Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study
Abstract
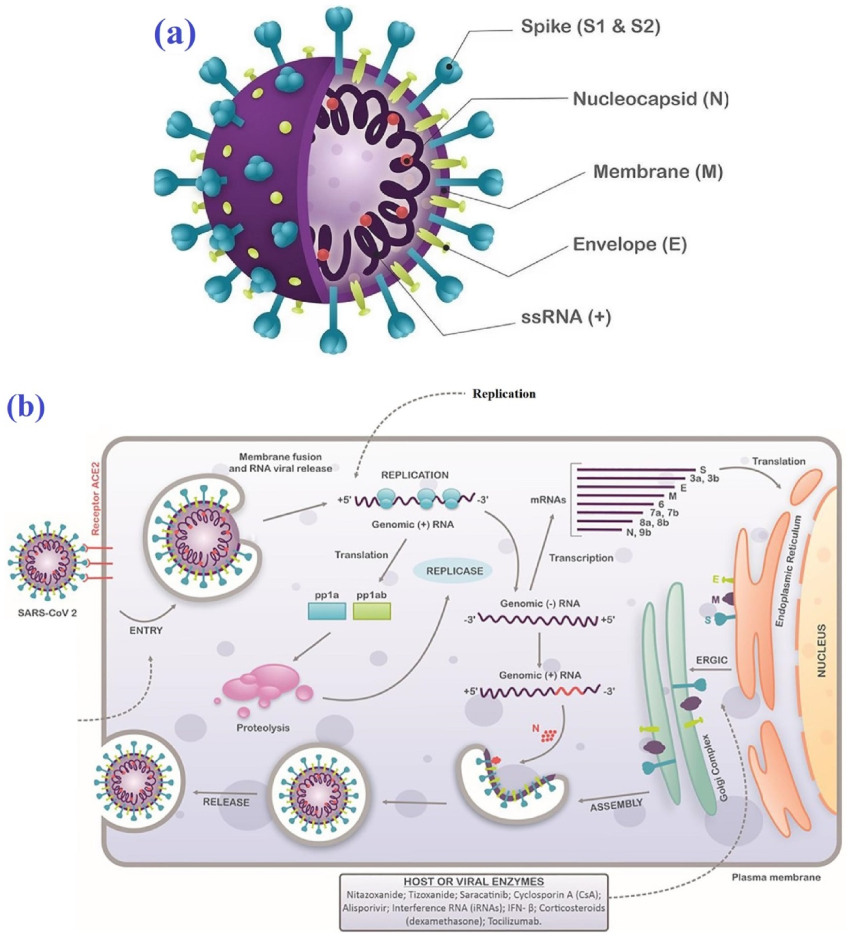
1. Introduction
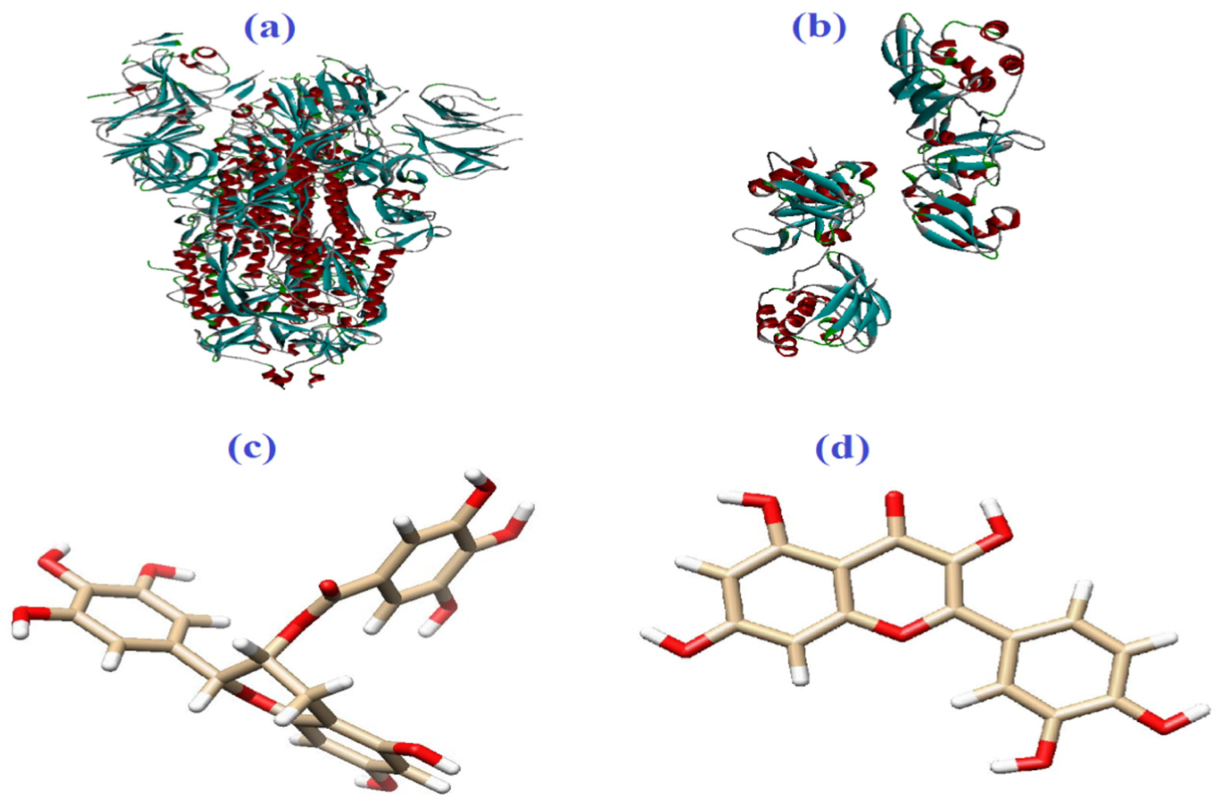
2. Materials and Methods
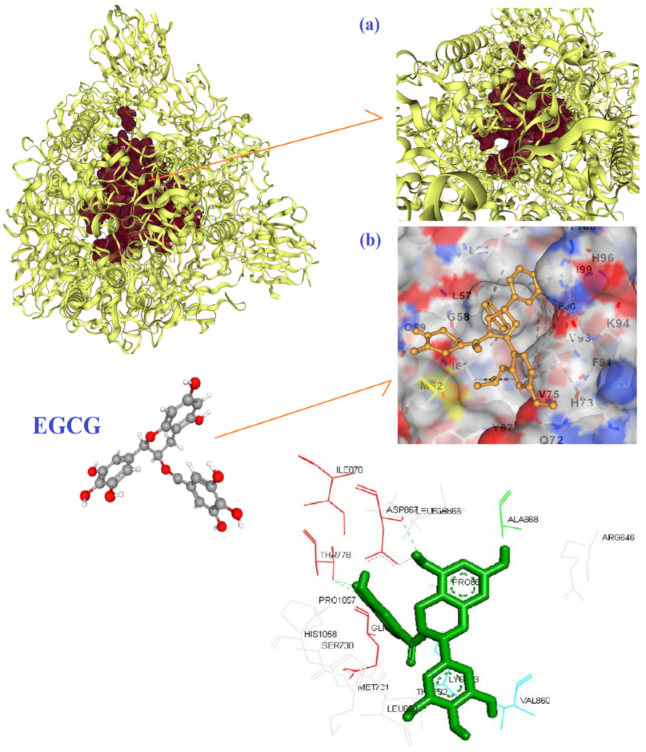
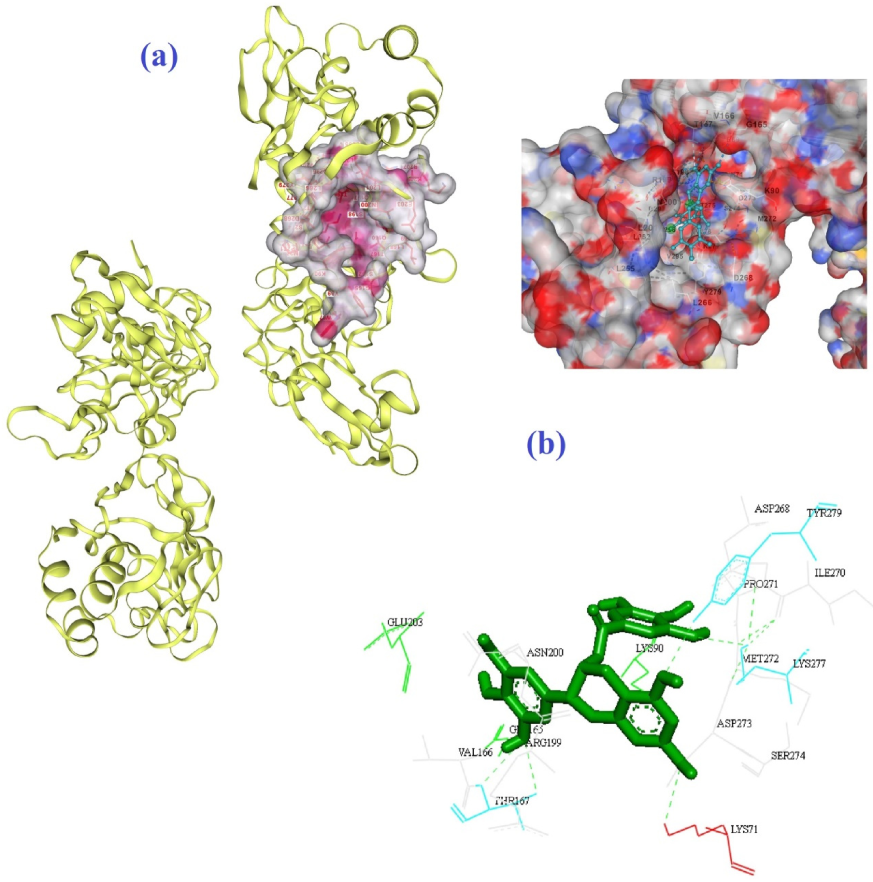
3. Results and Discussion
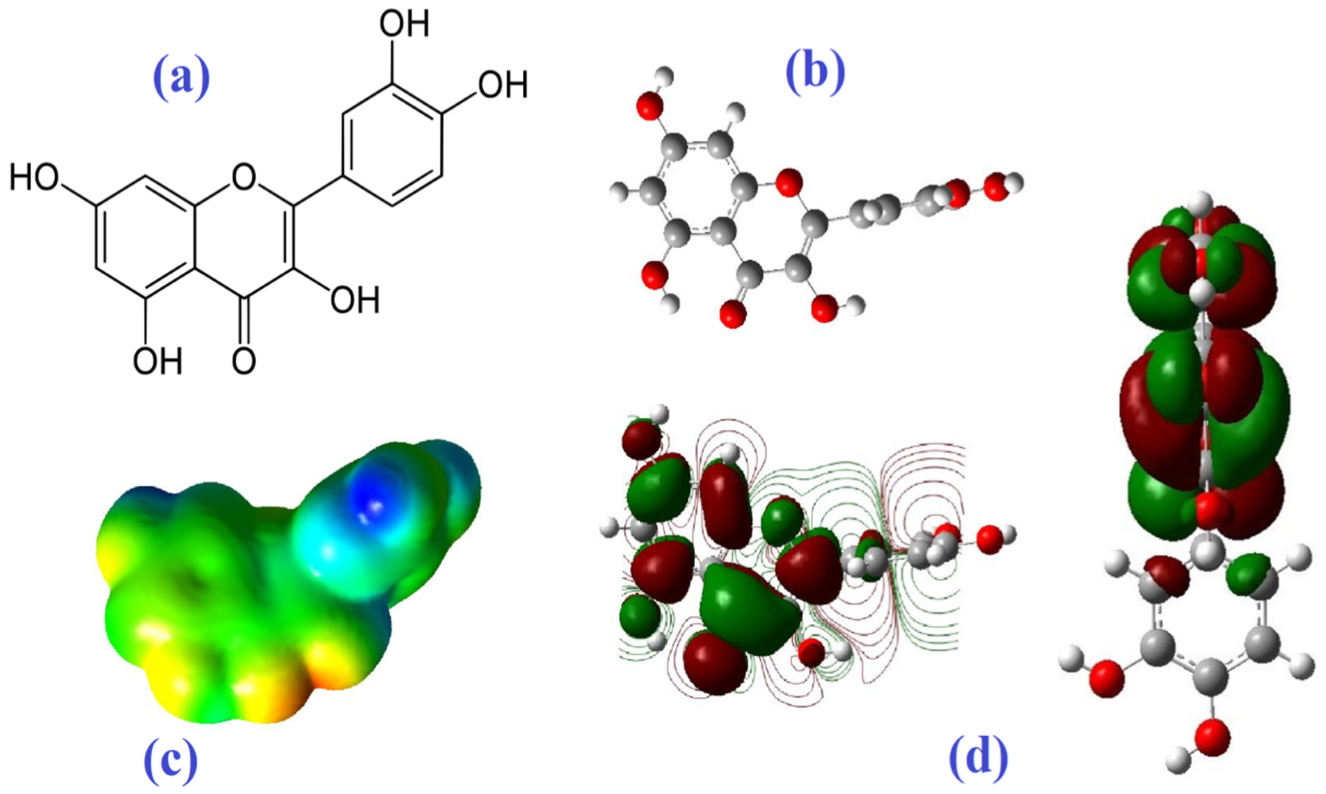
Molecular Electrostatic Potential
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aljelehawy, Q.H.A.; Alshaibah, L.H.H.; Khafaji, Z.K.A.A. Evaluation of virulence factors among Staphylococcus aureus strains isolated from patients with urinary tract infection in Al-Najaf Al-Ashraf teaching hospital. Cell. Mol. Biomed. Rep. 2021, 1, 78–87. [Google Scholar] [CrossRef]
- Ahmadi, S.; Ahmadi, G.; Ahmadi, H. A review on antifungal and antibacterial activities of some medicinal plants. Micro Nano Bio Asp. 2022, 1, 10–17. [Google Scholar]
- Alavi, M.; Rai, M. Antisense RNA, the modified CRISPR-Cas9, and metal/metal oxide nanoparticles to inactivate pathogenic bacteria. Cell. Mol. Biomed. Rep. 2021, 1, 52–59. [Google Scholar] [CrossRef]
- Amraei, S.; Ahmadi, S. Recent studies on antimicrobial and anticancer activities of saponins: A mini-review. Nano Micro Bios. 2022, 1, 22–26. [Google Scholar]
- Muhammad, I.; Sale, P.M.; Salisu, M.K.; Muhammad, T.M.; Abubakar, B.; Maidala, A.L.; Nuwanyada, E. Molecular analysis of Bio-makers of Chloroquine resistance in Plasmodium falciparum Isolate from Gombe Local Government Area, Gombe State, Nigeria. Cell. Mol. Biomed. Rep. 2022, 2, 42–55. [Google Scholar] [CrossRef]
- Baral, B.; Mozafari, M.R. Strategic Moves of “Superbugs” Against Available Chemical Scaffolds: Signaling, Regulation, and Challenges. ACS Pharmacol. Transl. Sci. 2020, 3, 373–400. [Google Scholar] [CrossRef]
- Mozafari, M.R.; Torkaman, S.; Karamouzian, F.M.; Rasti, B.; Baral, B. Antimicrobial Applications of Nanoliposome Encapsulated Silver Nanoparticles: A Potential Strategy to Overcome Bacterial Resistance. Curr. Nanosci. 2021, 17, 26–40. [Google Scholar] [CrossRef]
- Muhammad, I.; Abubakar, B.; Muhammad, T.M. Genetic resistance to human malaria. Cell. Mol. Biomed. Rep. 2022, 2, 116–128. [Google Scholar] [CrossRef]
- Alavi, M.; Martinez, F.; Delgado, D.R.; Tinjacá, D.A. Anticancer and antibacterial activities of embelin: Micro and nano aspects. Micro Nano Bio Asp. 2022, 1, 30–37. [Google Scholar]
- Alavi, M.; Hamblin, M.R.; Martinez, F.; Aghaie, E.; Khan, H.; Menezes, I.A. Micro and nanoformulations of insulin: New approaches. Nano Micro Bios. 2022, 1, 1–7. [Google Scholar]
- Alavi, M.; Rai, M.; Martinez, F.; Kahrizi, D.; Khan, H.; de Menezes, I.R.A.; Coutinho, H.D.M.; Costa, J.G.M. The efficiency of metal, metal oxide, and metalloid nanoparticles against cancer cells and bacterial pathogens: Different mechanisms of action. Cell. Mol. Biomed. Rep. 2022, 2, 10–21. [Google Scholar] [CrossRef]
- Almasian-Tehrani, N.; Alebouyeh, M.; Armin, S.; Soleimani, N.; Azimi, L.; Shaker-Darabad, R. Overview of typing techniques as molecular epidemiology tools for bacterial characterization. Cell. Mol. Biomed. Rep. 2021, 1, 69–77. [Google Scholar] [CrossRef]
