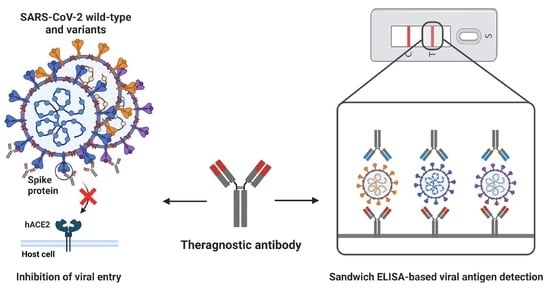
Development and Characterization of Phage-Display-Derived Novel Human Monoclonal Antibodies against the Receptor Binding Domain of SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of SARS-CoV-2 RBD-Specific Single-Chain Variable Fragments (scFvs) Using Phage-Display Technology
2.2. DNA Cloning
2.3. Cell Culture
2.4. Production of SARS-CoV-2 RBD IgG mAbs
2.5. Surface Plasmon Resonance (SPR) Binding Studies
2.6. Protein-Protein Interaction Inhibition Assay
2.7. In Vitro SARS-CoV-2 Pseudovirus Neutralization Assay
2.8. In Vitro SARS-CoV-2 Live Virus Neutralization Assay
2.9. In Vivo Mouse Study
2.10. Histology
2.11. Sandwich ELISA
2.12. Statistical Validation of Sandwich ELISA
3. Results
3.1. Isolation and Biochemical Characterization of SARS-CoV-2 RBD-Specific mAbs
3.2. Neutralization of Selected SARS-CoV-2 RBD-Specific mAbs against SARS-CoV-2
3.3. In Vivo Efficacy Evaluation of K102.1 in Wild-Type SARS-CoV-2-Infected Animal Model
3.4. Identification of SARS-CoV-2 RBD-Specific mAb Pair for Sandwich ELISA
3.5. Development and Characterization of Sandwich ELISA for Detection of Wild-Type SARS-CoV-2 RBD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.M.; Ketcheson, D.I.; Eguíluz, V.M.; Agustí, S.; Fernández-Gracia, J.; Jamil, T.; Laiolo, E.; Gojobori, T.; Alam, I. Rapid evolution of SARS-CoV-2 challenges human defenses. Sci. Rep. 2022, 12, 6457. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- World Health Organization (WHO). COVID-19 Weekly Epidemiological Update. 2022. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---17-september-2022 (accessed on 30 September 2022).
- Mehta, O.P.; Bhandari, P.; Raut, A.; Kacimi, S.E.O.; Huy, N.T. Coronavirus Disease (COVID-19): Comprehensive Review of Clinical Presentation. Front. Public Health 2020, 8, 582932. [Google Scholar] [CrossRef]
- Baj, J.; Karakuła-Juchnowicz, H.; Teresiński, G.; Buszewicz, G.; Ciesielka, M.; Sitarz, R.; Forma, A.; Karakuła, K.; Flieger, W.; Portincasa, P.; et al. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J. Clin. Med. 2020, 9, 1753. [Google Scholar] [CrossRef]
- Da Silva Ramos, F.J.; de Freitas, F.G.R.; Machado, F.R. Sepsis in patients hospitalized with coronavirus disease 2019: How often and how severe? Curr. Opin. Crit. Care 2021, 27, 474–479. [Google Scholar] [CrossRef]
- Olwal, C.O.; Nganyewo, N.N.; Tapela, K.; Djomkam Zune, A.L.; Owoicho, O.; Bediako, Y.; Duodu, S. Parallels in Sepsis and COVID-19 Conditions: Implications for Managing Severe COVID-19. Front. Immunol. 2021, 12, 602848. [Google Scholar] [CrossRef]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Manjunath, K.; Ranjan, R.K.; Kaushik, S.; Kumar, S.; Verma, V. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020, 16, e1008762. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, L.; Zhang, G.; Yao, Y.; Zhou, H.; Shen, S.; Shen, B.; Li, B.; Li, X.; Zhang, Q.; et al. A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes. Nat. Commun. 2021, 12, 2623. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Brown-Augsburger, P.L.; Corbett, K.S.; Westendorf, K.; Davies, J.; Cujec, T.P.; Wiethoff, C.M.; Blackbourne, J.L.; Heinz, B.A.; Foster, D.; et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021, 13, eabf1906. [Google Scholar] [CrossRef]
- Huo, J.; Le Bas, A.; Ruza, R.R.; Duyvesteyn, H.M.E.; Mikolajek, H.; Malinauskas, T.; Tan, T.K.; Rijal, P.; Dumoux, M.; Ward, P.N.; et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020, 27, 846–854. [Google Scholar] [CrossRef]
- Valenzuela-Nieto, G.; Miranda-Chacon, Z.; Salinas-Rebolledo, C.; Jara, R.; Cuevas, A.; Berking, A.; Rojas-Fernandez, A. Nanobodies: COVID-19 and Future Perspectives. Front. Drug Discov. 2022, 2, 927164. [Google Scholar] [CrossRef]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Silva Andrade, B.; Giovanetti, M.; Almeida Costa, E.; Kumavath, R.; Ghosh, P.; Góes-Neto, A.; Carlos Junior Alcantara, L.; Azevedo, V. Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain. F1000Research 2020, 9, 576. [Google Scholar] [CrossRef]
