The Immunological Epigenetic Landscape of the Human Life Trajectory
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of B-lymphocytes and Monocytes from Peripheral Blood
2.2. Genomic DNA Isolation from B-lymphocytes and Monocytes
2.3. Quantification of Global DNA Methylation
2.4. Bisulfite Conversion of Genomic DNA
2.5. Total RNA Isolation from B-lymphocytes and Monocytes
2.6. Real-Time PCR Quantification
2.7. HLA-DQA1*01 Promoter Cloning and Sequencing
2.8. HLA-DQA1*01 Promoter MS-HRM Analysis
2.9. Histone 3 and Histone 3 Lysine 27 Trimethylation (me3) Immunofluorescence
2.10. Protein Extraction
2.11. Electrophoresis and Western Blotting
2.12. Chromatin Immunoprecipitation (ChIP)-qPCR
2.13. Secondary Structure Prediction and Conservation of Long Non-Coding RNA TINA
2.14. Statistical Analysis
3. Results
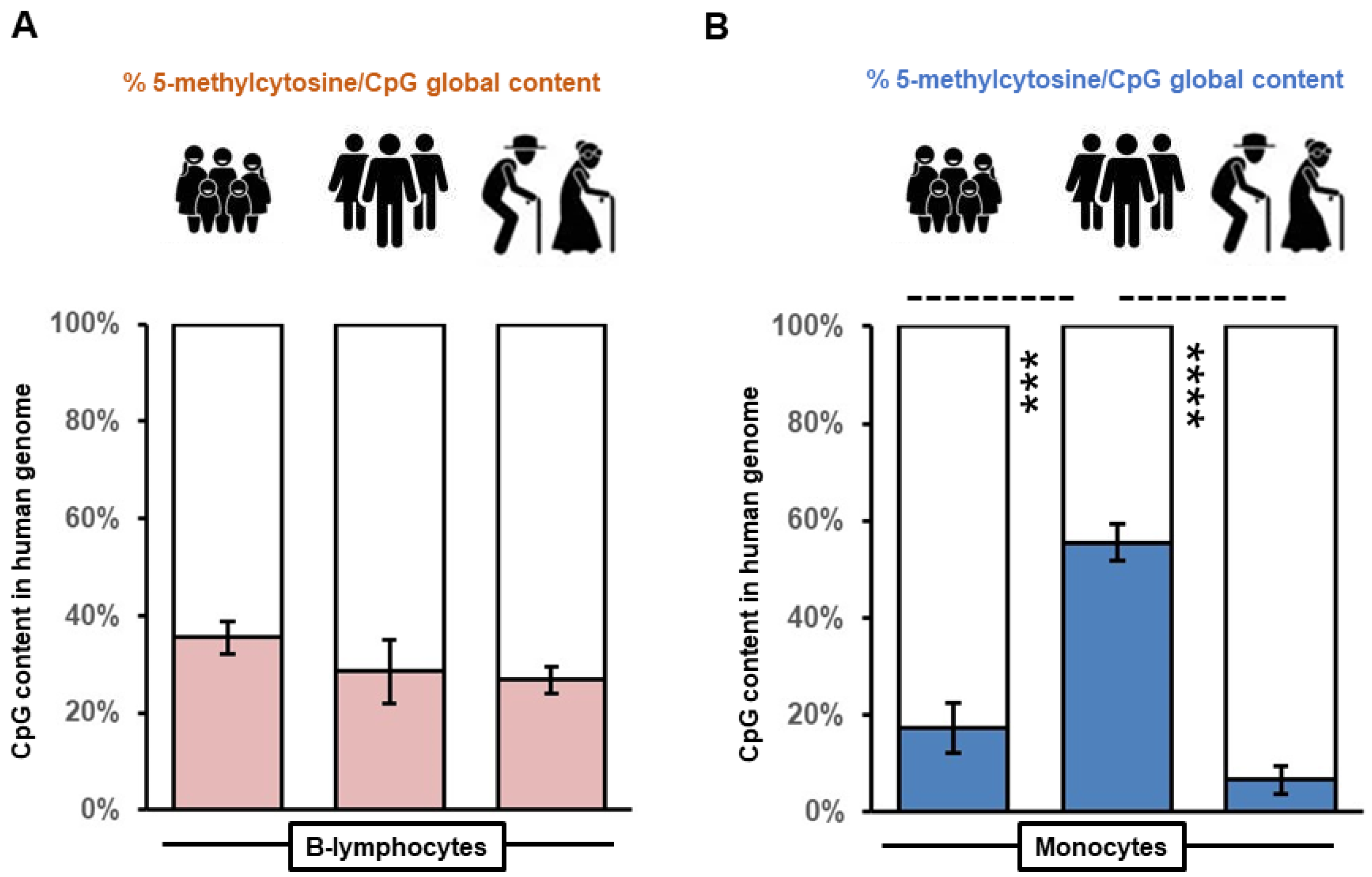
3.1. Monocytes Reveal Variation in Global Methylation of Genomic DNA across Age Groups as Determined from Long Interspersed Nucleotide Element 1 Repeats
3.2. HLA-DQA1*01 and HLA-DQB1*06 Elicit Distinctive Expression Profiles in Monocytes Relative to B-lymphocytes across Age Groups
3.3. Dynamic HLA-DQA1*01 Promoter Methylation through Representative Lifespan Stages
3.4. Identification by Bisulfite Sequencing of Methylated Cytosine Positioning within the HLA-DQA Promoter
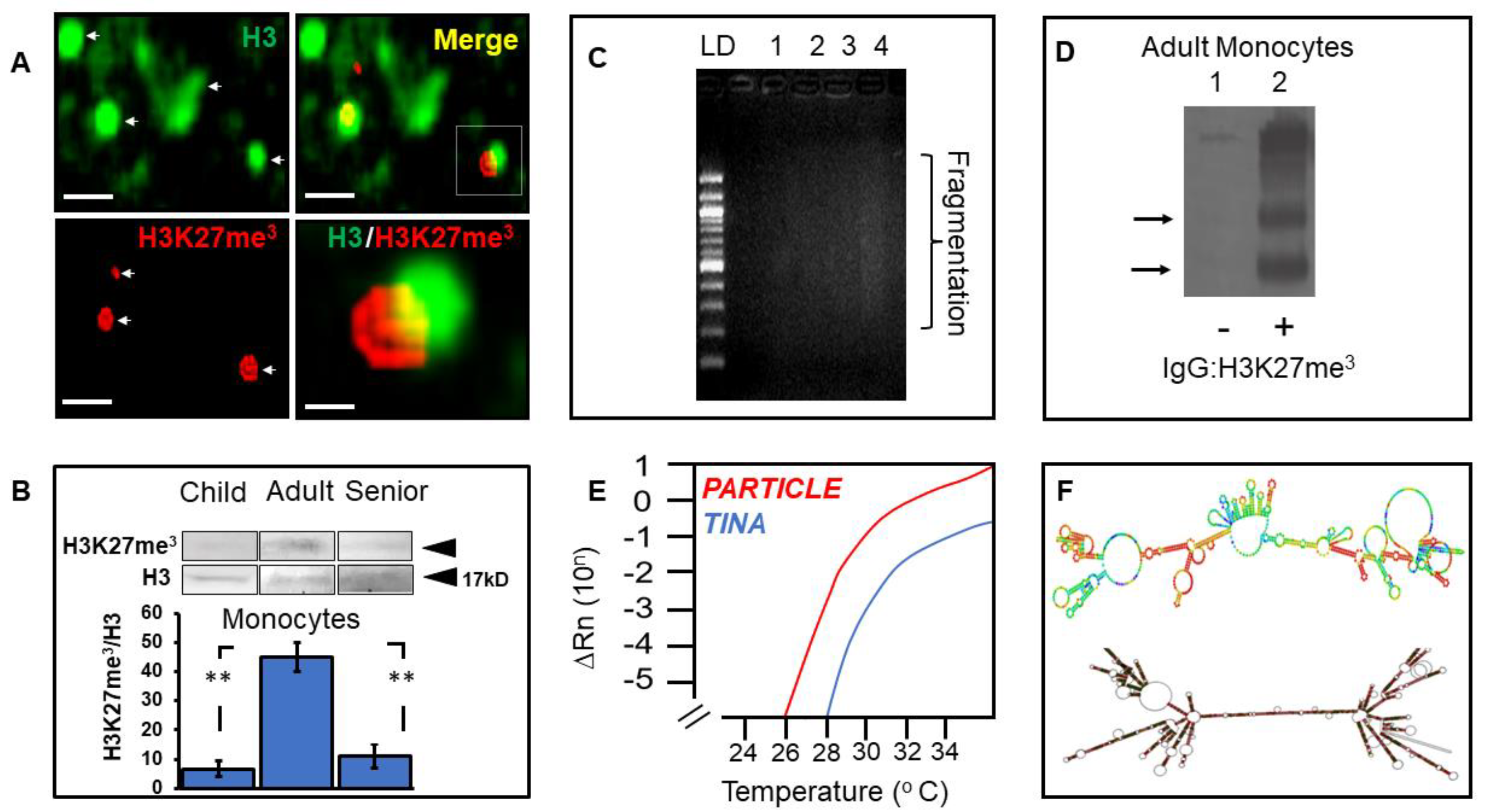
3.5. Elevated Histone Modification in Adult Monocytes Associates with the Long Non-Coding Transcriptome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ponnappan, S.; Ponnappan, U. Aging and immune function: Molecular mechanisms to interventions. Antioxid. Redox Signal. 2011, 14, 1551–1585. [Google Scholar] [CrossRef] [PubMed]
- Montecino-Rodriguez, E.; Berent-Maoz, B.; Dorshkind, K. Causes, consequences, and reversal of immune system aging. J. Clin. Investig. 2013, 123, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Laupeze, B.; Fardel, O.; Onno, M.; Bertho, N.; Drenou, B.; Fauchet, R.; Amiot, L. Differential expression of major histocompatibility complex class Ia, Ib, and II molecules on monocytes-derived dendritic and macrophagic cells. Hum. Immunol. 1999, 60, 591–597. [Google Scholar] [CrossRef]
