H19 and TUG1 lncRNAs as Novel Biomarkers for Irritable Bowel Syndrome in Diabetic Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Subjects
2.3. Sampling
2.4. Method
2.4.1. Laboratory Tests
2.4.2. LncRNA Expression
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Canavan, C.; West, J.; Card, T. The epidemiology of irritable bowel syndrome. Clin. Epidemiol. 2014, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Shoji, T.; Fukudo, S. Epidemiology of irritable bowel syndrome. Ann. Gastroenterol. 2015, 28, 158–159. [Google Scholar] [PubMed]
- Jahng, J.; Kim, Y.S. Irritable Bowel Syndrome: Is It Really a Functional Disorder? A New Perspective on Alteration of Enteric Nervous System. J. Neurogastroenterol. Motil. 2016, 22, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Bytzer, P.; Talley, N.J.; Leemon, M.; Young, L.J.; Jones, M.P.; Horowitz, M. Prevalence of Gastrointestinal Symptoms Associated With Diabetes Mellitus: A population-based survey of 15,000 adults. Arch. Intern. Med. 2001, 161, 1989–1996. [Google Scholar] [CrossRef]
- Chandrasekharan, B.; Srinivasan, S. Diabetes and the enteric nervous system. Neurogastroenterol. Motil. 2007, 19, 951–960. [Google Scholar] [CrossRef]
- Watkins, C.C.; Sawa, A.; Jaffrey, S.; Blackshaw, S.; Barrow, R.K.; Snyder, S.H.; Ferris, C.D. Insulin restores neuronal nitric oxide synthase expression and function that is lost in diabetic gastropathy. J. Clin. Investig. 2000, 106, 373–384. [Google Scholar] [CrossRef]
- Sabbagh, F.; Muhamad, I.I.; Niazmand, R.; Dikshit, P.K.; Kim, B.S. Recent progress in polymeric non-invasive insulin delivery. Int. J. Biol. Macromol. 2022, 203, 222–243. [Google Scholar] [CrossRef]
- Zhong, L.; Liu, P.; Fan, J.; Luo, Y. Long non-coding RNA H19: Physiological functions and involvements in central nervous system disorders. Neurochem. Int. 2021, 148, 105072. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, C.; He, B.; He, R.; Xu, L.; Zhang, S. The Role of lncRNAs in Regulating the Intestinal Mucosal Mechanical Barrier. BioMed Res. Int. 2021, 2021, 2294942. [Google Scholar] [CrossRef]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Abdelghany, A.A.; Toraih, E.A.; Mohamed, A.M. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn. J. Basic Med. Sci. 2020, 20, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Alfaifi, M.; Verma, A.K.; Alshahrani, M.Y.; Joshi, P.C.; Alkhathami, A.G.; Ahmad, I.; Hakami, A.R.; Beg, M.M.A. Assessment of Cell-Free Long Non-Coding RNA-H19 and miRNA-29a, miRNA-29b Expression and Severity of Diabetes. Diabetes Metab. Syndr. Obesity Targets Ther. 2020, 13, 3727–3737. [Google Scholar] [CrossRef] [PubMed]
- Chao, G.; Wang, Z.; Yang, Y.; Zhang, S. LncRNA H19 as a Competing Endogenous RNA to Regulate AQP Expression in the Intestinal Barrier of IBS-D Patients. Front. Physiol. 2021, 11, 602076. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Qi, Y.; Qu, J.; Gai, L.; Shi, Y.; Yuan, C. Pathophysiological Functions of the lncRNA TUG1. Curr. Pharm. Des. 2020, 26, 688–700. [Google Scholar] [CrossRef]
- Zhao, K.; Tan, J.-Y.; Mao, Q.-D.; Ren, K.-Y.; He, B.-G.; Zhang, C.-P.; Wei, L.-Z. Overexpression of long non-coding RNA TUG1 alleviates TNF-α-induced inflammatory injury in interstitial cells of Cajal. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 312–320. [Google Scholar]
- Wei, W.; Wang, X.; Wei, Y.; Liu, S.; Gao, S.; Tian, H.; Su, D. lncRNA TUG1 protects intestinal epithelial cells from damage induced by high glucose and high fat via AMPK/SIRT1. Mol. Med. Rep. 2022, 25, 139. [Google Scholar] [CrossRef]
- Kim, J.H.; Lin, E.; Pimentel, M. Biomarkers of Irritable Bowel Syndrome. J. Neurogastroenterol. Motil. 2017, 23, 20–26. [Google Scholar] [CrossRef]
- Nakov, R.; Snegarova, V.; Dimitrova-Yurukova, D.; Velikova, T. Biomarkers in Irritable Bowel Syndrome: Biological Rationale and Diagnostic Value. Dig. Dis. 2021, 40, 23–32. [Google Scholar] [CrossRef]
- Lacy, B.E.; Patel, N.K. Rome Criteria and a Diagnostic Approach to Irritable Bowel Syndrome. J. Clin. Med. 2017, 6, 99. [Google Scholar] [CrossRef]
- Messiah, S. Body Mass Index. In Encyclopedia of Behavioral Medicine; Gellman, M.D., Turner, J.R., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Jo, J.; Choi, S.; Oh, J.; Lee, S.-G.; Choi, S.Y.; Kim, K.K.; Park, C. Conventionally used reference genes are not outstanding for normalization of gene expression in human cancer research. BMC Bioinform. 2019, 20, 13–21. [Google Scholar] [CrossRef]
