Identification of Novel Metabolic Subtypes Using Multi-Trait Limited Mixed Regression in the Chinese Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Data Collection
2.3. Disease Definition
2.4. Genotyping and Quality Controls
2.5. Statistical Analysis
2.5.1. Clustering of Subtypes Using Multi-Trait Limited Mixed Regression
2.5.2. Characterizing Subtypes and Exploring the Associations between Subtypes and Cardiovascular Diseases
2.5.3. Exploring the Potential Genetic Bases of the Subtypes
3. Results
3.1. Characteristics of Five Inferred Metabolic Subtypes
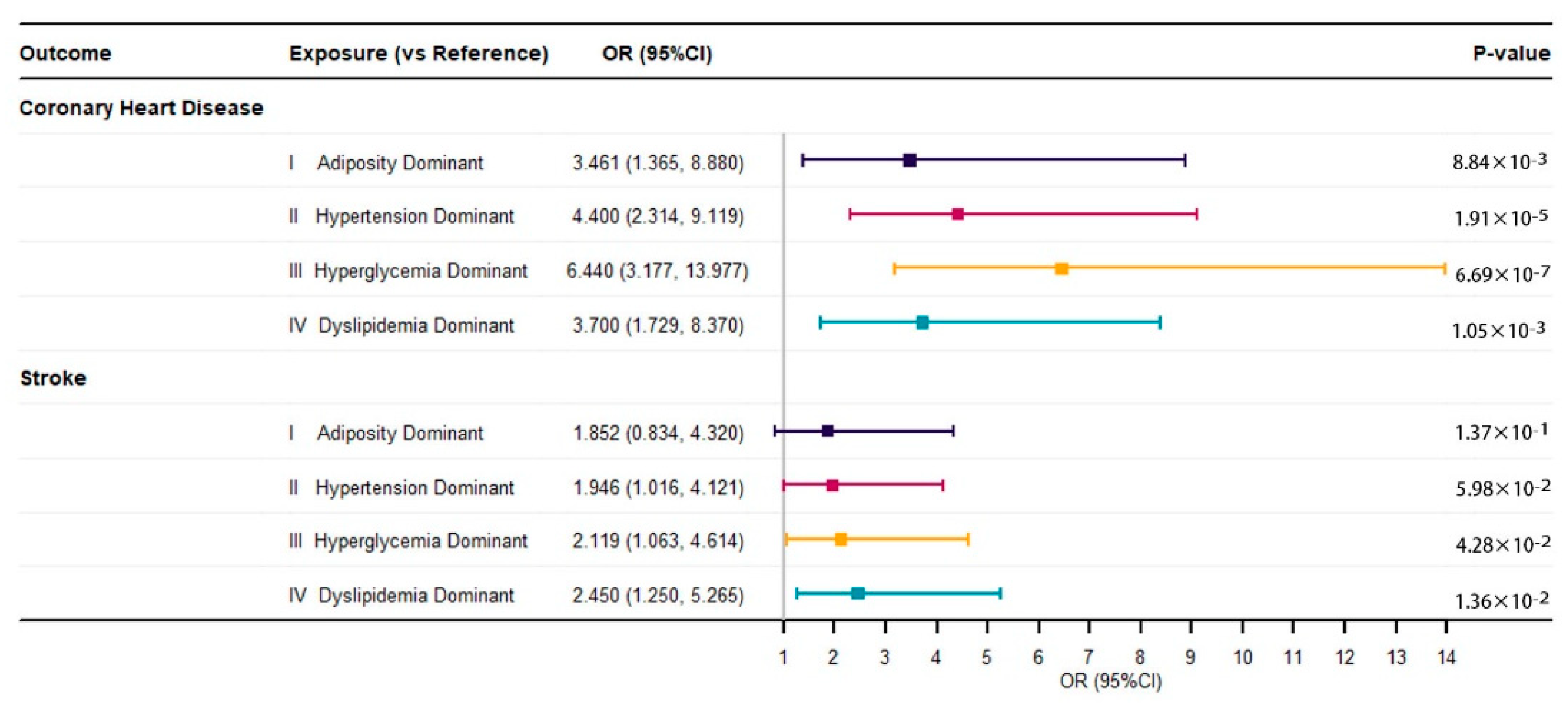
3.2. Associations between Metabolic Subtypes and Cardiovascular Diseases
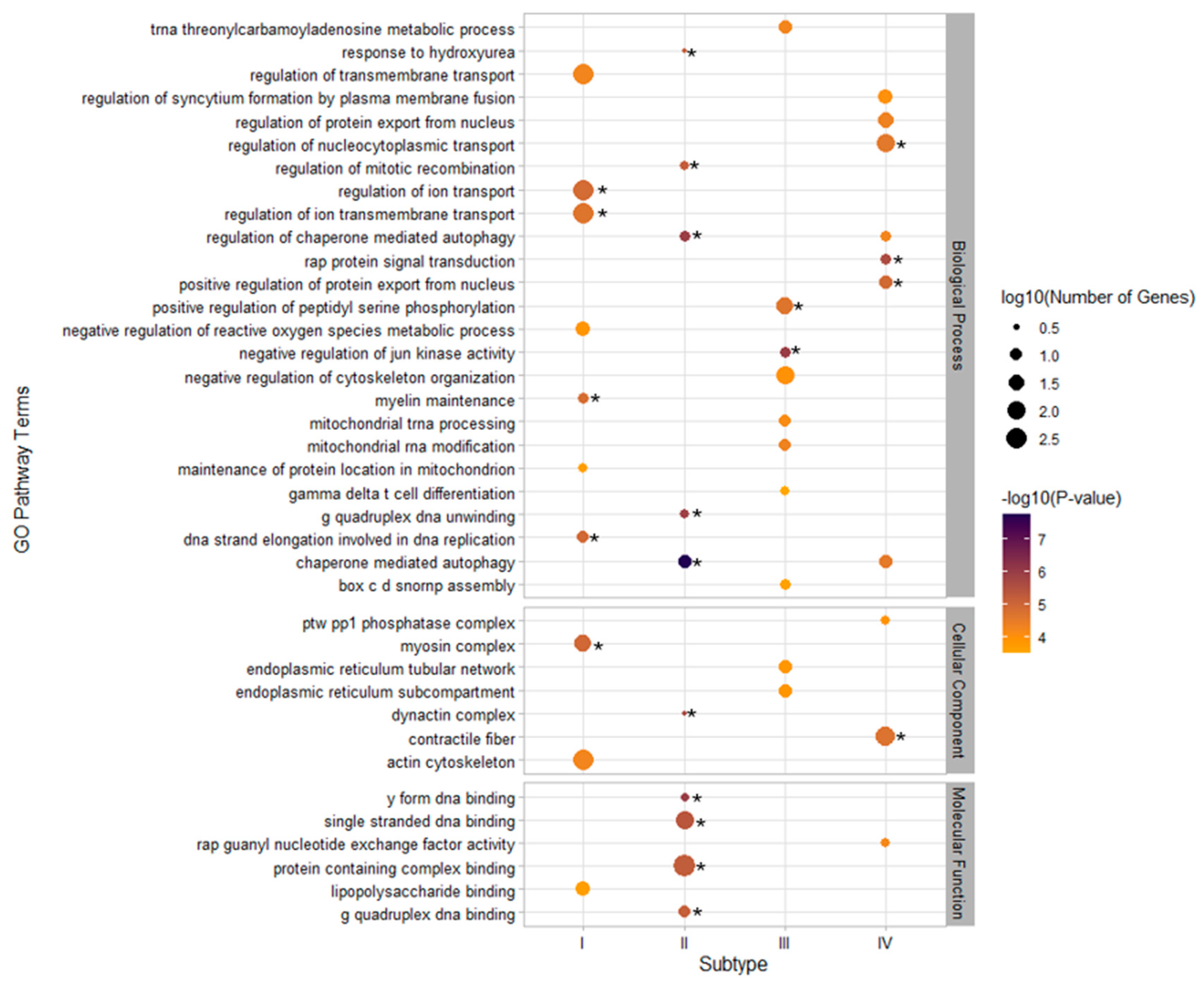
3.3. GO Pathway Enrichment and GTEx Tissue-Specific Enrichment for Metabolic Subtypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grundy, S.M.; Brewer, H.B., Jr.; Cleeman, J.I.; Smith, S.C., Jr.; Lenfant, C. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation 2004, 109, 433–438. [Google Scholar] [CrossRef] [Green Version]
- Sperling, L.S.; Mechanick, J.I.; Neeland, I.J.; Herrick, C.J.; Després, J.P.; Ndumele, C.E.; Vijayaraghavan, K.; Handelsman, Y.; Puckrein, G.A.; Araneta, M.R.; et al. The CardioMetabolic Health Alliance: Working Toward a New Care Model for the Metabolic Syndrome. J. Am. Coll. Cardiol. 2015, 66, 1050–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahlqvist, E.; Storm, P.; Käräjämäki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Castaldi, P.J.; Dy, J.; Ross, J.; Chang, Y.; Washko, G.R.; Curran-Everett, D.; Williams, A.; Lynch, D.A.; Make, B.J.; Crapo, J.D.; et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax 2014, 69, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Cheng, W.Y.; Glicksberg, B.S.; Gottesman, O.; Tamler, R.; Chen, R.; Bottinger, E.P.; Dudley, J.T. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med. 2015, 7, 311ra174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, A.; Cai, N.; Ko, A.; Laakso, M.; Pajukanta, P.; Flint, J.; Zaitlen, N. Reverse GWAS: Using genetics to identify and model phenotypic subtypes. PLoS Genet. 2019, 15, e1008009. [Google Scholar] [CrossRef] [Green Version]
