Label-Free Physical Techniques and Methodologies for Proteins Detection in Microfluidic Biosensor Structures
Abstract
:1. Introduction
2. Methods for Manufacturing Microfluidic Structures for Biosensors
2.1. Microfluidics: Basics and Materials
2.2. Technological Approaches for Microfluidics
2.2.1. Lithography as a Link between the Other Processes
2.2.2. Oxidation of Silicon
2.2.3. Chemical Vapour Deposition (CVD)
2.2.4. Wet and Dry Chemical Structuring of Substrate Material
2.2.5. 3D Printing of Microfluidic Systems
FDM or FFF Printing
Stereolithography (SLA) Printing
2.2.6. Sub-Micron Structuring
2.2.7. Molding and Bonding
2.2.8. Surface Functionalization to Avoid Protein Adsorption
3. Label-Free Optical Techniques of Protein Detection, Quantification, and Characterization in Microfluidics
- labeling-binding (constantly or temporarily) to a protein molecule a highly optically active (most often in the visible region) foreign molecule, quantum dot or molecular complex, e.g., by specific antibody-antigen chemical interaction [145];
- molecular sorting of different proteins in a mixture (using separation or focusing) on an analytical stage prior to detection, e.g., by electrophoresis, isoelectric focusing, acoustic waves, or chromatography;
- manyfold amplification of an optical signals emploing surface plasmon resonance, surface enhanced Raman spectroscopy, and surface-enhanced infrared absorption spectroscopy.
3.1. Optical Properties of Proteins in the UV, Visible and IR Regions
3.2. Overview of the Optical Techniques and Microfluidic Structures Design Strategies for Label-Free Protein Detection
3.3. Protein Detection by Absorption Spectroscopy
3.3.1. UV Absorption Spectroscopy
3.3.2. MIR Spectroscopy
3.4. Protein Detection by Intrinsic Fluorescence
3.5. Refractometry in Microfluidics
3.6. Plasmon Resonance for Protein Detection
3.6.1. Sensors Based on SPR
3.6.2. Sensors Based on LSPR
3.6.3. Microfluidics-Based Plasmonic Sensors
3.6.4. Protein Detection by Surface-Enhanced Raman Spectroscopy
3.7. Application of Diffusometric Methods for Protein Characterization
3.7.1. Dynamic Light Scattering
3.7.2. Nanoparticle Tracking Analysis
3.8. Optic Components in Optofluidics for Protein Detection
3.8.1. Microlenses
3.8.2. Waveguides
3.8.2.1. TIR-Based WGs
3.8.2.2. RI-Modulated WGs and Resonance WG Structures
3.8.2.3. ARROWs
3.8.3. Other Micron- and Submicron Scale Structures
4. Impedance Spectroscopy Microfluidic Techniques and Methods for Proteins Detection
4.1. Electrical Impedance Spectroscopy
4.2. Electrochemical Impedance Spectroscopy
| Structure | Detection Principle/Notes | Target Analyte | Limit of Detection (LOD) // Sensitivity | [Ref.]/Year |
|---|---|---|---|---|
| Flexible platinum electrodes | Voltammetric measurements, Electrochemical impedance spectroscopy | Dopamine, Parkinson’s disease protein 7 | 5.1 × 10−6 mol/L, 7.5 ng/mL // − | [390] 2020 |
| Capacitive sensor in a microfluidic channel | Dielectric spectroscopy 10 kHz to 100 MHz/sample volume < 10 μL | Blood coagulation factor, platelets | − | [378] 2018 |
| Microwave dielectric resonator–microfluidic system | Broadband microwave spectroscopy 200 MHz and 40 GHz/sample volume < 10 μL | Hemoglobin | SD ≈ 0.34 g/dL | [380] 2016 |
| Microfluidic impedance biosensor | Electrochemical impedance spectroscopy | Troponin I | 1 ng/mL // − | [63] 2021 |
| Microfluidic impedance biosensor | Electrochemical Impedance Spectroscopy | Prostate Specific Antigen | 1 ng/mL // − | [391] 2013 |
| Biomimetic sensors | Electrochemical Impedance Spectroscopy/Human serum analysis | Adiponectin, Leptin | 0.25 μg/mL, 0.110 ng/mL // − | [392] 2020 |
| Paper microfluidic biosensor | Electrochemical Impedance Spectroscopy/Functionalized multi-walled carbon nanotubes | Troponin I | 0.05 ng/mL // 1.85 mΩ/ng/mL | [393] 2019 |
| Multiwell microelectrode array | Electrochemical Impedance Spectroscopy | Tau protein | – | [394] 2016 |
| Microfluidic immunosensor | Electrochemical Impedance Spectroscopy/Polyethylenimine coated graphene electrode, wide dynamic range 1 pg/mL to 100 ng/mL | Glial fibrillary acidic protein | 1 pg/mL // − | [395] 2018 |