- Singh, R.; Chandel, S.; Dey, D.; Ghosh, A.; Roy, S.; Ravichandiran, V.; Ghosh, D. Epigenetic modification and therapeutic targets of diabetes mellitus. Biosci. Rep. 2020, 40, BSR20202160. [Google Scholar] [CrossRef]
- Krishnan, B.; Babu, S.; Walker, J.; Walker, A.B.; Pappachan, J.M. Gastrointestinal complications of diabetes mellitus. World J. Diabetes 2013, 4, 51–63. [Google Scholar] [CrossRef]
- Nagasako, C.K.; Montes, C.G.; Lorena, S.L.S.; Mesquita, M.A. Irritable bowel syndrome subtypes: Clinical and psychological features, body mass index and comorbidities. Rev. Española Enferm. Dig. 2015, 108, 59–64. [Google Scholar] [CrossRef]
- Javadekar, N.S.; Oka, G.A.; Joshi, A.S.; Vaste, P.; Tamane, S.; Lawate, P.S. Prevalence of irritable bowel syndrome and metabolic syndrome among young adults in an annual health check-up setting. JGH Open Access J. Gastroenterol. Hepatol. 2021, 5, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Gulcan, E.; Taser, F.; Toker, A.; Korkmaz, U.; Alcelik, A. Increased Frequency of Prediabetes in Patients With Irritable Bowel Syndrome. Am. J. Med. Sci. 2009, 338, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Andalib, I.; Hsueh, W.; Shope, T.R.; Brebbia, J.S.; Koch, T.R. Prevalence of Irritable Bowel Syndrome in Morbidly Obese Individuals Seeking Bariatric Surgery. J. Gastroenterol. Hepatol. Res. 2018, 7, 2516–2520. [Google Scholar] [CrossRef]
- Lee, C.G.; Lee, J.K.; Kang, Y.-S.; Shin, S.; Kim, J.H.; Lim, Y.J.; Koh, M.-S.; Lee, J.H.; Kang, H.W. Visceral Abdominal Obesity Is Associated With an Increased Risk of Irritable Bowel Syndrome. Am. J. Gastroenterol. 2015, 110, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.-J.; Guan, Q.-K.; Meng, L.; Qin, N.; Zhao, J.; Jin, B.-Z. Long non-coding RNA TUG1 as a potential prognostic biomarker in human cancers: A meta-analysis. Oncotarget 2017, 8, 62454–62462. [Google Scholar] [CrossRef]
- Li, Z.; Shen, J.; Chan, M.T.; Wu, W.K.K. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016, 49, 471–475. [Google Scholar] [CrossRef]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Kamiya, S.; Ishiwata, T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front. Biosci. 2018, 23, 614–625. [Google Scholar] [CrossRef]