- Dahl, A.; Zaitlen, N. Genetic Influences on Disease Subtypes. Annu. Rev. Genom. Hum. Genet. 2020, 21, 413–435. [Google Scholar] [CrossRef]
- Dapas, M.; Lin, F.T.J.; Nadkarni, G.N.; Sisk, R.; Legro, R.S.; Urbanek, M.; Hayes, M.G.; Dunaif, A. Distinct subtypes of polycystic ovary syndrome with novel genetic associations: An unsupervised, phenotypic clustering analysis. PLoS Med. 2020, 17, e1003132. [Google Scholar] [CrossRef]
- Manipur, I.; Granata, I.; Maddalena, L.; Guarracino, M.R. Clustering analysis of tumor metabolic networks. BMC Bioinform. 2020, 21, 349. [Google Scholar] [CrossRef]
- Yang, C.; Huang, X.; Liu, Z.; Qin, W.; Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020, 14, 896–913. [Google Scholar] [CrossRef]
- Yu, T.J.; Ma, D.; Liu, Y.Y.; Xiao, Y.; Gong, Y.; Jiang, Y.Z.; Shao, Z.M.; Hu, X.; Di, G.H. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021, 29, 2350–2365. [Google Scholar] [CrossRef]
- Ding, M.; Li, F.; Wang, B.; Chi, G.; Liu, H. A comprehensive analysis of WGCNA and serum metabolomics manifests the lung cancer-associated disordered glucose metabolism. J. Cell. Biochem. 2019, 120, 10855–10863. [Google Scholar] [CrossRef]
- Bidkhori, G.; Benfeitas, R.; Klevstig, M.; Zhang, C.; Nielsen, J.; Uhlen, M.; Boren, J.; Mardinoglu, A. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl. Acad. Sci. USA 2018, 115, e11874–e11883. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Hu, Y.; Chen, D.; Zhan, S.; Zhang, Z.; Dou, H. The Fangshan/Family-based Ischemic Stroke Study In China (FISSIC) protocol. BMC Med. Genet 2007, 8, 60. [Google Scholar] [CrossRef] [Green Version]
- Fan, F.; Qi, L.; Jia, J.; Xu, X.; Liu, Y.; Yang, Y.; Qin, X.; Li, J.; Li, H.; Zhang, Y.; et al. Noninvasive Central Systolic Blood Pressure Is More Strongly Related to Kidney Function Decline Than Peripheral Systolic Blood Pressure in a Chinese Community-Based Population. Hypertension 2016, 67, 1166–1172. [Google Scholar] [CrossRef]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith Jr, S.C.