- Hudec, M.; Riegerova, K.; Pala, J.; Kutna, V.; Cerna, M.; O’Leary, V.B. Celiac Disease Defined by Over-Sensitivity to Gliadin Activation and Superior Antigen Presentation of Dendritic Cells. Int. J. Mol. Sci. 2021, 22, 9982. [Google Scholar] [CrossRef] [PubMed]
- Krensky, A.M. The HLA system, antigen processing and presentation. Kidney Int. Suppl. 1997, 58, S2–S7. [Google Scholar] [PubMed]
- Hudec, M.; Juříčková, I.; Riegerová, K.; Ovsepian, S.V.; Černá, M.; O’Leary, V.B. Enhanced Extracellular Transfer of HLA-DQ Activates CD3(+) Lymphocytes towards Compromised Treg Induction in Celiac Disease. Int. J. Mol. Sci. 2022, 23, 6102. [Google Scholar] [CrossRef]
- Williams, F.; Meenagh, A.; Darke, C.; Acosta, A.; Daar, A.S.; Gorodezky, C.; Hammond, M.; Nascimento, E.; Middleton, D. Analysis of the distribution of HLA-B alleles in populations from five continents. Hum. Immunol. 2001, 62, 645–650. [Google Scholar] [CrossRef]
- Takata, H.; Suzuki, M.; Ishii, T.; Sekiguchi, S.; Iri, H. Influence of major histocompatibility complex region genes on human longevity among Okinawan-Japanese centenarians and nonagenarians. Lancet 1987, 2, 824–826. [Google Scholar] [CrossRef]
- Caruso, C.; Candore, G.; Colonna Romano, G.; Lio, D.; Bonafe, M.; Valensin, S.; Franceschi, C. HLA, aging, and longevity: A critical reappraisal. Hum. Immunol. 2000, 61, 942–949. [Google Scholar] [CrossRef]
- Franceschi, C.; Valensin, S.; Fagnoni, F.; Barbi, C.; Bonafe, M. Biomarkers of immunosenescence within an evolutionary perspective: The challenge of heterogeneity and the role of antigenic load. Exp. Gerontol. 1999, 34, 911–921. [Google Scholar] [CrossRef]