- Cheng, X.-W.; Chen, Z.-F.; Wan, Y.-F.; Zhou, Q.; Wang, H.; Zhu, H.-Q. Long Non-coding RNA H19 Suppression Protects the Endothelium Against Hyperglycemic-Induced Inflammation via Inhibiting Expression of miR-29b Target Gene Vascular Endothelial Growth Factor a Through Activation of the Protein Kinase B/Endothelial Nitric Oxide Synthase Pathway. Front. Cell Dev. Biol. 2019, 7, 263. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Morris, J.R. Genes, Genetics, and Epigenetics: A Correspondence. Science 2001, 293, 1103–1105. [Google Scholar] [CrossRef] [PubMed]
- Mahurkar-Joshi, S.; Chang, L. Epigenetic Mechanisms in Irritable Bowel Syndrome. Front. Psychiatry 2020, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Prandi, F.R.; Lecis, D.; Illuminato, F.; Milite, M.; Celotto, R.; Lerakis, S.; Romeo, F.; Barillà, F. Epigenetic Modifications and Non-Coding RNA in Diabetes-Mellitus-Induced Coronary Artery Disease: Pathophysiological Link and New Therapeutic Frontiers. Int. J. Mol. Sci. 2022, 23, 4589. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.; Tiwary, S.; Kesharwani, D.; Datta, M. Long non-coding RNA H19 inhibition promotes hyperglycemia in mice by upregulating hepatic FoxO1 levels and promoting gluconeogenesis. Klin. Wochenschr. 2018, 97, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Tello-Flores, V.A.; Valladares-Salgado, A.; Ramírez-Vargas, M.A.; Cruz, M.; Del-Moral-Hernández, O.; Cahua-Pablo, J.; Ramírez-Ruano, M.; Hernández-Sotelo, D.; Armenta-Solis, A.; Flores-Alfaro, E. Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type-2 diabetes. Non-Coding RNA Res. 2020, 5, 71–76. [Google Scholar] [CrossRef]
- Chen, S.-W.; Wang, P.-Y.; Liu, Y.-C.; Sun, L.; Zhu, J.; Zuo, S.; Ma, J.; Li, T.-Y.; Zhang, J.-L.; Chen, G.-W.; et al. Effect of Long Noncoding RNA H19 Overexpression on Intestinal Barrier Function and Its Potential Role in the Pathogenesis of Ulcerative Colitis. Inflamm. Bowel Dis. 2016, 22, 2582–2592. [Google Scholar] [CrossRef]