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation 2005, 112, 2735–2752. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.S.; Zhang, H.; Cheung, C.Y.; Xu, M.; Ho, J.C.; Zhou, W.; Cherny, S.S.; Zhang, Y.; Holmen, O.; Au, K.W.; et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun. 2015, 6, 10206. [Google Scholar] [CrossRef] [Green Version]
- Delaneau, O.; Marchini, J.; Zagury, J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2011, 9, 179–181. [Google Scholar] [CrossRef]
- Delaneau, O.; Zagury, J.F.; Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 2013, 10, 5–6. [Google Scholar] [CrossRef]
- Howie, B.N.; Donnelly, P.; Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, L.; Lu, L.; Tan, Y.; Pan, X.; Cai, Y.; Wang, X.; Hong, J.; Zhong, C.; Wang, F.; Zhang, X.Y.; et al. Genome-wide association discoveries of alcohol dependence. Am. J. Addict. 2014, 23, 526–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.Q.; Cai, W.J.; Hou, X.H.; Cui, M.; Tan, L.; Yu, J.T.; Dong, Q. Genome-Wide Association Study of Cerebral Microbleeds on MRI. Neurotox. Res. 2020, 37, 146–155. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udler, M.S.; Kim, J.; von Grotthuss, M.; Bonàs-Guarch, S.; Cole, J.B.; Chiou, J.; Boehnke, M.; Laakso, M.; Atzmon, G.; Glaser, B.; et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLoS Med. 2018, 15, e1002654. [Google Scholar] [CrossRef] [Green Version]
- Broeks, A.; Schmidt, M.K.; Sherman, M.E.; Couch, F.J.; Hopper, J.L.; Dite, G.S.; Apicella, C.; Smith, L.D.; Hammet, F.; Southey, M.C.; et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: Findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2011, 20, 3289–3303. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.K.; Hogervorst, F.; van Hien, R.; Cornelissen, S.; Broeks, A.; Adank, M.A.; Meijers, H.; Waisfisz, Q.; Hollestelle, A.; Schutte, M.; et al. Age- and Tumor Subtype-Specific Breast Cancer Risk Estimates for CHEK2*1100delC Carriers. J. Clin. Oncol. 2016, 34, 2750–2760. [Google Scholar] [CrossRef] [Green Version]