- Ivanova, R.; Henon, N.; Lepage, V.; Charron, D.; Vicaut, E.; Schachter, F. HLA-DR alleles display sex-dependent effects on survival and discriminate between individual and familial longevity. Hum. Mol. Genet. 1998, 7, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Mays-Hoopes, L.; Chao, W.; Butcher, H.C.; Huang, R.C. Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Dev. Genet. 1986, 7, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Casillas, M.A., Jr.; Lopatina, N.; Andrews, L.G.; Tollefsbol, T.O. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol. Cell. Biochem. 2003, 252, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cheung, T.H.; Charville, G.W.; Hurgo, B.M.; Leavitt, T.; Shih, J.; Brunet, A.; Rando, T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013, 4, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Ebata, A.; Alipanahiramandi, E.; Lee, S.S. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 2012, 11, 315–325. [Google Scholar] [CrossRef]
- McCartney, D.L.; Zhang, F.; Hillary, R.F.; Zhang, Q.; Stevenson, A.J.; Walker, R.M.; Bermingham, M.L.; Boutin, T.; Morris, S.W.; Campbell, A.; et al. An epigenome-wide association study of sex-specific chronological ageing. Genome Med. 2019, 12, 1. [Google Scholar] [CrossRef]
- Gaaib, J.N.; Nassief, A.F.; Al-Assi, A. Simple salting—Out method for genomic DNA extraction from whole blood. Tikrit J. Pure Sci. 2011, 16, 9–11. [Google Scholar]
- Zajacova, M.; Kotrbova-Kozak, A.; Cepek, P.; Cerna, M. Differences in promoter DNA methylation and mRNA expression of individual alleles of the HLA class II DQA1 gene. Immunol. Lett. 2015, 167, 147–154. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Hain, S.; Maugg, D.; Smida, J.; Azimzadeh, O.; Tapio, S.; Ovsepian, S.V.; Atkinson, M.J. Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. 2017, 7, 1790. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA 2012, 18, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Reiche, K.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed]