| Disposable microfluidic amperometric dual-sensor | Electrocatalytic reduction/Human blood analysis | Glycated hemoglobin, total hemoglobin | 3.7 nM, 82 nM // − | [396] 2017 |
| Microfluidic immunosensor | Pulse voltammetry, Electrochemical impedance spectroscopy | Epidermal growth factor receptor 2 | 1.0 fM, 1.0 pM // 0.585 μA/μM × cm2, 43.7 kΩ/μM × cm2 | [397] 2016 |
| Plastic-paper microfluidic chip | Impedance spectroscopy in the frequency range of 100 Hz to 100 kHz/Human serum analysis | Alpha-fetoprotein | 10 ng/mL // − | [379] 2018 |
| Molecular imprinted polymer (MIP)-based impedimetric sensor | Electrochemical impedance spectroscopy | NS1 (non-structural protein 1—a specific and sensitive biomarker for dengue virus infection) | 0.3 ng/mL | [398] 2020 |
| Symmetrical split ring resonator (SSRR) based microwave sensor | Microwave spectroscopy | Drugs | − | [381] 2017 |
| Interdigitated electrode sensor | Microwave spectroscopy/cerebrospinal fluid analysis, wide dynamic range 0 to 100 g/L | Albumin | − | [382] 2015 |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copeland, R.A. Methods for Protein Analysis; Springer: Boston, MA, USA, 1994; ISBN 978-1-4757-1507-1. [Google Scholar]
- Zhu, Z.; Lu, J.J.; Liu, S. Protein separation by capillary gel electrophoresis: A review. Anal. Chim. Acta 2012, 709, 21–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Descalzo, L.; Garca-Lopez, E.; Alcazar, A.; Baquero, F.; Ci, C. Gel Electrophoresis of Proteins. In Gel Electrophoresis-Principles and Basics; Magdeldin, S., Ed.; InTech: Rijeka, Croatia, 2012; pp. 57–68. ISBN 978-953-51-0458-2. [Google Scholar]
- Dunn, M.J. (Ed.) Gel Electrophoresis of Proteins; Butterworth-Heinemann: Oxford, UK, 1986; ISBN 9780723608820. [Google Scholar]
- Pergande, M.R.; Cologna, S.M. Isoelectric Point Separations of Peptides and Proteins. Proteomes 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righetti, P.G.; Sebastiano, R.; Citterio, A. Capillary electrophoresis and isoelectric focusing in peptide and protein analysis. Proteomics 2013, 13, 325–340. [Google Scholar] [CrossRef]
- Aguilar, M.-I. (Ed.) HPLC of Peptides and Proteins; Humana Press: Totowa, NJ, USA, 2003; ISBN 1-59259-742-4. [Google Scholar]
- Tarasova, I.A.; Masselon, C.D.; Gorshkov, A.V.; Gorshkov, M.V. Predictive chromatography of peptides and proteins as a complementary tool for proteomics. Analyst 2016, 141, 4816–4832. [Google Scholar] [CrossRef]
- Donnelly, D.P.; Rawlins, C.M.; DeHart, C.J.; Fornelli, L.; Schachner, L.F.; Lin, Z.; Lippens, J.L.; Aluri, K.C.; Sarin, R.; Chen, B.; et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat. Methods 2019, 16, 587–594. [Google Scholar] [CrossRef]
- Gingras, A.-C.; Gstaiger, M.; Raught, B.; Aebersold, R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007, 8, 645–654. [Google Scholar] [CrossRef]
- Noor, Z.; Ahn, S.B.; Baker, M.S.; Ranganathan, S.; Mohamedali, A. Mass spectrometry-based protein identification in proteomics—A review. Brief. Bioinform. 2021, 22, 1620–1638. [Google Scholar] [CrossRef] [PubMed]
- Low, T.Y.; Syafruddin, S.E.; Mohtar, M.A.; Vellaichamy, A.; A Rahman, N.S.; Pung, Y.-F.; Tan, C.S.H. Recent progress in mass spectrometry-based strategies for elucidating protein-protein interactions. Cell. Mol. Life Sci. 2021, 78, 5325–5339. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai-Kastoori, L.; Schutz-Geschwender, A.R.; Harford, J.A. A systematic approach to quantitative Western blot analysis. Anal. Biochem. 2020, 593, 113608. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Tiwari, S.; Gomes, A.V. Protein purification and analysis: Next generation Western blotting techniques. Expert Rev. Proteom. 2017, 14, 1037–1053. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Gupta, A.; Bharti, C.; Sharma, H. Emerging techniques of western blotting for purification and analysis of protein. Future J. Pharm. Sci. 2021, 7, 1–14. [Google Scholar] [CrossRef]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Hnasko, R. (Ed.) ELISA; Springer: New York, NY, USA, 2015; ISBN 978-1-4939-2741-8. [Google Scholar]
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. Enzyme-Linked Immunosorbent Assay (ELISA): From A to Z; Springer: Singapore, 2018; ISBN 978-981-10-6765-5. [Google Scholar]