- Bønnelykke, K.; Ober, C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J. Allergy Clin. Immunol. 2016, 137, 667–679. [Google Scholar] [CrossRef]
- Woodruff, P.G.; Modrek, B.; Choy, D.F.; Jia, G.; Abbas, A.R.; Ellwanger, A.; Koth, L.L.; Arron, J.R.; Fahy, J.V. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 388–395. [Google Scholar] [CrossRef] [Green Version]
- Bernier, R.; Golzio, C.; Xiong, B.; Stessman, H.A.; Coe, B.P.; Penn, O.; Witherspoon, K.; Gerdts, J.; Baker, C.; Vulto-van Silfhout, A.T.; et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 2014, 158, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Jeste, S.S.; Geschwind, D.H. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat. Rev. Neurol. 2014, 10, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Arnedo, J.; Svrakic, D.M.; Del Val, C.; Romero-Zaliz, R.; Hernández-Cuervo, H.; Fanous, A.H.; Pato, M.T.; Pato, C.N.; de Erausquin, G.A.; Cloninger, C.R.; et al. Uncovering the hidden risk architecture of the schizophrenias: Confirmation in three independent genome-wide association studies. Am. J. Psychiatry 2015, 172, 139–153. [Google Scholar] [CrossRef] [Green Version]
- Huckins, L.M.; Dobbyn, A.; Ruderfer, D.M.; Hoffman, G.; Wang, W.; Pardiñas, A.F.; Rajagopal, V.M.; Als, T.D.; Hoang, T.N.; Girdhar, K.; et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 2019, 51, 659–674. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, X.; Sui, Z.; Niu, H.; Chen, L.; Hu, C.; Xuan, Q.; Hou, X.; Zhang, R.; Zhou, L.; et al. A multi-omics investigation of the molecular characteristics and classification of six metabolic syndrome relevant diseases. Theranostics 2020, 10, 2029–2046. [Google Scholar] [CrossRef]
- Vaura, F.C.; Salomaa, V.V.; Kantola, I.M.; Kaaja, R.; Lahti, L.; Niiranen, T.J. Unsupervised hierarchical clustering identifies a metabolically challenged subgroup of hypertensive individuals. J. Clin. Hypertens 2020, 22, 1546–1553. [Google Scholar] [CrossRef]