- Alavi, M.; Hamblin, M.R.; Mozafari, M.R.; Rose Alencar de Menezes, I.; Douglas Melo Coutinho, H. Surface modification of SiO2 nanoparticles for bacterial decontaminations of blood products. Cell. Mol. Biomed. Rep. 2022, 2, 87–97. [Google Scholar] [CrossRef]
- Alavi, M.; Kowalski, R.; Capasso, R.; Douglas Melo Coutinho, H.; Rose Alencar de Menezes, I. Various novel strategies for functionalization of gold and silver nanoparticles to hinder drug-resistant bacteria and cancer cells. Micro Nano Bio Asp. 2022, 1, 38–48. [Google Scholar]
- Ahmadi, S. Antibacterial and antifungal activities of medicinal plant species and endophytes. Cell. Mol. Biomed. Rep. 2022, 2, 109–115. [Google Scholar] [CrossRef]
- Alavi, M.; Hamblin, M.R.; Martinez, F.; Kennedy, J.F.; Khan, H. Synergistic combinations of metal, metal oxide, or metalloid nanoparticles plus antibiotics against resistant and non-resistant bacteria. Micro Nano Bio Asp. 2022, 1, 1–9. [Google Scholar]
- Alavi, M.; Thomas, S.; Sreedharan, M. Modification of silica nanoparticles for antibacterial activities: Mechanism of action. Micro Nano Bio Asp. 2022, 1, 49–58. [Google Scholar]
- Sabbagh, F.; Kiarostami, K.; Mahmoudi Khatir, N.; Rezania, S.; Muhamad, I.I. Green Synthesis of Mg0.99 Zn0.01O Nanoparticles for the Fabrication of κ-Carrageenan/NaCMC Hydrogel in order to Deliver Catechin. Polymers 2020, 12, 861. [Google Scholar] [CrossRef]
- Sabbagh, F.; Kiarostami, K.; Khatir, N.M.; Rezania, S.; Muhamad, I.I.; Hosseini, F. Effect of zinc content on structural, functional, morphological, and thermal properties of kappa-carrageenan/NaCMC nanocomposites. Polym. Test. 2021, 93, 106922. [Google Scholar] [CrossRef]
- Shehata, A.A.; Attia, Y.A.; Rahman, M.T.; Basiouni, S.; El-Seedi, H.R.; Azhar, E.I.; Khafaga, A.F.; Hafez, H.M. Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2. Animals 2022, 12, 378. [Google Scholar] [CrossRef]
- Tamimi, F.; Altigani, S.; Sanz, M. Periodontitis and coronavirus disease 2019. Periodontol. 2000 2022, 89, 207–214. [Google Scholar] [CrossRef]
- Mohammadi, M.R.; Sabati, H. When Successive Viral Mutations Prevent Definitive Treatment of COVID-19. Cell. Mol. Biomed. Rep. 2022, 2, 98–108. [Google Scholar] [CrossRef]
- Rahbar-Karbasdehi, E.; Rahbar-Karbasdehi, F. Clinical challenges of stress cardiomyopathy during coronavirus 2019 epidemic. Cell. Mol. Biomed. Rep. 2021, 1, 88–90. [Google Scholar] [CrossRef]
- Alavi, M.; Asare-Addo, K.; Nokhodchi, A. Lectin Protein as a Promising Component to Functionalize Micelles, Liposomes and Lipid NPs against Coronavirus. Biomedicines 2020, 8, 580. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Santos, I.A.; Grosche, V.R.; Bergamini, F.R.G.; Sabino-Silva, R.; Jardim, A.C.G. Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment? Front. Microbiol. 2020, 11, 1818. [Google Scholar] [CrossRef]
- Casas-Sanchez, A.; Romero-Ramirez, A.; Hargreaves, E.; Ellis, C.C.; Grajeda, B.I.; Estevao, I.L.; Patterson, E.I.; Hughes, G.L.; Almeida, I.C.; Zech, T.; et al. Inhibition of Protein N-Glycosylation Blocks SARS-CoV-2 Infection. mBio 2022, 13, e03718-21. [Google Scholar] [CrossRef]
