerman, O.Z.; Bhatt, K.A.; Lin, S.E.; Voss, J.; Holmes, J.W.; Levine, J.P.; Gurtner, G.C. Mechanical strain alters gene expression in an in vitro model of hypertrophic scarring. Ann. Plast. Surg. 2005, 55, 69–75. [Google Scholar] [CrossRef]
- Corrò, C.; Novellasdemunt, L.; Li, V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imai, Y.; Mori, N.; Nihashi, Y.; Kumagai, Y.; Shibuya, Y.; Oshima, J.; Sasaki, M.; Sasaki, K.; Aihara, Y.; Sekido, M.; et al. Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model. Biomedicines 2022, 10, 2388. https://doi.org/10.3390/biomedicines10102388
Imai Y, Mori N, Nihashi Y, Kumagai Y, Shibuya Y, Oshima J, Sasaki M, Sasaki K, Aihara Y, Sekido M, et al. Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model. Biomedicines. 2022; 10(10):2388. https://doi.org/10.3390/biomedicines10102388
Chicago/Turabian StyleImai, Yukiko, Nobuhito Mori, Yuma Nihashi, Yutaro Kumagai, Yoichiro Shibuya, Junya Oshima, Masahiro Sasaki, Kaoru Sasaki, Yukiko Aihara, Mitsuru Sekido, and et al. 2022. "Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model" Biomedicines 10, no. 10: 2388. https://doi.org/10.3390/biomedicines10102388
APA StyleImai, Y., Mori, N., Nihashi, Y., Kumagai, Y., Shibuya, Y., Oshima, J., Sasaki, M., Sasaki, K., Aihara, Y., Sekido, M., & Kida, Y. S. (2022). Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model. Biomedicines, 10(10), 2388. https://doi.org/10.3390/biomedicines10102388