- Yang, D.Y.; Nie, Z.Q.; Liao, L.Z.; Zhang, S.Z.; Zhou, H.M.; Sun, X.T.; Zhong, X.B.; Du, Z.M.; Zhuang, X.D.; Liao, X.X. Phenomapping of subgroups in hypertensive patients using unsupervised data-driven cluster analysis: An exploratory study of the SPRINT trial. Eur. J. Prev. Cardiol. 2019, 26, 1693–1706. [Google Scholar] [CrossRef]
- Guo, Q.; Lu, X.; Gao, Y.; Zhang, J.; Yan, B.; Su, D.; Song, A.; Zhao, X.; Wang, G. Cluster analysis: A new approach for identification of underlying risk factors for coronary artery disease in essential hypertensive patients. Sci. Rep. 2017, 7, 43965. [Google Scholar] [CrossRef] [Green Version]
- Slieker, R.C.; Donnelly, L.A.; Fitipaldi, H.; Bouland, G.A.; Giordano, G.N.; Åkerlund, M.; Gerl, M.J.; Ahlqvist, E.; Ali, A.; Dragan, I.; et al. Distinct Molecular Signatures of Clinical Clusters in People With Type 2 Diabetes: An IMI-RHAPSODY Study. Diabetes 2021, 70, 2683–2693. [Google Scholar] [CrossRef]
- Li, J.K.Y.; Ng, M.C.Y.; So, W.Y.; Chiu, C.K.P.; Ozaki, R.; Tong, P.C.Y.; Cockram, C.S.; Chan, J.C.N. Phenotypic and genetic clustering of diabetes and metabolic syndrome in Chinese families with type 2 diabetes mellitus. Diabetes/Metab. Res. Rev. 2006, 22, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Guardiola, M.; Ribalta, J. Update on APOA5 Genetics: Toward a Better Understanding of Its Physiological Impact. Curr. Atheroscler. Rep. 2017, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Aung, L.H.; Yin, R.X.; Wu, D.F.; Wang, W.; Liu, C.W.; Pan, S.L. Association of the variants in the BUD13-ZNF259 genes and the risk of hyperlipidaemia. J. Cell. Mol. Med. 2014, 18, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Anwar, M.A.; Choi, S. Association of BUD13-ZNF259-APOA5-APOA1-SIK3 cluster polymorphism in 11q23.3 and structure of APOA5 with increased plasma triglyceride levels in a Korean population. Sci. Rep. 2019, 9, 8296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahed, G.; Aoun, L.; Bou Zerdan, M.; Allam, S.; Bou Zerdan, M.; Bouferraa, Y.; Assi, H.I. Metabolic Syndrome: Updates on Pathophysiology and Management in 2021. Int. J. Mol. Sci. 2022, 23, 786. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, G.; Vastrad, B.; Tengli, A.; Vastrad, C.; Kotturshetti, I. Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules. BMC Endocr. Disord. 2021, 21, 80. [Google Scholar] [CrossRef]