- Raden, M.; Ali, S.M.; Alkhnbashi, O.S.; Busch, A.; Costa, F.; Davis, J.A.; Eggenhofer, F.; Gelhausen, R.; Georg, J.; Heyne, S.; et al. Freiburg RNA tools: A central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018, 46, W25–W29. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Soman, N.S.; Verdine, G.L.; Bestor, T.H. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 1997, 270, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Briggs, E.M.; Ha, S.; Mita, P.; Brittingham, G.; Sciamanna, I.; Spadafora, C.; Logan, S.K. Long interspersed nuclear element-1 expression and retrotransposition in prostate cancer cells. Mob. DNA 2018, 9, 1. [Google Scholar] [CrossRef]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef]
- Erichsen, L.; Beermann, A.; Arauzo-Bravo, M.J.; Hassan, M.; Dkhil, M.A.; Al-Quraishy, S.; Hafiz, T.A.; Fischer, J.C.; Santourlidis, S. Genome-wide hypomethylation of LINE-1 and Alu retroelements in cell-free DNA of blood is an epigenetic biomarker of human aging. Saudi J. Biol. Sci. 2018, 25, 1220–1226. [Google Scholar] [CrossRef]
- Huen, K.; Calafat, A.M.; Bradman, A.; Yousefi, P.; Eskenazi, B.; Holland, N. Maternal phthalate exposure during pregnancy is associated with DNA methylation of LINE-1 and Alu repetitive elements in Mexican-American children. Environ. Res. 2016, 148, 55–62. [Google Scholar] [CrossRef]
- Zajacova, M.; Kotrbova-Kozak, A.; Cerna, M. Expression of HLA-DQA1 and HLA-DQB1 genes in B lymphocytes, monocytes and whole blood. Int. J. Immunogenet. 2018, 45, 128–137. [Google Scholar] [CrossRef]
- Harton, J.A.; Ting, J.P. Class II transactivator: Mastering the art of major histocompatibility complex expression. Mol. Cell. Biol. 2000, 20, 6185–6194. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Brunet, A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012, 22, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.A.; Zhang, Y.; Fullerton, J.N.; Boelen, L.; Rongvaux, A.; Maini, A.A.; Bigley, V.; Flavell, R.A.; Gilroy, D.W.; Asquith, B.; et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017, 214, 1913–1923. [Google Scholar] [CrossRef]