- Li, C.; Zhuang, M.; Zhu, B.; Li, Y.; Zhang, W.; Yan, H.; Zhang, P.; Li, D.; Yang, J.; Sun, Y.; et al. Epidermal growth factor regulation by autophagy-mediated lncRNA H19 in murine intestinal tract after severe burn. J. Cell. Mol. Med. 2020, 24, 5878–5887. [Google Scholar] [CrossRef]
- Juan, V.; Crain, C.; Wilson, C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000, 28, 1221–1227. [Google Scholar] [CrossRef]
- Liang, J.W.; Bai, W.J.; Wang, X.Y.; Chi, L.L. Integrated Analysis of lncRNA-miRNA-mRNA ceRNA Network in Human Diarrhea Irritable Bowel Syndrome. Res. Sq. 2021. Preprint V1. [Google Scholar] [CrossRef]
- Dieter, C.; Lemos, N.E.; Corrêa, N.R.d.F.; Assmann, T.S.; Crispim, D. The Impact of lncRNAs in Diabetes Mellitus: A Systematic Review and In Silico Analyses. Front. Endocrinol. 2021, 12, 602597. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; Benshoshan, M.; Di Segni, A.; Dexheimer, P.; Braun, T.; Weiss, B.; Walters, T.D.; Baldassano, R.N.; Noe, J.D.; Markowitz, J.; et al. Long ncRNA Landscape in the Ileum of Treatment-Naive Early-Onset Crohn Disease. Inflamm. Bowel Dis. 2018, 24, 346–360. [Google Scholar] [CrossRef] [PubMed]
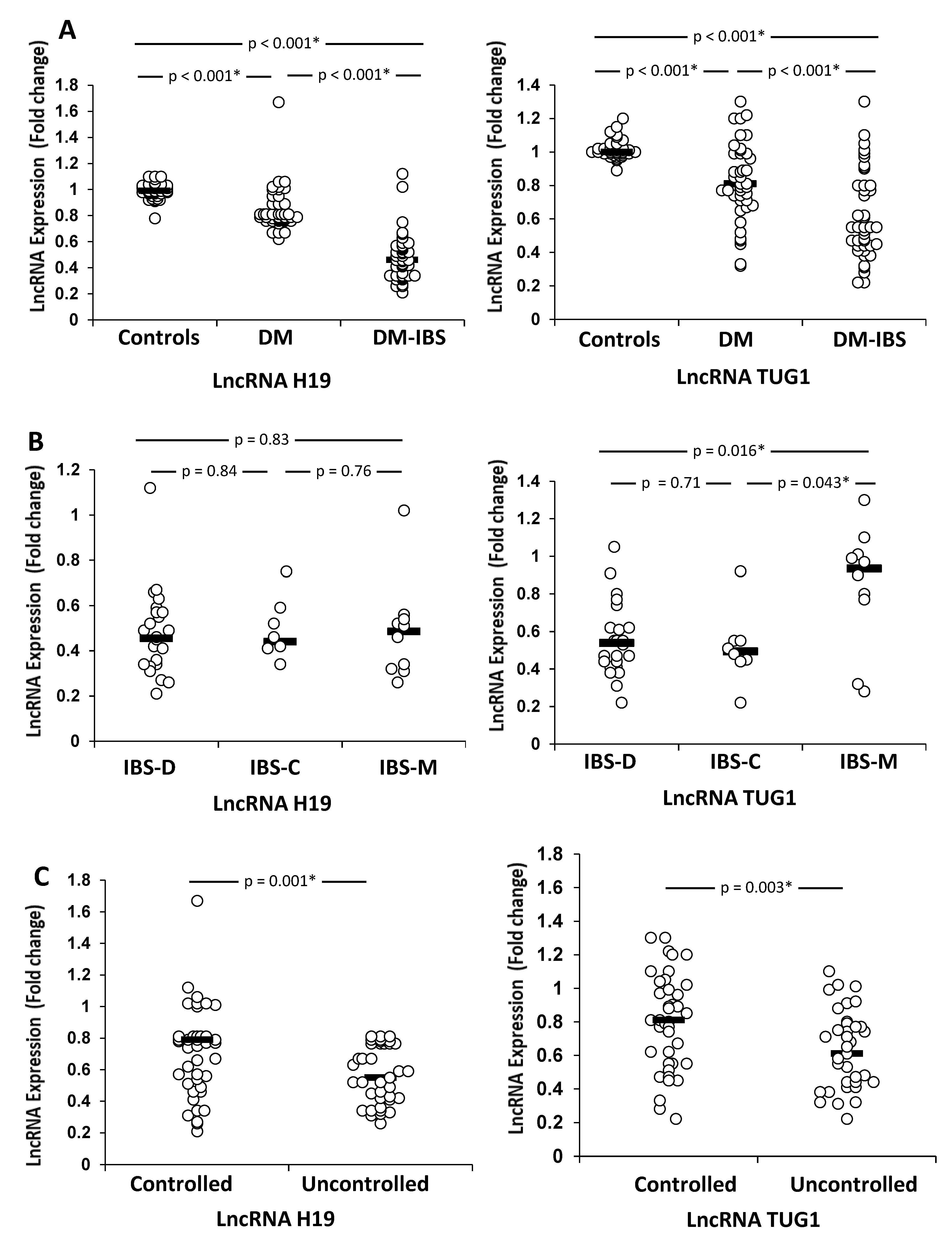
| Parameter | Control Group (No. = 42) | Diabetes Group (No. = 42) | Diabetic IBS Group (No. = 42) | p |
|---|---|---|---|---|
| Age (years) | 36 (21–60) | 37.5 (22–5) | 36 (22–56) | 0.35 |
| Sex Male/female | 20/22 (47.6/52.4) | 22/20 (52.4/47.6) | 17/25 (40.5/59.5) | 0.75 |
| Duration of IBS symptoms (months) | ------- | ------ | 7 [1–30] | |
| Smoking | 20 (47.6) | 20 (47.6) | 26 (61.9) | 0.32 |
| BMI (kg/m2) | 25.4 (23.3–29.5) | 27 (23.3–29.6) a | 27.4 (22.6–33.2) a | 0.002 * |
| IBS Type: | ||||
| IBS—diarrhea | 24 (57.1) | |||
| IBS—constipation | 8 (19) | |||
| IBS—mixed | 10 (23.9) | |||
| Severity: | ||||
| IBS-SSS | 289 (80–430) | |||
| Mild | 6 (14.3) | |||
| Moderate | 17 (40.5) | |||
| Severe | 19 (45.2) | |||
| Laboratory Tests: | ||||
| Fasting glucose (mg/dL) | 83.4 (74.8–101) | 120.3 (108.3–224.3) a | 132.3 (98–313.6) a | <0.001 * |
| HbA1c (%) | 4.1 (3.2–5) | 6.1 (4.9–8.9) a | 7.2 (5.5–15.2) a, b | <0.001 * |
| Total cholesterol (mg/dL) | 127.9 (76.4–188) | 117 (88–171.3) | 160.8 (100.9–289.5) a, b | <0.001 * |
| Triglyceride (mg/dL) | 82 (65.3–145.6) | 84 (74–124.2) | 98.6 (72.4–199.4) b | 0.036 * |