- Pillon, M.C.; Frazier, M.N.; Dillard, L.B.; Williams, J.G.; Kocaman, S.; Krahn, J.M.; Perera, L.; Hayne, C.K.; Gordon, J.; Stewart, Z.D.; et al. Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15. Biorxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Kabarkouhi, Z.; Mehrarya, M.; Gharehchelou, B.; Jalilian, Z.; Jalili, R.; Wintrasiri, M.N.; Mozafari, M.R. Liposome, Nanoliposome and Allied Technologies in COVID-19 Vaccines: Key Roles and Functionalities. Curr. Drug Deliv. 2022, 20, 3–7. [Google Scholar] [CrossRef]
- Mirtaleb, M.S.; Mirtaleb, A.H.; Nosrati, H.; Heshmatnia, J.; Falak, R.; Zolfaghari Emameh, R. Potential therapeutic agents to COVID-19: An update review on antiviral therapy, immunotherapy, and cell therapy. Biomed. Pharmacother. 2021, 138, 111518. [Google Scholar] [CrossRef]
- Taguchi, Y.-H.; Turki, T. A new advanced in silico drug discovery method for novel coronavirus (SARS-CoV-2) with tensor decomposition-based unsupervised feature extraction. PLoS ONE 2020, 15, e0238907. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Brierley, S.; Gandhi, M.J.; Cohen, M.A.; Moschella, P.C.; Declan, A.B.L. Repurposing Therapeutics for Potential Treatment of SARS-CoV-2: A Review. Viruses 2020, 12, 705. [Google Scholar] [CrossRef] [PubMed]
- Kivrak, A.; Ulaş, B.; Kivrak, H. A comparative analysis for anti-viral drugs: Their efficiency against SARS-CoV-2. Int. Immunopharmacol. 2021, 90, 107232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Guo, X.; Geary, K.; Zhang, D. Emerging Therapeutic Strategies for COVID-19 patients. Discoveries 2020, 8, e105. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Parkar, J.; Ansari, A.; Vora, A.; Talwar, D.; Tiwaskar, M.; Patil, S.; Barkate, H. Role of favipiravir in the treatment of COVID-19. Int. J. Infect. Dis. 2021, 102, 501–508. [Google Scholar] [CrossRef]
- Arévalo, A.P.; Pagotto, R.; Pórfido, J.L.; Daghero, H.; Segovia, M.; Yamasaki, K.; Varela, B.; Hill, M.; Verdes, J.M.; Duhalde Vega, M.; et al. Ivermectin reduces in vivo coronavirus infection in a mouse experimental model. Sci. Rep. 2021, 11, 7132. [Google Scholar] [CrossRef]
- Mettelman, R.C.; Allen, E.K.; Thomas, P.G. Mucosal immune responses to infection and vaccination in the respiratory tract. Immunity 2022, 55, 749–780. [Google Scholar] [CrossRef]
- Tripathy, S.; Dassarma, B.; Roy, S.; Chabalala, H.; Matsabisa, M.G. A review on possible modes of action of chloroquine/hydroxychloroquine: Repurposing against SAR-CoV-2 (COVID-19) pandemic. Int. J. Antimicrob. Agents 2020, 56, 106028. [Google Scholar] [CrossRef]
- Kournoutou, G.G.; Dinos, G. Azithromycin through the Lens of the COVID-19 Treatment. Antibiotics 2022, 11, 1063. [Google Scholar] [CrossRef]
- Uzunova, K.; Filipova, E.; Pavlova, V.; Vekov, T. Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS-CoV-2. Biomed. Pharmacother. 2020, 131, 110668. [Google Scholar] [CrossRef]