- Rajpoot, S.; Ohishi, T.; Kumar, A.; Pan, Q.; Banerjee, S.; Zhang, K.Y.J.; Baig, M.S. A Novel Therapeutic Peptide Blocks SARS-CoV-2 Spike Protein Binding with Host Cell ACE2 Receptor. Drugs RD 2021, 21, 273–283. [Google Scholar] [CrossRef]
- Zhang, X.; Han, P.; Wang, H.; Xu, Y.; Li, F.; Li, M.; Fan, L.; Zhang, H.; Dai, Q.; Lin, H.; et al. Engineering mesenchymal stromal cells with neutralizing and anti-inflammatory capability against SARS-CoV-2 infection. Mol. Ther. Methods Clin. Dev. 2021, 21, 754–764. [Google Scholar] [CrossRef]
- Chaouat, A.E.; Achdout, H.; Kol, I.; Berhani, O.; Roi, G.; Vitner, E.B.; Melamed, S.; Politi, B.; Zahavy, E.; Brizic, I.; et al. SARS-CoV-2 receptor binding domain fusion protein efficiently neutralizes virus infection. PLoS Pathog. 2021, 17, e1010175. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef] [PubMed]
- Dickey, T.H.; Tang, W.K.; Butler, B.; Ouahes, T.; Orr-Gonzalez, S.; Salinas, N.D.; Lambert, L.E.; Tolia, N.H. Design of the SARS-CoV-2 RBD vaccine antigen improves neutralizing antibody response. Sci. Adv. 2022, 8, eabq8276. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zheng, J.; Zhang, X.; Tai, W.; Odle, A.E.; Perlman, S.; Du, L. RBD-mRNA vaccine induces broadly neutralizing antibodies against Omicron and multiple other variants and protects mice from SARS-CoV-2 challenge. Transl. Res. J. Lab. Clin. Med. 2022, 248, 11–21. [Google Scholar] [CrossRef]
- Castelli, M.S.; McGonigle, P.; Hornby, P.J. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacol. Res. Perspect. 2019, 7, e00535. [Google Scholar] [CrossRef]
- Tsumoto, K.; Isozaki, Y.; Yagami, H.; Tomita, M. Future perspectives of therapeutic monoclonal antibodies. Immunotherapy 2019, 11, 119–127. [Google Scholar] [CrossRef]
- Parray, H.A.; Shukla, S.; Samal, S.; Shrivastava, T.; Ahmed, S.; Sharma, C.; Kumar, R.J.I.I. Hybridoma technology a versatile method for isolation of monoclonal antibodies, its applicability across species, limitations, advancement and future perspectives. Int. Immunopharm. 2020, 85, 106639. [Google Scholar] [CrossRef]
- Harding, F.A.; Stickler, M.M.; Razo, J.; DuBridge, R.B. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. mAbs 2010, 2, 256–265. [Google Scholar] [CrossRef]
- Pansri, P.; Jaruseranee, N.; Rangnoi, K.; Kristensen, P.; Yamabhai, M. A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 2009, 9, 6. [Google Scholar] [CrossRef]
- Frenzel, A.; Schirrmann, T.; Hust, M.J.M. Phage display-derived human antibodies in clinical development and therapy. mAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, J.W.; Yang, H.R.; Song, S.W.; Lee, S.J.; Jeon, Y.; Ju, A.; Lee, N.; Kim, M.G.; Kim, M.; et al. A Fully-Human Antibody Specifically Targeting a Membrane-Bound Fragment of CADM1 Potentiates the T Cell-Mediated Death of Human Small-Cell Lung Cancer Cells. Int. J. Mol. Sci. 2022, 23, 6895. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.B.; Kim, J.W.; Heo, K.; Kim, H.J.; Yun, S.; Lee, H.S.; Shin, H.G.; Shim, H.; Yu, H.; Kim, Y.H.; et al. An internalizing antibody targeting of cell surface GRP94 effectively suppresses tumor angiogenesis of colorectal cancer. Biomed. Pharmacother. 2022, 150, 113051. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, N.; Kumar, J.S.; Yadav, P.; Shete, A.M.; Jain, R.; Shrivastava, A.; Dash, P.K. Development of double antibody sandwich ELISA as potential diagnostic tool for rapid detection of Crimean-Congo hemorrhagic fever virus. Sci. Rep. 2021, 11, 14699. [Google Scholar] [CrossRef] [PubMed]
- Zai, J.; Yi, K.; Xie, L.; Zhu, J.; Feng, X.; Li, Y. Dual monoclonal antibody-based sandwich ELISA for detection of in vitro packaged Ebola virus. Diagn. Pathol. 2018, 13, 96. [Google Scholar] [CrossRef]
- Rajna, M.; Irena, Z. Optimization, Validation and standardization of ELISA. In Norovirus; Gyula, M., Ed.; IntechOpen: Rijeka, Croatia, 2020; Chapter 2. [Google Scholar]