| HDL-C (mg/dL) | 42.7 (32.1–62.3) | 42.1 (26.9–55.2) | 42.4 (29.8–57.4) | 0.78 |
| LDL -C(mg/dL) | 61.7 (26–122.5) | 63.8 (23.2–115.9) | 101.5 (41.9–224.6) a, b | <0.001 * |
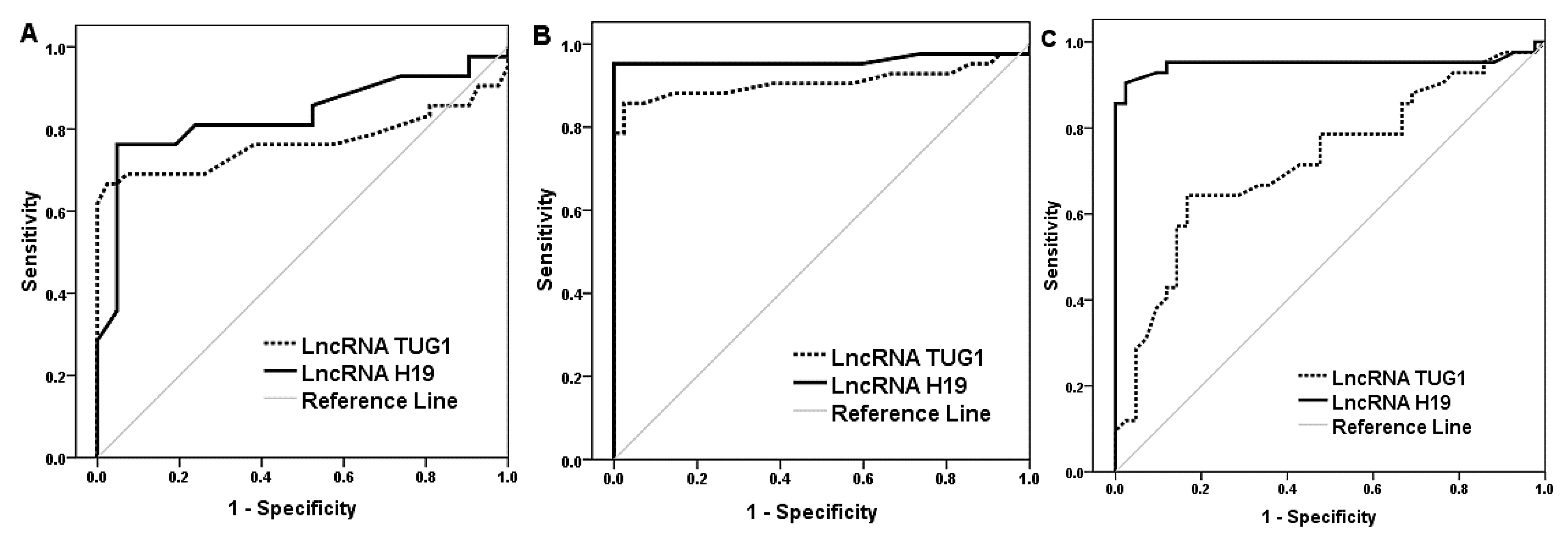
| Marker | AUC [95% CI] | Cutoff | Youden’s Index | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| Differentiation between healthy controls and diabetic-only patients | ||||||||
| LncRNA H19 | 0.832 [0.737–0.927] | 0.9 | 0.71 | 76.2 | 95.2 | 94.1 | 80 | 85.7 |
| LncRNA TUG1 | 0.768 [0.653–0.882] | 0.92 | 0.64 | 66.6 | 97.6 | 96.6 | 74.5 | 82.1 |
| Differentiation between healthy controls and diabetic patients with IBS | ||||||||
| LncRNA H19 | 0.960 [0.905–1.015] | 0.76 | 0.95 | 95.2 | 100 | 100 | 95.5 | 97.6 |
| LncRNA TUG1 | 0.908 [0.830–0.985] | 0.94 | 0.83 | 85.7 | 97.6 | 97.3 | 87.2 | 91.7 |
| Differentiation between diabetic patients and diabetic patients with IBS | ||||||||
| LncRNA H19 | 0.950 [0.889–1.011] | 0.66 | 0.88 | 90.5 | 97.6 | 97.4 | 91.1 | 94 |
| LncRNA TUG1 | 0.722 [0.611–0.833] | 0.64 | 0.47 | 64.3 | 83.3 | 79.4 | 70 | 73.8 |
| Parameter | LncRNA H19 | LncRNA TUG1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Diabetes Group (No. = 42) | Diabetic IBS Group (No. = 42) | Diabetes Group (no. = 42) | Diabetic IBS Group (no. = 42) | |||||
| r | p | r | p | r | p | r | p | |
| Age | −0.13 | 0.4 | −0.02 | 0.88 | −0.1 | 0.52 | −0.3 | 0.05 |
| BMI | −0.25 | 0.11 | 0.3 | 0.85 | −0.27 | 0.09 | 0.19 | 0.22 |
| Symptoms duration | ---- | ---- | 0.05 | 0.73 | ---- | ---- | 0.16 | 0.31 |
| IBS-SSS | ---- | ---- | −0.46 | 0.002 * | ---- | ---- | −0.37 | 0.017 * |
| Fasting glucose | 0.07 | 0.66 | 0.24 | 0.12 | 0.04 | 081 | 0.23 | 0.14 |
| HbA1c | −0.39 | 0.01 * | −0.28 | 0.07 | −0.31 | 0.04 * | −0.07 | 0.66 |
| Cholesterol | −0.42 | 0.006 * | 0.19 | 0.91 | 0.09 | 0.57 | 0.01 | 0.98 |
| LDL-C | −0.46 | 0.002 * | 0.05 | 0.75 | 0.07 | 0.66 | −0.01 | 0.93 |
| Triglycerides | −0.36 | 0.02 * | −0.24 | 0.13 | 0.02 | 0.9 | −0.16 | 0.33 |
| HDL-C | 0.18 | 0.24 | 0.14 | 0.37 | −0.11 | 0.5 | 0.21 | 0.85 |
| LncRNA H19 | 1 | 1 | 0.13 | 0.4 | 0.24 | 0.12 | ||
| LncRNA TUG1 | 0.13 | 0.4 | 0.24 | 0.12 | 1 | 1 | ||