- Seidler, S.; Zimmermann, H.W.; Bartneck, M.; Trautwein, C.; Tacke, F. Age-dependent alterations of monocyte subsets and monocyte-related chemokine pathways in healthy adults. BMC Immunol. 2010, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyo, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P. Aging and epigenetic drift: A vicious cycle. J. Clin. Investig. 2014, 124, 24–29. [Google Scholar] [CrossRef]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Bermick, J.R.; Lambrecht, N.J.; denDekker, A.D.; Kunkel, S.L.; Lukacs, N.W.; Hogaboam, C.M.; Schaller, M.A. Neonatal monocytes exhibit a unique histone modification landscape. Clin. Epigenetics 2016, 8, 99. [Google Scholar] [CrossRef]
- Sun, D.; Luo, M.; Jeong, M.; Rodriguez, B.; Xia, Z.; Hannah, R.; Wang, H.; Le, T.; Faull, K.F.; Chen, R.; et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 2014, 14, 673–688. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, H.; Cai, Y.; Wang, H.; Niu, K.; Wu, X.; Ma, H.; Yang, Y.; Tong, W.; Liu, F.; et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife 2018, 7, e35368. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juříčková, I.; Hudec, M.; Votava, F.; Vosáhlo, J.; Ovsepian, S.V.; Černá, M.; O’Leary, V.B. The Immunological Epigenetic Landscape of the Human Life Trajectory. Biomedicines 2022, 10, 2894. https://doi.org/10.3390/biomedicines10112894
Juříčková I, Hudec M, Votava F, Vosáhlo J, Ovsepian SV, Černá M, O’Leary VB. The Immunological Epigenetic Landscape of the Human Life Trajectory. Biomedicines. 2022; 10(11):2894. https://doi.org/10.3390/biomedicines10112894
Chicago/Turabian StyleJuříčková, Iva, Michael Hudec, Felix Votava, Jan Vosáhlo, Saak Victor Ovsepian, Marie Černá, and Valerie Bríd O’Leary. 2022. "The Immunological Epigenetic Landscape of the Human Life Trajectory" Biomedicines 10, no. 11: 2894. https://doi.org/10.3390/biomedicines10112894
APA StyleJuříčková, I., Hudec, M., Votava, F., Vosáhlo, J., Ovsepian, S. V., Černá, M., & O’Leary, V. B. (2022). The Immunological Epigenetic Landscape of the Human Life Trajectory. Biomedicines, 10(11), 2894. https://doi.org/10.3390/biomedicines10112894