- Vicenti, I.; Zazzi, M.; Saladini, F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021, 31, 325–337. [Google Scholar] [CrossRef]
- Yang, L.; Guo, J.; Yu, N.; Liu, Y.; Song, H.; Niu, J.; Gu, Y. Tocilizumab mimotope alleviates kidney injury and fibrosis by inhibiting IL-6 signaling and ferroptosis in UUO model. Life Sci. 2020, 261, 118487. [Google Scholar] [CrossRef]
- Fischer, M.; Müller, P.; Scheidt, H.A.; Luck, M. Drug–Membrane Interactions: Effects of Virus-Specific RNA-Dependent RNA Polymerase Inhibitors Remdesivir and Favipiravir on the Structure of Lipid Bilayers. Biochemistry 2022, 61, 1392–1403. [Google Scholar] [CrossRef]
- Komeno, T.; Furuta, Y.; Nakajima, N.; Tani, H.; Morinaga, Y. Analysis of the responsible site for favipiravir resistance in RNA-dependent RNA polymerase of influenza virus A/PR/8/34 (H1N1) using site-directed mutagenesis. Antivir. Res. 2022, 205, 105387. [Google Scholar] [CrossRef]
- Han, Y.J.; Ren, Z.G.; Li, X.X.; Yan, J.L.; Ma, C.Y.; Wu, D.D.; Ji, X.Y. Advances and challenges in the prevention and treatment of COVID-19. Int. J. Med. Sci. 2020, 17, 1803–1810. [Google Scholar] [CrossRef]
- Singh, P.K. Chapter 11—Molecular modeling studies of fused pyrimidine derivatives at various receptors. In Fused Pyrimidine-Based Drug Discovery; Kumar, R., Vardanyan, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 273–332. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Alessandri, F.; Oliva, A.; Borrazzo, C.; Dell'Isola, S.; Ialungo, A.M.; Rastrelli, E.; Pelli, M.; Raponi, G.; Turriziani, O.; et al. The role of teicoplanin in the treatment of SARS-CoV-2 infection: A retrospective study in critically ill COVID-19 patients (Tei-COVID study). J. Med. Virol. 2021, 93, 4319–4325. [Google Scholar] [CrossRef]
- Yang, S.N.Y.; Atkinson, S.C.; Wang, C.; Lee, A.; Bogoyevitch, M.A.; Borg, N.A.; Jans, D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res. 2020, 177, 104760. [Google Scholar] [CrossRef]
- Mahmud-Al-Rafat, A.; Majumder, A.; Taufiqur Rahman, K.M.; Mahedi Hasan, A.M.; Didarul Islam, K.M.; Taylor-Robinson, A.W.; Billah, M.M. Decoding the enigma of antiviral crisis: Does one target molecule regulate all? Cytokine 2019, 115, 13–23. [Google Scholar] [CrossRef]
- Narendrakumar, L.; Joseph, I.; Thomas, S. Potential effectiveness and adverse implications of repurposing doxycycline in COVID-19 treatment. Expert Rev. Anti-Infect. Ther. 2021, 19, 1001–1008. [Google Scholar] [CrossRef]
- Chamkhi, I.; Benali, T.; Aanniz, T.; El Menyiy, N.; Guaouguaou, F.E.; El Omari, N.; El-Shazly, M.; Zengin, G.; Bouyahya, A. Plant-microbial interaction: The mechanism and the application of microbial elicitor induced secondary metabolites biosynthesis in medicinal plants. Plant Physiol. Biochem. 2021, 167, 269–295. [Google Scholar] [CrossRef]
- Choudhary, S.; Zehra, A.; Mukarram, M.; Wani, K.I.; Naeem, M.; Hakeem, K.R.; Aftab, T. Potential Uses of Bioactive Compounds of Medicinal Plants and Their Mode of Action in Several Human Diseases. In Medicinal and Aromatic Plants: Healthcare and Industrial Applications; Aftab, T., Hakeem, K.R., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 143–158. [Google Scholar] [CrossRef]