- Mondaca-Ruff, D.; Quiroga, C.; Norambuena-Soto, I.; Riquelme, J.A.; San Martin, A.; Bustamante, M.; Lavandero, S.; Chiong, M. Regulation of total LC3 levels by angiotensin II in vascular smooth muscle cells. J. Cell. Mol. Med. 2022, 26, 1710–1713. [Google Scholar] [CrossRef]
- Weston, C.R.; Davis, R.J. The JNK signal transduction pathway. Curr. Opin. Cell. Biol. 2007, 19, 142–149. [Google Scholar] [CrossRef]
- Zhao, Q.; Xu, H.; Hong, S.; Song, N.; Xie, J.; Yan, Z.; Wang, R.; Yang, P.; Jiang, X. Rapeseed Protein-Derived Antioxidant Peptide RAP Ameliorates Nonalcoholic Steatohepatitis and Related Metabolic Disorders in Mice. Mol. Pharm. 2019, 16, 371–381. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell. Biol. 2015, 17, 759–770. [Google Scholar] [CrossRef]
- Qiao, L.; Ma, J.; Zhang, Z.; Sui, W.; Zhai, C.; Xu, D.; Wang, Z.; Lu, H.; Zhang, M.; Zhang, C.; et al. Deficient Chaperone-Mediated Autophagy Promotes Inflammation and Atherosclerosis. Circ. Res. 2021, 129, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.D.; Chen-Plotkin, A.S. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, S.E.; Booms, A.; Prahl, J.; van der Schans, E.J.C.; Tyson, T.; Coetzee, G.A. Post-GWAS knowledge gap: The how, where, and when. NPJ Parkinsons Dis. 2020, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Farh, K.K.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]
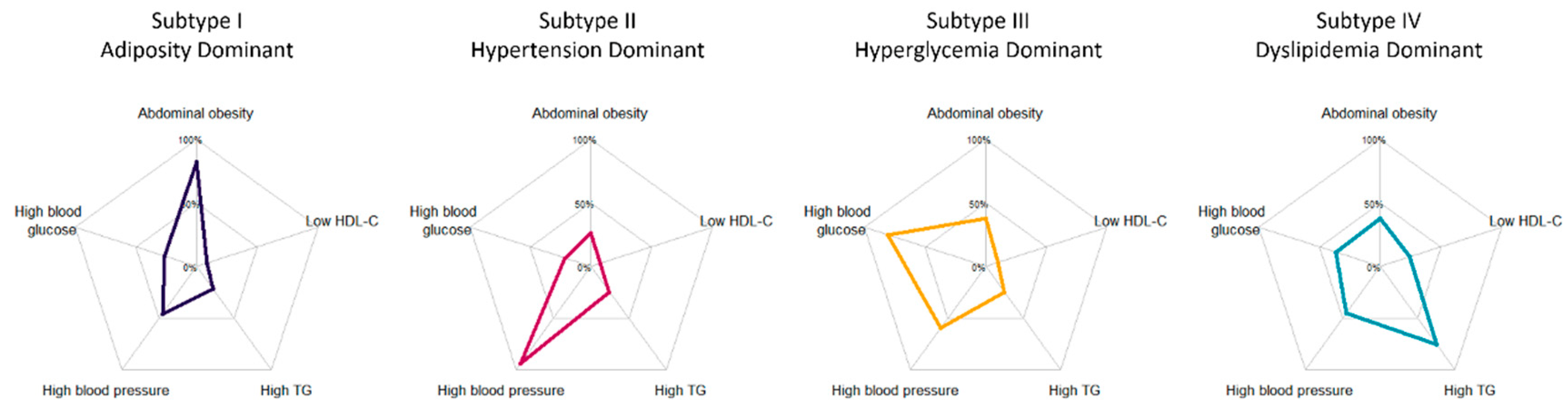
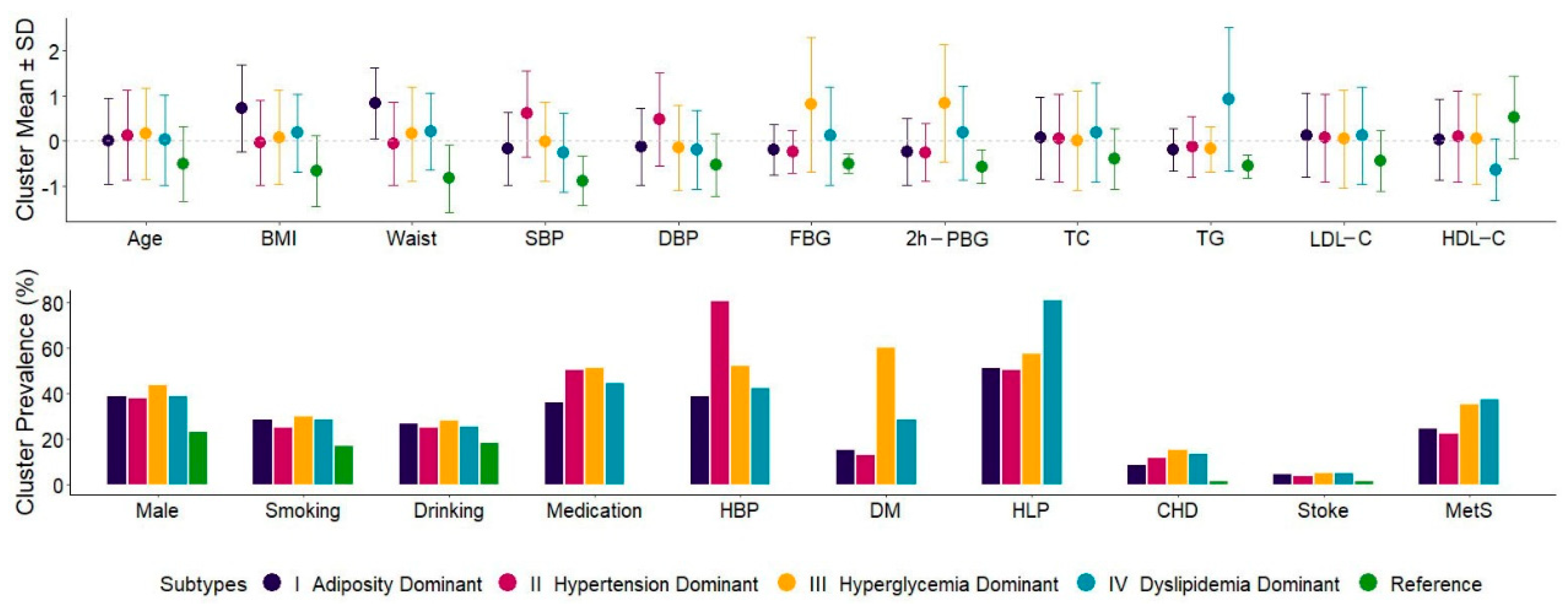

| Overall (N = 4632) | Subtype I (N = 428) | Subtype II (N = 1617) | Subtype III (N = 919) | Subtype IV (N = 936) | Reference (N = 732) | p-Values among Subtypes | |
|---|---|---|---|---|---|---|---|
| Age, years, mean ± sd | 57.1 ± 8.9 | 57.1 ± 8.6 | 58.2 ± 8.9 | 58.6 ± 9.0 | 57.4 ± 9.0 | 52.6 ± 7.5 | 0.003 |
