TNBC Therapeutics Based on Combination of Fusarochromanone with EGFR Inhibitors
Abstract
1. Introduction
1.1. Cancer Statistics and Treatment Modalities
1.2. Triple Negative Breast Cancer (TNBC)
1.3. The Potential of Fusarochromanone as an mTOR Inhibitor
1.4. Drug Panel
2. Materials and Methods
2.1. Drug Treatments
2.2. Cell Lines and Mammalian Cell Culture
2.3. Crystal Violet Staining Assay
2.4. MTT Viability Assay
2.5. Drug Washout Assay
2.6. Statistical Analysis
3. Results
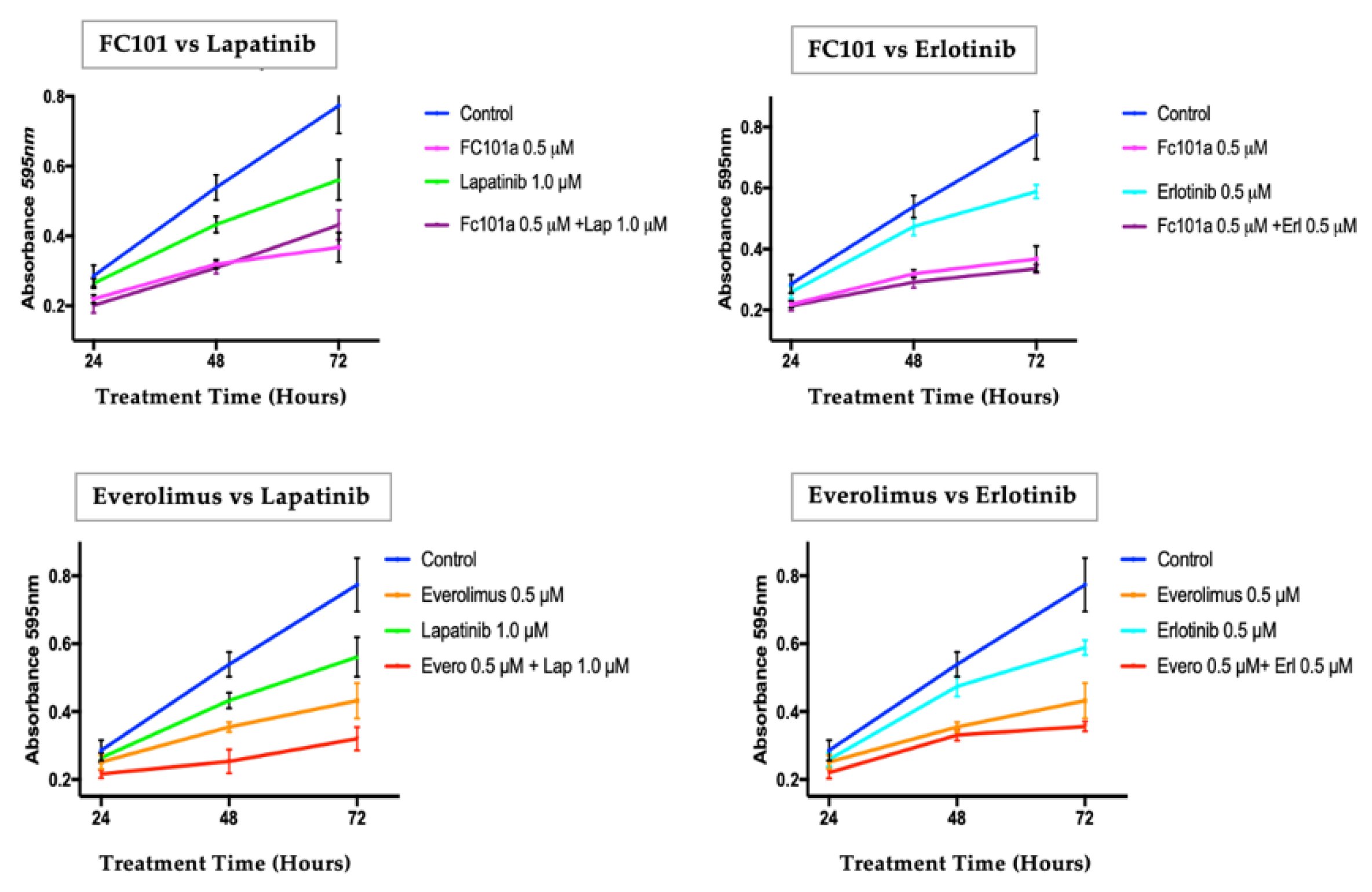
3.1. Crystal Violet Proliferation Assay
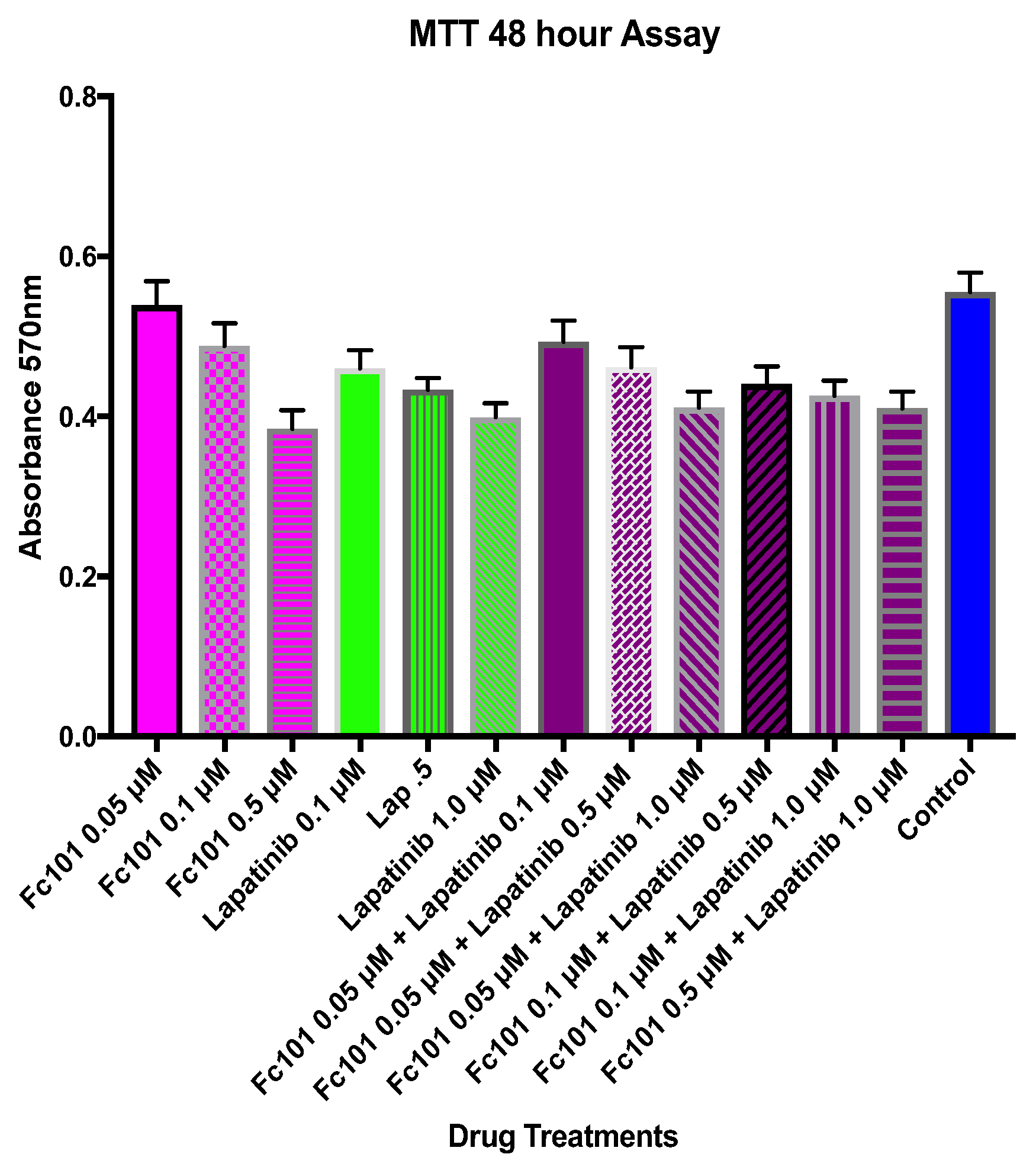
3.2. MTT Viability Assay
3.3. Drug Washout Survival Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breast Cancer|Breast Cancer Information & Overview. Available online: https://www.cancer.org/cancer/breast-cancer.html (accessed on 16 December 2021).
- Cancer Facts & Figures 2021|American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html (accessed on 15 December 2021).
- Metastatic Cancer: When Cancer Spreads—National Cancer Institute. Available online: https://www.cancer.gov/types/metastatic-cancer (accessed on 15 December 2021).
- Huang, C.-Y.; Ju, D.-T.; Chang, C.-F.; Muralidhar Reddy, P.; Velmurugan, B.K. A Review on the Effects of Current Chemotherapy Drugs and Natural Agents in Treating Non-Small Cell Lung Cancer. Biomedicine 2017, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Targeted Cancer Therapies Fact Sheet—National Cancer Institute. Available online: https://www.cancer.gov/about-cancer/treatment/types/targeted-therapies/targeted-therapies-fact-sheet (accessed on 15 December 2021).