- Albuquerque, B.R.; Heleno, S.A.; Oliveira, M.B.P.P.; Barros, L.; Ferreira, I.C.F.R. Phenolic compounds: Current industrial applications, limitations and future challenges. Food Funct. 2021, 12, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Machin, L.; Monzote, L.; Sharifi-Rad, J.; Ezzat, S.M.; Salem, M.A.; Merghany, R.M.; El Mahdy, N.M.; Kılıç, C.S.; Sytar, O.; et al. Therapeutic Potential of Quercetin: New Insights and Perspectives for Human Health. ACS Omega 2020, 5, 11849–11872. [Google Scholar] [CrossRef] [PubMed]
- Hashemzaei, M.; Delarami Far, A.; Yari, A.; Heravi, R.E.; Tabrizian, K.; Taghdisi, S.M.; Sadegh, S.E.; Tsarouhas, K.; Kouretas, D.; Tzanakakis, G.; et al. Anticancer and apoptosis-inducing effects of quercetin in vitro and in vivo. Oncol. Rep. 2017, 38, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A.; Mujawdiya, P.K.; Lysiuk, R.; Shanaida, M.; Peana, M.; Gasmi Benahmed, A.; Beley, N.; Kovalska, N.; Bjørklund, G. Quercetin in the Prevention and Treatment of Coronavirus Infections: A Focus on SARS-CoV-2. Pharmaceuticals 2022, 15, 1049. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Adulrahman, N.A.; Haleem, A.A.; Al-Râwanduzi, A.D.H.; Khusro, A.; Abdelgawad, M.A.; Ghoneim, M.M.; Batiha, G.E.; Kahrizi, D.; Martinez, F.; et al. Nanoformulations of curcumin and quercetin with silver nanoparticles for inactivation of bacteria. Cell. Mol. Biol. 2022, 67, 151–156. [Google Scholar] [CrossRef]
- Yi, H.; Peng, H.; Wu, X.; Xu, X.; Kuang, T.; Zhang, J.; Du, L.; Fan, G. The Therapeutic Effects and Mechanisms of Quercetin on Metabolic Diseases: Pharmacological Data and Clinical Evidence. Oxid. Med. Cell. Longev. 2021, 2021, 6678662. [Google Scholar] [CrossRef]
- Maiti, S.; Banerjee, A. Epigallocatechin gallate and theaflavin gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction: Bioinformatics and molecular docking study. Drug Dev. Res. 2021, 82, 86–96. [Google Scholar] [CrossRef]
- Isbrucker, R.A.; Edwards, J.A.; Wolz, E.; Davidovich, A.; Bausch, J. Safety studies on epigallocatechin gallate (EGCG) preparations. Part 2: Dermal, acute and short-term toxicity studies. Food Chem. Toxicol. 2006, 44, 636–650. [Google Scholar] [CrossRef]
- Hong, S.; Seo, S.H.; Woo, S.J.; Kwon, Y.; Song, M.; Ha, N.C. Epigallocatechin Gallate Inhibits the Uridylate-Specific Endoribonuclease Nsp15 and Efficiently Neutralizes the SARS-CoV-2 Strain. J. Agric. Food Chem. 2021, 69, 5948–5954. [Google Scholar] [CrossRef]
- Ntamo, Y.; Jack, B.; Ziqubu, K.; Mazibuko-Mbeje, S.E.; Nkambule, B.B.; Nyambuya, T.M.; Mabhida, S.E.; Hanser, S.; Orlando, P.; Tiano, L.; et al. Epigallocatechin gallate as a nutraceutical to potentially target the metabolic syndrome: Novel insights into therapeutic effects beyond its antioxidant and anti-inflammatory properties. Crit. Rev. Food Sci. Nutr. 2022, 1–23. [Google Scholar] [CrossRef]