| BMI, kg/m2, mean ± sd | 26.0 ± 3.4 | 28.5 ± 3.2 | 25.9 ± 3.2 | 26.3 ± 3.5 | 26.6 ± 2.9 | 23.8 ± 2.6 | <0.001 |
| Waist circumference, cm, mean ± sd | 83.0 ± 8.5 | 90.2 ± 6.7 | 82.5 ± 7.8 | 84.4 ± 8.9 | 84.8 ± 7.2 | 76.0 ± 6.4 | <0.001 |
| SBP, mmHg, mean ± sd | 133.7 ± 16.5 | 130.9 ± 13.3 | 143.7 ± 15.7 | 133.5 ± 14.6 | 129.5 ± 14.4 | 119.1 ± 9.2 | <0.001 |
| DBP, mmHg, mean ± sd | 74.9 ± 9.9 | 73.6 ± 8.5 | 79.7 ± 10.3 | 73.3 ± 9.3 | 72.9 ± 8.7 | 69.6 ± 7.0 | <0.001 |
| FBG, mmol/L, mean ± sd | 6.1 ± 1.7 | 5.8 ± 0.9 | 5.7 ± 0.8 | 7.4 ± 2.5 | 6.3 ± 1.8 | 5.3 ± 0.4 | <0.001 |
| 2h-PBG, mmol/L, mean ± sd | 8.4 ± 3.8 | 7.5 ± 2.8 | 7.5 ± 2.4 | 11.6 ± 4.9 | 9.1 ± 4.0 | 6.3 ± 1.4 | <0.001 |
| TC, mmol/L, mean ± sd | 5.3 ± 1.0 | 5.4 ± 0.9 | 5.4 ± 1.0 | 5.3 ± 1.1 | 5.5 ± 1.1 | 4.9 ± 0.7 | 0.002 |
| TG, mmol/L, mean ± sd | 1.6 ± 1.1 | 1.4 ± 0.5 | 1.4 ± 0.8 | 1.4 ± 0.6 | 2.6 ± 1.8 | 0.9 ± 0.3 | <0.001 |
| LDL-C, mmol/L, mean ± sd | 3.2 ± 0.8 | 3.3 ± 0.8 | 3.3 ± 0.8 | 3.3 ± 0.9 | 3.3 ± 0.9 | 2.9 ± 0.6 | 0.342 |
| HDL-C, mmol/L, mean ± sd | 1.4 ± 0.4 | 1.4 ± 0.3 | 1.5 ± 0.4 | 1.4 ± 0.4 | 1.2 ± 0.3 | 1.6 ± 0.3 | <0.001 |
| Male, n (%) | 1713 (37.0%) | 166 (38.8%) | 615 (38.0%) | 399 (43.4%) | 362 (38.7%) | 171 (23.4%) | 0.054 |
| Smoking, n (%) | 1192 (25.7%) | 122 (28.5%) | 401 (24.8%) | 276 (30.0%) | 269 (28.7%) | 124 (16.9%) | 0.019 |
| Drinking, n (%) | 1153 (24.9%) | 114 (26.6%) | 407 (25.2%) | 259 (28.2%) | 237 (25.3%) | 136 (18.6%) | 0.370 |
| Medication, n (%) | |||||||
| Antihypertensive drugs | 1488 (32.1%) | 115 (26.9%) | 736 (45.5%) | 303 (33.0%) | 334 (35.7%) | - | <0.001 |
| Antidiabetic drugs | 537 (11.6%) | 52 (12.1%) | 127 (7.9%) | 211 (23.0%) | 147 (15.7%) | - | <0.001 |
| Lipid-lowing drugs | 495 (10.7%) | 64 (15.0%) | 173 (10.7%) | 130 (14.1%) | 128 (13.7%) | - | <0.001 |
| Hypertension, n (%) | 2335 (50.4%) | 166 (38.8%) | 1296 (80.1%) | 479 (52.1%) | 394 (42.1%) | - | <0.001 |
| Type 2 diabetes, n (%) | 1096 (23.7%) | 66 (15.4%) | 212 (13.1%) | 551 (60.0%) | 267 (28.5%) | - | <0.001 |
| Dyslipidemia, n (%) | 2317 (50.0%) | 218 (50.9%) | 816 (50.5%) | 525 (57.1%) | 758 (81.0%) | - | <0.001 |