- Purslow, J.A.; Khatiwada, B.; Bayro, M.J.; Venditti, V. NMR Methods for Structural Characterization of Protein-Protein Complexes. Front. Mol. Biosci. 2020, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, N.; Malmendal, A.; Vosegaard, T. Techniques and applications of NMR to membrane proteins. Mol. Membr. Biol. 2004, 21, 129–141. [Google Scholar] [CrossRef]
- Marion, D. An introduction to biological NMR spectroscopy. Mol. Cell. Proteom. 2013, 12, 3006–3025. [Google Scholar] [CrossRef] [Green Version]
- Beynon, R.J.; Pratt, J.M. Metabolic labeling of proteins for proteomics. Mol. Cell. Proteom. 2005, 4, 857–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugarinov, V.; Kanelis, V.; Kay, L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006, 1, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Permentier, H.P.; Bischoff, R. Chemical isotope labeling for quantitative proteomics. Mass Spectrom. Rev. 2021, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Recent applications of light scattering measurement in the biological and biopharmaceutical sciences. Anal. Biochem. 2016, 501, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Some, D. Light-scattering-based analysis of biomolecular interactions. Biophys. Rev. 2013, 5, 147–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, H.K.; Kartanas, T.; Saar, K.L.; Luxhøj, C.M.; Devenish, S.; Knowles, T.P.J. Rapid highly sensitive general protein quantification through on-chip chemiluminescence. Biomicrofluidics 2021, 15, 024113. [Google Scholar] [CrossRef]
- Calabretta, M.M.; Zangheri, M.; Calabria, D.; Lopreside, A.; Montali, L.; Marchegiani, E.; Trozzi, I.; Guardigli, M.; Mirasoli, M.; Michelini, E. Paper-Based Immunosensors with Bio-Chemiluminescence Detection. Sensors 2021, 21, 4309. [Google Scholar] [CrossRef] [PubMed]
- Speletas, M.; Kyritsi, M.A.; Vontas, A.; Theodoridou, A.; Chrysanthidis, T.; Hatzianastasiou, S.; Petinaki, E.; Hadjichristodoulou, C. Evaluation of Two Chemiluminescent and Three ELISA Immunoassays for the Detection of SARS-CoV-2 IgG Antibodies: Implications for Disease Diagnosis and Patients’ Management. Front. Immunol. 2020, 11, 609242. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and methods for circular dichroism spectroscopy of proteins: A tutorial review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef] [Green Version]
- Colón, W. Analysis of protein structure by solution optical spectroscopy. In Amyloid, Prions, and Other Protein Aggregates; Elsevier: Amsterdam, The Netherlands, 1999; pp. 605–632. ISBN 9780121822101. [Google Scholar]
- Christov, C.Z. (Ed.) Introduction: Biomolecular Spectroscopy: Advances from Integrating Experiments and Theory; Elsevier: Oxford, UK, 2013; Volume 93, pp. 1–336. ISBN 978-0-12-416596-0. [Google Scholar]
- Narhi, L.O.; Li, C.H.; Ramachander, R.; Svitel, J.; Jiang, Y. Optical Spectroscopic Methods for the Analysis of Biological Macromolecules. In Molecular Biophysics for the Life Sciences; Allewell, N., Narhi, L.O., Rayment, I., Eds.; Springer: New York, NY, USA, 2013; pp. 33–90. ISBN 978-1-4614-8547-6. [Google Scholar]
- Legrain, P.; Aebersold, R.; Archakov, A.; Bairoch, A.; Bala, K.; Beretta, L.; Bergeron, J.; Borchers, C.H.; Corthals, G.L.; Costello, C.E.; et al. The human proteome project: Current state and future direction. Mol. Cell. Proteom. 2011, 10, M111.009993. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omenn, G.S. Reflections on the HUPO Human Proteome Project, the Flagship Project of the Human Proteome Organization, at 10 Years. Mol. Cell. Proteom. 2021, 20, 100062. [Google Scholar] [CrossRef]
- Adhikari, S.; Nice, E.C.; Deutsch, E.W.; Lane, L.; Omenn, G.S.; Pennington, S.R.; Paik, Y.-K.; Overall, C.M.; Corrales, F.J.; Cristea, I.M.; et al. A high-stringency blueprint of the human proteome. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Silberring, J.; Ciborowski, P. Biomarker discovery and clinical proteomics. Trends Analyt. Chem. 2010, 29, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrebaeck, C.A.K. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef]