- Hassanpour, S.H.; Dehghani, M. Review of Cancer from Perspective of Molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Sams-Dodd, F. Target-Based Drug Discovery: Is Something Wrong? Drug Discov. Today 2005, 10, 139–147. [Google Scholar] [CrossRef]
- Zhang, J.; Späth, S.S.; Marjani, S.L.; Zhang, W.; Pan, X. Characterization of Cancer Genomic Heterogeneity by Next-Generation Sequencing Advances Precision Medicine in Cancer Treatment. Precis. Clin. Med. 2018, 1, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.S.; Ryan, M.J.; Dhruv, H.D.; Gill, S.; Eichhoff, O.M.; Seashore-Ludlow, B.; Kaffenberger, S.D.; Eaton, J.K.; Shimada, K.; Aguirre, A.J.; et al. Dependency of a Therapy-Resistant State of Cancer Cells on a Lipid Peroxidase Pathway. Nature 2017, 547, 453–457. [Google Scholar] [CrossRef]
- Raghavendra, N.M.; Pingili, D.; Kadasi, S.; Mettu, A.; Prasad, S.V.U.M. Dual or Multi-Targeting Inhibitors: The next Generation Anticancer Agents. Eur. J. Med. Chem. 2018, 143, 1277–1300. [Google Scholar] [CrossRef]
- Tosi, D.; Pérez-Gracia, E.; Atis, S.; Vié, N.; Combès, E.; Gabanou, M.; Larbouret, C.; Jarlier, M.; Mollevi, C.; Torro, A.; et al. Rational Development of Synergistic Combinations of Chemotherapy and Molecular Targeted Agents for Colorectal Cancer Treatment. BMC Cancer 2018, 18, 812. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primers 2019, 5, 1–31. [Google Scholar] [CrossRef]
- Lawrence, R.T.; Perez, E.M.; Hernández, D.; Miller, C.P.; Haas, K.M.; Irie, H.Y.; Lee, S.-I.; Blau, C.A.; Villén, J. The Proteomic Landscape of Triple-Negative Breast Cancer. Cell Rep. 2015, 11, 630–644. [Google Scholar] [CrossRef]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of Epidermal Growth Factor Receptor in Breast Cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef]
- Madden, J.M.; Mueller, K.L.; Bollig-Fischer, A.; Stemmer, P.; Mattingly, R.R.; Boerner, J.L. Abrogating Phosphorylation of EIF4B Is Required for EGFR and MTOR Inhibitor Synergy in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2014, 147, 283–293. [Google Scholar] [CrossRef][Green Version]
- Zhou, X.; Tan, M.; Stone Hawthorne, V.; Klos, K.S.; Lan, K.-H.; Yang, Y.; Yang, W.; Smith, T.L.; Shi, D.; Yu, D. Activation of the Akt/Mammalian Target of Rapamycin/4E-BP1 Pathway by ErbB2 Overexpression Predicts Tumor Progression in Breast Cancers. Clin. Cancer Res. 2004, 10, 6779–6788. [Google Scholar] [CrossRef]
- Buck, E.; Eyzaguirre, A.; Brown, E.; Petti, F.; McCormack, S.; Haley, J.D.; Iwata, K.K.; Gibson, N.W.; Griffin, G. Rapamycin Synergizes with the Epidermal Growth Factor Receptor Inhibitor Erlotinib in Non-Small-Cell Lung, Pancreatic, Colon, and Breast Tumors. Mol. Cancer Ther. 2006, 5, 2676–2684. [Google Scholar] [CrossRef]
- Gadgeel, S.M.; Lew, D.L.; Synold, T.W.; LoRusso, P.; Chung, V.; Christensen, S.D.; Smith, D.C.; Kingsbury, L.; Hoering, A.; Kurzrock, R. Phase I Study Evaluating the Combination of Lapatinib (a Her2/Neu and EGFR Inhibitor) and Everolimus (an MTOR Inhibitor) in Patients with Advanced Cancers: South West Oncology Group (SWOG) Study S0528. Cancer Chemother. Pharmacol. 2013, 72, 1089–1096. [Google Scholar] [CrossRef]
- Lee, Y.W.; Mirocha, C.J.; Shroeder, D.J.; Walser, M.M. TDP-1, a Toxic Component Causing Tibial Dyschondroplasia in Broiler Chickens, and Trichothecenes from Fusarium Roseum “Graminearum”. Appl. Environ. Microbiol. 1985, 50, 102–107. [Google Scholar] [CrossRef]