- Nundy, S.; Ghosh, A.; Mesloub, A.; Albaqawy, G.A.; Alnaim, M.M. Impact of COVID-19 pandemic on socio-economic, energy-environment and transport sector globally and sustainable development goal (SDG). J. Clean. Prod. 2021, 312, 127705. [Google Scholar] [CrossRef]
- Hwang, Y.C.; Lu, R.M.; Su, S.C.; Chiang, P.Y.; Ko, S.H.; Ke, F.Y.; Liang, K.H.; Hsieh, T.Y.; Wu, H.C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 2022, 29, 1. [Google Scholar] [CrossRef]
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 2021, 184, 1821–1835.e1816. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020, 182, 73–84.e16. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Ryu, D.-K.; Lee, J.; Kim, Y.-I.; Seo, J.-M.; Kim, Y.-G.; Jeong, J.-H.; Kim, M.; Kim, J.-I.; Kim, P.; et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.A.; Zhou, D.; Tan, T.K.; Chen, C.; Duyvesteyn, H.M.E.; Zhao, Y.; Ginn, H.M.; Qin, L.; Rijal, P.; Schimanski, L.; et al. Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2. Theranostics 2022, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chandele, A.; Sharma, A. Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog. 2021, 17, e1009885. [Google Scholar] [CrossRef]
- Chen, X.; Li, R.; Pan, Z.; Qian, C.; Yang, Y.; You, R.; Zhao, J.; Liu, P.; Gao, L.; Li, Z.; et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020, 17, 647–649. [Google Scholar] [CrossRef]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Ryman, J.T.; Meibohm, B. Pharmacokinetics of Monoclonal Antibodies. CPT Pharmacomet. Syst. Pharm. 2017, 6, 576–588. [Google Scholar] [CrossRef]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis—A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]







| Antibody | KD (nM) | Ka (1/M−1 s−1) | Kd (s−1) |
|---|---|---|---|
| K102.1 | 1.11 | 5.95 105 | 6.61 10−4 |
| K102.2 | 2.45 | 4.03 105 | 9.89 10−4 |
| K102.3 | 11.3 | 3.43 104 | 3.89 10−4 |
| K102.4 | 3.16 | 3.15 105 | 9.96 10−4 |
| RBD Type | IC50 (nM) | |||
|---|---|---|---|---|
| K102.1 | K102.2 | K102.3 | K102.4 | |
| Wild-type | 0.89 0.15 | 1.71 0.23 | ND | 8.15 0.82 |
| B.1.1.7 (Alpha) | 7.27 1.01 | 18.85 2.41 | ND | 21.83 2.55 |
| B.1.617.2 (Delta) | 1.57 0.52 | 2.99 0.51 | ND | 12.54 1.42 |
| Spiked Level (ng/mL) | Intra-Assay (n = 6) | Inter-Assay (n = 6) | ||||
|---|---|---|---|---|---|---|
| SD (ng/mL) | Recovery (%) | CV (%) | SD (ng/mL) | Recovery (%) | CV (%) | |
| 5 | 5.28 0.45 | 105.57 | 8.46 | 4.93 0.47 | 98.56 | 9.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.W.; Min, S.W.; Lee, J.; Shin, H.G.; Choi, H.L.; Yang, H.R.; Lee, J.H.; Cho, Y.B.; Shim, H.; Lee, S. Development and Characterization of Phage-Display-Derived Novel Human Monoclonal Antibodies against the Receptor Binding Domain of SARS-CoV-2. Biomedicines 2022, 10, 3274. https://doi.org/10.3390/biomedicines10123274
Kim JW, Min SW, Lee J, Shin HG, Choi HL, Yang HR, Lee JH, Cho YB, Shim H, Lee S. Development and Characterization of Phage-Display-Derived Novel Human Monoclonal Antibodies against the Receptor Binding Domain of SARS-CoV-2. Biomedicines. 2022; 10(12):3274. https://doi.org/10.3390/biomedicines10123274
Chicago/Turabian StyleKim, Ji Woong, Sung Won Min, Jichul Lee, Ha Gyeong Shin, Hye Lim Choi, Ha Rim Yang, Ji Hyun Lee, Yea Bin Cho, Hyunbo Shim, and Sukmook Lee. 2022. "Development and Characterization of Phage-Display-Derived Novel Human Monoclonal Antibodies against the Receptor Binding Domain of SARS-CoV-2" Biomedicines 10, no. 12: 3274. https://doi.org/10.3390/biomedicines10123274
APA StyleKim, J. W., Min, S. W., Lee, J., Shin, H. G., Choi, H. L., Yang, H. R., Lee, J. H., Cho, Y. B., Shim, H., & Lee, S. (2022). Development and Characterization of Phage-Display-Derived Novel Human Monoclonal Antibodies against the Receptor Binding Domain of SARS-CoV-2. Biomedicines, 10(12), 3274. https://doi.org/10.3390/biomedicines10123274