| Coronary Heart Disease, n (%) | 505 (10.9%) | 37 (8.6%) | 192 (11.9%) | 140 (15.2%) | 125 (13.4%) | 11 (1.5%) | 0.004 |
| Stroke, n (%) | 185 (4.0%) | 19 (4.4%) | 63 (3.9%) | 45 (4.9%) | 48 (5.1%) | 10 (1.4%) | 0.444 |
| Metabolic syndrome, n (%) | 1142 (24.7%) | 105 (24.5%) | 365 (22.6%) | 322 (35.0%) | 350 (37.4%) | - | <0.001 |
| Metabolic syndrome components, n (%) | |||||||
| Abdominal obesity | 1477 (31.9%) | 350 (81.8%) | 424 (26.2%) | 349 (38.0%) | 354 (37.8%) | - | <0.001 |
| High blood pressure | 2679 (57.8%) | 195 (45.6%) | 1515 (93.7%) | 548 (59.6%) | 421 (45.0%) | - | <0.001 |
| High blood glucose | 1546 (33.4%) | 112 (26.2%) | 349 (21.6%) | 742 (80.7%) | 343 (36.6%) | - | <0.001 |
| High blood TG | 1416 (30.6%) | 93 (21.7%) | 395 (24.4%) | 225 (24.5%) | 703 (75.1%) | - | <0.001 |
| Low blood HDL-C | 479 (10.3%) | 37 (8.6%) | 121 (7.5%) | 94 (10.2%) | 227 (24.3%) | - | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, K.; Zhou, Z.; Ma, Y.; Li, X.; Xiao, H.; Wu, Y.; Wu, T.; Chen, D. Identification of Novel Metabolic Subtypes Using Multi-Trait Limited Mixed Regression in the Chinese Population. Biomedicines 2022, 10, 3093. https://doi.org/10.3390/biomedicines10123093
Ding K, Zhou Z, Ma Y, Li X, Xiao H, Wu Y, Wu T, Chen D. Identification of Novel Metabolic Subtypes Using Multi-Trait Limited Mixed Regression in the Chinese Population. Biomedicines. 2022; 10(12):3093. https://doi.org/10.3390/biomedicines10123093
Chicago/Turabian StyleDing, Kexin, Zechen Zhou, Yujia Ma, Xiaoyi Li, Han Xiao, Yiqun Wu, Tao Wu, and Dafang Chen. 2022. "Identification of Novel Metabolic Subtypes Using Multi-Trait Limited Mixed Regression in the Chinese Population" Biomedicines 10, no. 12: 3093. https://doi.org/10.3390/biomedicines10123093
APA StyleDing, K., Zhou, Z., Ma, Y., Li, X., Xiao, H., Wu, Y., Wu, T., & Chen, D. (2022). Identification of Novel Metabolic Subtypes Using Multi-Trait Limited Mixed Regression in the Chinese Population. Biomedicines, 10(12), 3093. https://doi.org/10.3390/biomedicines10123093