- Dréau, D.; Foster, M.; Hogg, M.; Culberson, C.; Nunes, P.; Wuthier, R.E. Inhibitory Effects of Fusarochromanone on Melanoma Growth. Anticancer Drugs 2007, 18, 897–904. [Google Scholar] [CrossRef]
- Mahdavian, E.; Palyok, P.; Adelmund, S.; Williams-Hart, T.; Furmanski, B.D.; Kim, Y.-J.; Gu, Y.; Barzegar, M.; Wu, Y.; Bhinge, K.N.; et al. Biological Activities of Fusarochromanone: A Potent Anti-Cancer Agent. BMC Res. Notes 2014, 7, 601. [Google Scholar] [CrossRef]
- Mahdavian, E.; Marshall, M.; Martin, P.M.; Cagle, P.; Salvatore, B.A.; Quick, Q.A. Caspase-Dependent Signaling Underlies Glioblastoma Cell Death in Response to the Fungal Metabolite, Fusarochromanone. Int. J. Mol. Med. 2014, 34, 880–885. [Google Scholar] [CrossRef]
- Kuang, Y.-H.; Shen, T.; Chen, X.; Sodani, K.; Hopper-Borge, E.; Tiwari, A.K.; Lee, J.W.K.K.; Fu, L.-W.; Chen, Z.-S. Lapatinib and Erlotinib Are Potent Reversal Agents for MRP7 (ABCC10)-Mediated Multidrug Resistance. Biochem. Pharmacol. 2010, 79, 154–161. [Google Scholar] [CrossRef]
- Iyer, R.; Bharthuar, A. A Review of Erlotinib--an Oral, Selective Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor. Expert Opin. Pharmacother. 2010, 11, 311–320. [Google Scholar] [CrossRef]
- Wainberg, Z.A.; Anghel, A.; Desai, A.J.; Ayala, R.; Luo, T.; Safran, B.; Fejzo, M.S.; Hecht, J.R.; Slamon, D.J.; Finn, R.S. Lapatinib, a Dual EGFR and HER2 Kinase Inhibitor, Selectively Inhibits HER2-Amplified Human Gastric Cancer Cells and Is Synergistic with Trastuzumab in Vitro and in Vivo. Clin. Cancer Res. 2010, 16, 1509–1519. [Google Scholar] [CrossRef]
- Cohen, M.H.; Johnson, J.R.; Chen, Y.-F.; Sridhara, R.; Pazdur, R. FDA Drug Approval Summary: Erlotinib (Tarceva) Tablets. Oncologist 2005, 10, 461–466. [Google Scholar] [CrossRef]
- Voigtlaender, M.; Schneider-Merck, T.; Trepel, M. Lapatinib. Recent Results Cancer Res. 2018, 211, 19–44. [Google Scholar] [CrossRef]
- Xia, W.; Mullin, R.J.; Keith, B.R.; Liu, L.H.; Ma, H.; Rusnak, D.W.; Owens, G.; Alligood, K.J.; Spector, N.L. Anti-tumor activity of GW572016: A dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene 2002, 21, 6255–6263. [Google Scholar] [CrossRef]
- Hasskarl, J. Everolimus. Recent Results Cancer Res. 2018, 211, 101–123. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carroll, N.; Youngblood, R.; Smith, A.; Dragoi, A.-M.; Salvatore, B.A.; Mahdavian, E. TNBC Therapeutics Based on Combination of Fusarochromanone with EGFR Inhibitors. Biomedicines 2022, 10, 2906. https://doi.org/10.3390/biomedicines10112906
Carroll N, Youngblood R, Smith A, Dragoi A-M, Salvatore BA, Mahdavian E. TNBC Therapeutics Based on Combination of Fusarochromanone with EGFR Inhibitors. Biomedicines. 2022; 10(11):2906. https://doi.org/10.3390/biomedicines10112906
Chicago/Turabian StyleCarroll, Natalie, Reneau Youngblood, Alena Smith, Ana-Maria Dragoi, Brian A. Salvatore, and Elahe Mahdavian. 2022. "TNBC Therapeutics Based on Combination of Fusarochromanone with EGFR Inhibitors" Biomedicines 10, no. 11: 2906. https://doi.org/10.3390/biomedicines10112906
APA StyleCarroll, N., Youngblood, R., Smith, A., Dragoi, A.-M., Salvatore, B. A., & Mahdavian, E. (2022). TNBC Therapeutics Based on Combination of Fusarochromanone with EGFR Inhibitors. Biomedicines, 10(11), 2906. https://doi.org/10.3390/biomedicines10112906