| Covariate | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p | AOR (95% CI) | p | |
| Age | 1.1 (0.99–1.24) | 0.07 | 1.53 (0.91–2.58) | 0.11 |
| Sex (male) | 0.33 (0.09–1.21) | 0.09 | 0.42 (0.01–20.6) | 0.66 |
| Smoking | 1.1 (0.32–3.8) | 0.89 | 6.39 (0–699) | 0.63 |
| Symptoms duration | 1.02 (0.95–1.09) | 0.58 | 1.04 (0.81–1.35) | 0.75 |
| BMI | 0.73 (0.53–1.01) | 0.05 | 0.35 (0.07–1.6) | 0.18 |
| Fasting glucose | 1 (0.99–1.01) | 0.49 | 1.03 (0.98–1.08) | 0.17 |
| Diabetes control | 0.69 (0.2–2.35) | 0.55 | 0.06(0–406) | 0.63 |
| Total cholesterol | 0.99 (0.98–1) | 0.38 | 0.02 (0–7.4) | 0.14 |
| Triglyceride | 1.02 (0.99–1.06) | 0.19 | 2.2 (0.79–6.5) | 0.13 |
| HDL-C | 0.98 (0.9–1.07) | 0.68 | 381 (0.13–1089) | 0.14 |
| LDL -C | 0.99 (0.98–1.1) | 0.39 | 452 (0.12–1602) | 0.14 |
| LncRNA H19 | 0.001 (0–0.33) | 0.019 * | 0.00001 (0–0.5) | 0.045 * |
| LncRNA TUG1 | 0.02 (0.001–0.45) | 0.01 * | 0.01(0–8.3) | 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esawy, M.M.; Saleh Al-Sowayan, N.; Mobasher, M.A.; Abd-elhameed, A.; Abd elbaser, E.S.; Baioumy, S.A.; Shabana, M.A. H19 and TUG1 lncRNAs as Novel Biomarkers for Irritable Bowel Syndrome in Diabetic Patients. Biomedicines 2022, 10, 2978. https://doi.org/10.3390/biomedicines10112978
Esawy MM, Saleh Al-Sowayan N, Mobasher MA, Abd-elhameed A, Abd elbaser ES, Baioumy SA, Shabana MA. H19 and TUG1 lncRNAs as Novel Biomarkers for Irritable Bowel Syndrome in Diabetic Patients. Biomedicines. 2022; 10(11):2978. https://doi.org/10.3390/biomedicines10112978
Chicago/Turabian StyleEsawy, Marwa M., Noorah Saleh Al-Sowayan, Maysa A. Mobasher, Amir Abd-elhameed, Elsayed S. Abd elbaser, Shereen A. Baioumy, and Marwa A. Shabana. 2022. "H19 and TUG1 lncRNAs as Novel Biomarkers for Irritable Bowel Syndrome in Diabetic Patients" Biomedicines 10, no. 11: 2978. https://doi.org/10.3390/biomedicines10112978
APA StyleEsawy, M. M., Saleh Al-Sowayan, N., Mobasher, M. A., Abd-elhameed, A., Abd elbaser, E. S., Baioumy, S. A., & Shabana, M. A. (2022). H19 and TUG1 lncRNAs as Novel Biomarkers for Irritable Bowel Syndrome in Diabetic Patients. Biomedicines, 10(11), 2978. https://doi.org/10.3390/biomedicines10112978