- Yang, Q.Q.; Wei, X.L.; Fang, Y.P.; Gan, R.Y.; Wang, M.; Ge, Y.Y.; Zhang, D.; Cheng, L.Z.; Corke, H. Nanochemoprevention with therapeutic benefits: An updated review focused on epigallocatechin gallate delivery. Crit. Rev. Food Sci. Nutr. 2020, 60, 1243–1264. [Google Scholar] [CrossRef]
- de Maat, M.P.; Pijl, H.; Kluft, C.; Princen, H.M. Consumption of black and green tea had no effect on inflammation, haemostasis and endothelial markers in smoking healthy individuals. Eur. J. Clin. Nutr. 2000, 54, 757–763. [Google Scholar] [CrossRef]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Quinn, T.R.; Patel, H.N.; Koh, K.H.; Haines, B.E.; Norrby, P.O.; Helquist, P.; Wiest, O. Automated fitting of transition state force fields for biomolecular simulations. PLoS ONE 2022, 17, e0264960. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L. Improved protein-ligand binding affinity prediction by using a curvature-dependent surface-area model. Bioinformatics 2014, 30, 1674–1680. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.T.; Hou, M.C.; Xiao, Z.X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Guedes, I.A.; Costa, L.S.C.; Dos Santos, K.B.; Karl, A.L.M.; Rocha, G.K.; Teixeira, I.M.; Galheigo, M.M.; Medeiros, V.; Krempser, E.; Custódio, F.L.; et al. Drug design and repurposing with DockThor-VS web server focusing on SARS-CoV-2 therapeutic targets and their non-synonym variants. Sci. Rep. 2021, 11, 1–20. [Google Scholar] [CrossRef]
- Zhang, W.; Bell, E.W.; Yin, M.; Zhang, Y. EDock: Blind protein–ligand docking by replica-exchange monte carlo simulation. J. Cheminform. 2020, 12, 37. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Andijani, N.; Wazzan, N.A. The effect of electron-donating substituents on tuning the nonlinear optical properties of pyrene-core arylamine derivatives: DFT calculations. Results Phys. 2018, 11, 605–616. [Google Scholar] [CrossRef]
- Lopes, B.R.P.; da Costa, M.F.; Genova Ribeiro, A.; da Silva, T.F.; Lima, C.S.; Caruso, I.P.; de Araujo, G.C.; Kubo, L.H.; Iacovelli, F.; Falconi, M.; et al. Quercetin pentaacetate inhibits in vitro human respiratory syncytial virus adhesion. Virus Res. 2020, 276, 197805. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Patel, C.N.; Chandra, M. Identification of novel inhibitors of SARS-CoV-2 main protease (Mpro) from Withania sp. by molecular docking and molecular dynamics simulation. J. Comput. Chem. 2021, 42, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Samy, M.N.; Attia, E.Z.; Shoman, M.E.; Khalil, H.E.; Sugimoto, S.; Matsunami, K.; Fahim, J.R. Phytochemical investigation of Amphilophium paniculatum; an underexplored Bignoniaceae species as a source of SARS-CoV-2 Mpro inhibitory metabolites: Isolation, identification, and molecular docking study. S. Afr. J. Bot. 2021, 141, 421–430. [Google Scholar] [CrossRef]
- Mpiana, P.T.; Ngbolua, K.-T.; Tshibangu, D.S.; Kilembe, J.T.; Gbolo, B.Z.; Mwanangombo, D.T.; Inkoto, C.L.; Lengbiye, E.M.; Mbadiko, C.M.; Matondo, A.; et al. Identification of potential inhibitors of SARS-CoV-2 main protease from Aloe vera compounds: A molecular docking study. Chem. Phys. Lett. 2020, 754, 137751. [Google Scholar] [CrossRef] [PubMed]
- Puttaswamy, H.; Gowtham, H.G.; Ojha, M.D.; Yadav, A.; Choudhir, G.; Raguraman, V.; Kongkham, B.; Selvaraju, K.; Shareef, S.; Gehlot, P.; et al. In silico studies evidenced the role of structurally diverse plant secondary metabolites in reducing SARS-CoV-2 pathogenesis. Sci. Rep. 2020, 10, 20584. [Google Scholar] [CrossRef]
- Rahman, F.; Tabrez, S.; Ali, R.; Alqahtani, A.S.; Ahmed, M.Z.; Rub, A. Molecular docking analysis of rutin reveals possible inhibition of SARS-CoV-2 vital proteins. J. Tradit. Complement. Med. 2021, 11, 173–179. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Dahab, M.A.; Metwaly, A.M.; Elhady, S.S.; Elkaeed, E.B.; Eissa, I.H.; Darwish, K.M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021, 9, 661230. [Google Scholar] [CrossRef]
- Eweas, A.F.; Alhossary, A.A.; Abdel-Moneim, A.S. Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2. Front. Microbiol. 2021, 11, 592908. [Google Scholar] [CrossRef]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Woo, H.J.; Kang, H.K.; Nguyen, V.D.; Kim, Y.M.; Kim, D.W.; Ahn, S.A.; Xia, Y.; Kim, D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012, 34, 831–838. [Google Scholar] [CrossRef]
- Du, A.; Zheng, R.; Disoma, C.; Li, S.; Chen, Z.; Li, S.; Liu, P.; Zhou, Y.; Shen, Y.; Liu, S.; et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 176, 1–12. [Google Scholar] [CrossRef]
- Golonka, I.; Wilk, S.; Musiał, W. The Influence of UV Radiation on the Degradation of Pharmaceutical Formulations Containing Quercetin. Molecules 2020, 25, 5454. [Google Scholar] [CrossRef]
- Schmitz, J.; Gilberg, E.; Löser, R.; Bajorath, J.; Bartz, U.; Gütschow, M. Cathepsin B: Active site mapping with peptidic substrates and inhibitors. Biorg. Med. Chem. 2019, 27, 1–15. [Google Scholar] [CrossRef]
- Kawano, M.; Hwang, J. Influence of Guanidine, Imidazole, and Some Heterocyclic Compounds on Dissolution Rates of Amorphous Silica. Clays Clay Miner. 2010, 58, 757–765. [Google Scholar] [CrossRef]
- Murad, H.A.S.; Alqurashi, T.M.A.; Hussien, M.A. Interactions of selected cardiovascular active natural compounds with CXCR4 and CXCR7 receptors: A molecular docking, molecular dynamics, and pharmacokinetic/toxicity prediction study. BMC Complement. Med. Ther. 2022, 22, 35. [Google Scholar] [CrossRef]
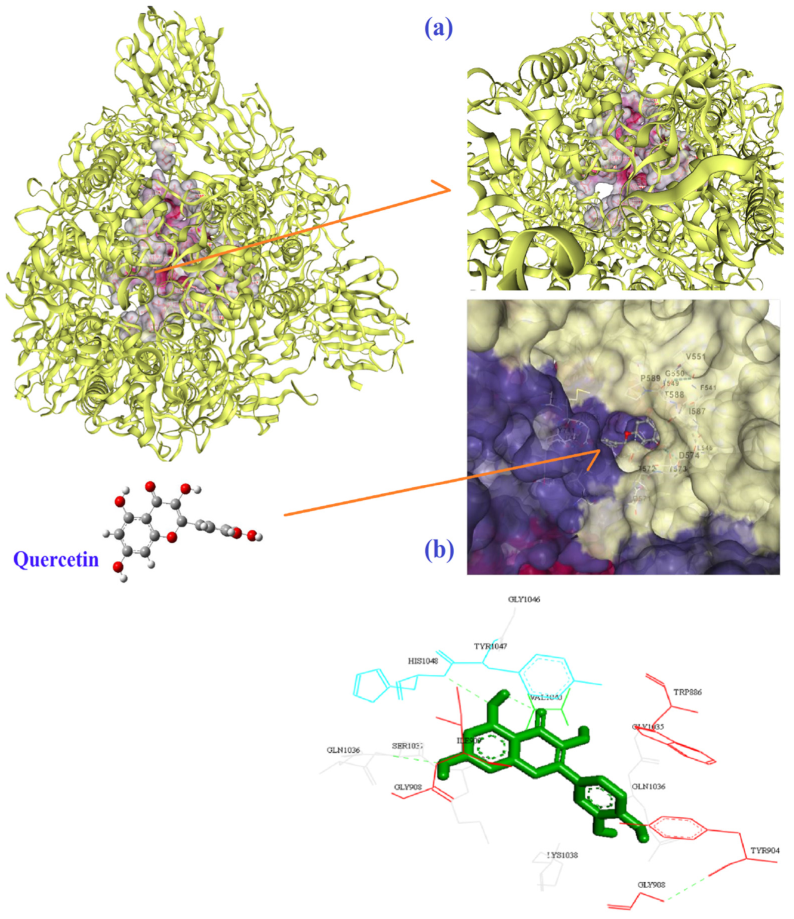
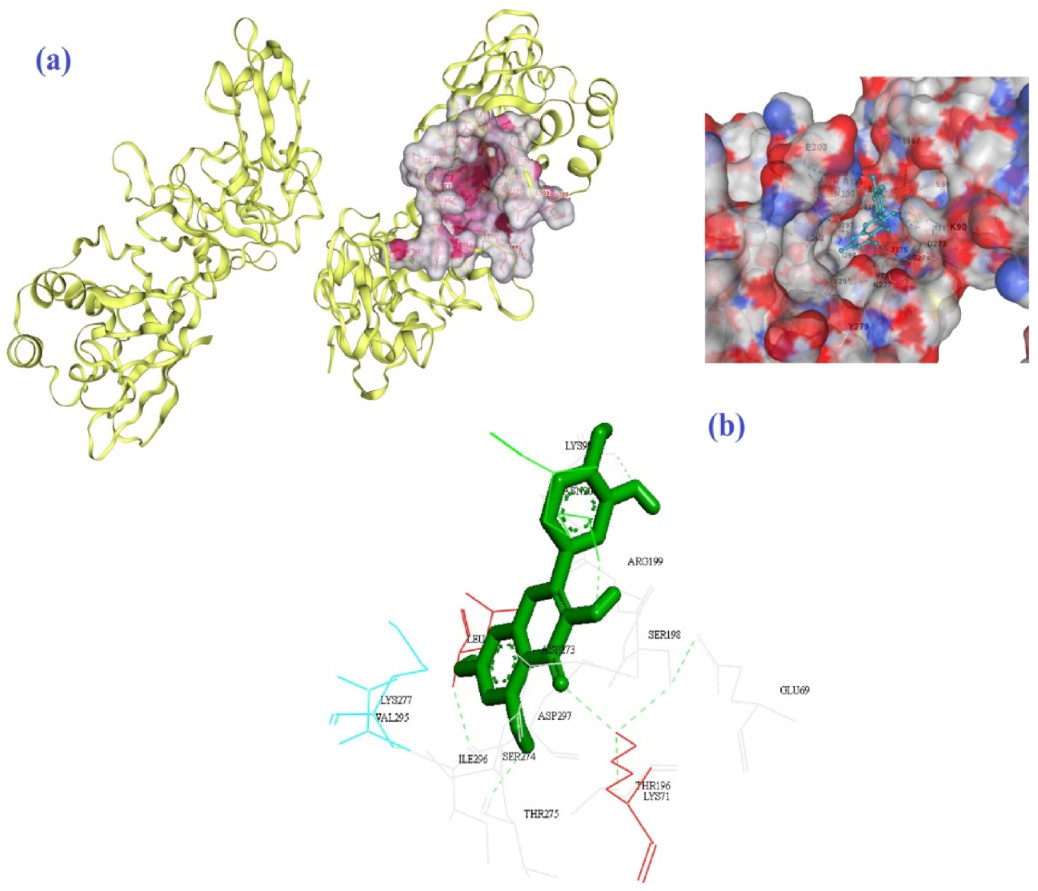
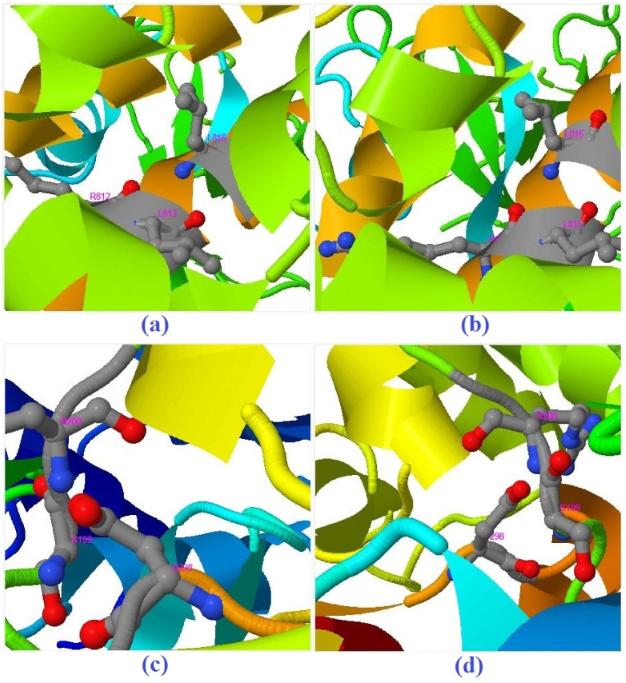
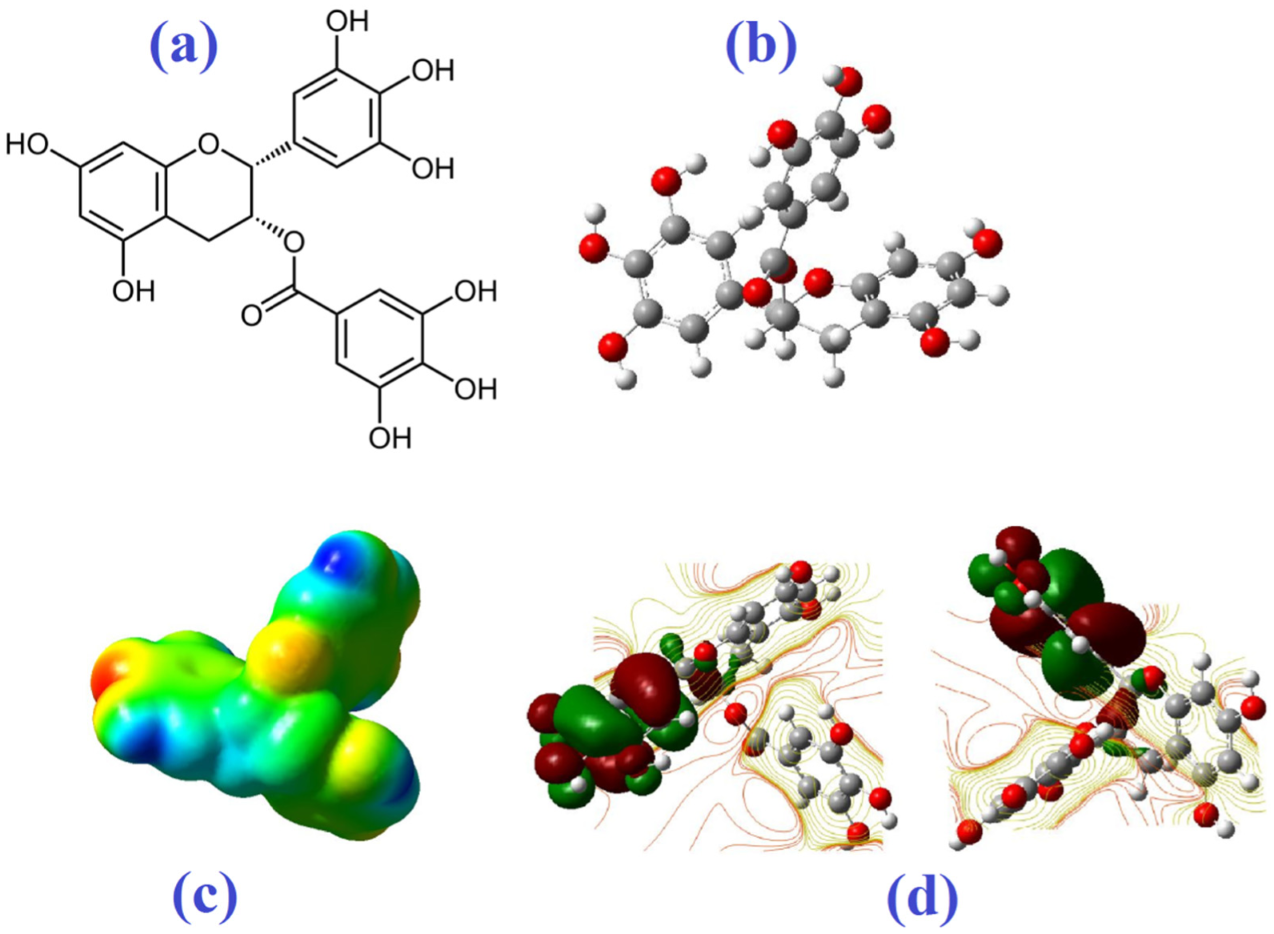
| Molecule | Mode of Action | Ref. |
|---|---|---|
| chloroquine/hydroxychloroquine | inhibiting glycosylation of host receptors | [38] |
| azithromycin | antibiotic/anti-inflammatory activities | [39] |
| lopinavir/ritonavir | inactivation of the viral 3CL protease | [40] |
| ribavirin | blocking RNA-dependent RNA polymerase | [41] |
| tocilizumab | inhibition of IL-6 signaling | [42] |
| baricitinib/remdesivir | blocking RNA-dependent RNA polymerase | [43] |
| favipiravir | selective inhibition of viral RNA polymerase | [44] |
| abidol | broad-spectrum antiviral compound | [45] |
| ruxolitinib | competitively blocking the ATP-binding catalytic site on Janus kinases 1 and 2 | [46] |
| teicoplanin | an antibiotic applied in the prophylaxis and therapy of bacterial infections caused by Gram-positive bacteria | [47] |
| ivermectin | inhibition of the nuclear transport of viral proteins | [48] |
| corticosteroid | immunosuppressive and anti-inflammatory activities | [49] |
| doxycycline | blocking bacterial protein synthesis via binding to the 30S ribosomal subunit | [50] |
| Vina Score | Cavity Volume (Å3) | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| −9.9 | 5396 | 207 | 244 | 243 | 35 | 23 | 35 |
| −9.1 | 8798 | 252 | 231 | 233 | 32 | 35 | 35 |
| −9 | 11,401 | 227 | 228 | 172 | 35 | 33 | 35 |
| −8.9 | 7201 | 224 | 221 | 215 | 31 | 35 | 34 |
| −8.4 | 2780 | 225 | 249 | 213 | 29 | 23 | 35 |
| Vina Score | Cavity Volume (Å3) | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| −8.8 | 1021 | −68 | 28 | 25 | 23 | 23 | 23 |
| −8.7 | 1074 | −73 | 26 | −30 | 23 | 23 | 23 |
| −8.2 | 627 | −56 | 24 | 20 | 23 | 23 | 23 |
| −7.7 | 615 | −81 | 18 | −21 | 23 | 23 | 23 |
| −7.6 | 666 | −53 | 32 | −4 | 23 | 23 | 23 |
| Vina Score | Cavity Volume (Å3) | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| −8.3 | 2780 | 225 | 250 | 213 | 29 | 21 | 35 |
| −8.2 | 8798 | 253 | 232 | 233 | 32 | 35 | 35 |
| −8.1 | 11,401 | 227 | 229 | 172 | 35 | 33 | 35 |
| −8.1 | 5396 | 207 | 245 | 243 | 35 | 28 | 35 |
| −7.7 | 7201 | 225 | 222 | 215 | 31 | 35 | 34 |
| Vina Score | Cavity Volume (Å3) | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| −8.2 | 1021 | −74 | 26 | −30 | 21 | 21 | 21 |
| −7.7 | 631 | −56 | 24 | 20 | 21 | 21 | 21 |
| −7.3 | 605 | −53 | 21 | −13 | 21 | 21 | 21 |
| −6.6 | 641 | −82 | 18 | −21 | 21 | 21 | 21 |
| −6.1 | 667 | −53 | 32 | −4 | 21 | 21 | 21 |
| Ligand | Binding Affinity (kcal/mol) for 6VSB | Binding Affinity (kcal/mol) for 6VWW |
|---|---|---|
| EGCG | −9.9 | −7.3 |
| −9.8 | −7.2 | |
| Quercetin | −7.6 | −6.1 |
| −7.4 | −5.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alavi, M.; Mozafari, M.R.; Ghaemi, S.; Ashengroph, M.; Hasanzadeh Davarani, F.; Mohammadabadi, M. Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study. Biomedicines 2022, 10, 3074. https://doi.org/10.3390/biomedicines10123074
Alavi M, Mozafari MR, Ghaemi S, Ashengroph M, Hasanzadeh Davarani F, Mohammadabadi M. Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study. Biomedicines. 2022; 10(12):3074. https://doi.org/10.3390/biomedicines10123074
Chicago/Turabian StyleAlavi, Mehran, M. R. Mozafari, Saba Ghaemi, Morahem Ashengroph, Fatemeh Hasanzadeh Davarani, and Mohammadreza Mohammadabadi. 2022. "Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study" Biomedicines 10, no. 12: 3074. https://doi.org/10.3390/biomedicines10123074
APA StyleAlavi, M., Mozafari, M. R., Ghaemi, S., Ashengroph, M., Hasanzadeh Davarani, F., & Mohammadabadi, M. (2022). Interaction of Epigallocatechin Gallate and Quercetin with Spike Glycoprotein (S-Glycoprotein) of SARS-CoV-2: In Silico Study. Biomedicines, 10(12), 3074. https://doi.org/10.3390/biomedicines10123074