- Sallam, R.M. Proteomics in cancer biomarkers discovery: Challenges and applications. Dis. Markers 2015, 2015, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.W.; Jo, H.-S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. 2021, 8, 747333. [Google Scholar] [CrossRef]
- Chen, L.; Gu, H.; Li, J.; Yang, Z.-Y.; Sun, X.; Zhang, L.; Shan, L.; Wu, L.; Wei, X.; Zhao, Y.; et al. Comprehensive maternal serum proteomics identifies the cytoskeletal proteins as non-invasive biomarkers in prenatal diagnosis of congenital heart defects. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, G.; He, P.; Du, Y.; Zhang, S. Identification of Biomarkers by Proteomics for Prenatal Screening for Neural Tube Defects. Tohoku J. Exp. Med. 2016, 238, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, H.S.; Lee, S.M.; Jung, Y.M.; Oh, S.; Park, J.K.; Lee, E.B.; Park, C.-W.; Park, J.S.; Han, D.; Jun, J.K. Proteomic biomarkers in mid-trimester amniotic fluid associated with adverse pregnancy outcomes in patients with systemic lupus erythematosus. PLoS ONE 2020, 15, e0235838. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, N. Accelerating protein biomarker discovery and translation from proteomics research for clinical utility. Bioanalysis 2020, 12, 1469–1481. [Google Scholar] [CrossRef]
- Mejía-Salazar, J.R.; Rodrigues Cruz, K.; Materón Vásques, E.M.; Novais de Oliveira, O. Microfluidic Point-of-Care Devices: New Trends and Future Prospects for eHealth Diagnostics. Sensors 2020, 20, 1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Segal, E. Lab-on-a-Chip Devices for Point-of-Care Medical Diagnostics. Adv. Biochem. Eng. Biotechnol. 2020, 1–19. [Google Scholar] [CrossRef]
- Zimmerman, W.B.J. Microfluidics: History, Theory and Applications; Springer: Vienna, Austria, 2006; ISBN 978-3-211-32994-8. [Google Scholar]
- Gökaltun, A.; Kang, Y.B.A.; Yarmush, M.L.; Usta, O.B.; Asatekin, A. Simple Surface Modification of Poly(dimethylsiloxane) via Surface Segregating Smart Polymers for Biomicrofluidics. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Sugioka, K.; Cheng, Y. Femtosecond laser three-dimensional micro- and nanofabrication. Appl. Phys. Rev. 2014, 1, 041303. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.; Kim, S.; Lee, J.; Choi, J.; Kim, R.-K.; Lee, S.-J.; Sul, O.; Lee, S.-B. Clogging-free microfluidics for continuous size-based separation of microparticles. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Enders, A.; Preuss, J.-A.; Bahnemann, J. 3D Printed Microfluidic Spiral Separation Device for Continuous, Pulsation-Free and Controllable CHO Cell Retention. Micromachines 2021, 12, 1060. [Google Scholar] [CrossRef]
- Liao, Y.; Mechulam, Y.; Lassalle-Kaiser, B. A millisecond passive micromixer with low flow rate, low sample consumption and easy fabrication. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Krevelen, D.W.; Nijenhuis, K.T. Properties of Polymers; Elsevier: Amsterdam, The Netherlands, 2009; ISBN 9780080548197. [Google Scholar]
- Hu, H.; Eustace, D.; Merten, C.A. Efficient cell pairing in droplets using dual-color sorting. Lab. Chip 2015, 15, 3989–3993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horrer, A.; Haas, J.; Freudenberger, K.; Gauglitz, G.; Kern, D.P.; Fleischer, M. Compact plasmonic optical biosensors based on nanostructured gradient index lenses integrated into microfluidic cells. Nanoscale 2017, 9, 17378–17386. [Google Scholar] [CrossRef]
- Alsabbagh, K.; Hornung, T.; Voigt, A.; Sadir, S.; Rajabi, T.; Länge, K. Microfluidic Impedance Biosensor Chips Using Sensing Layers Based on DNA-Based Self-Assembled Monolayers for Label-Free Detection of Proteins. Biosensors 2021, 11, 80. [Google Scholar] [CrossRef]
- Khodayari Bavil, A.; Sticker, D.; Rothbauer, M.; Ertl, P.; Kim, J. A microfluidic microparticle-labeled impedance sensor array for enhancing immunoassay sensitivity. Analyst 2021, 146, 3289–3298. [Google Scholar] [CrossRef]
- Liao, H.; Zhou, Y.; Chai, Y.; Yuan, R. An ultrasensitive electrochemiluminescence biosensor for detection of MicroRNA by in-situ electrochemically generated copper nanoclusters as luminophore and TiO2 as coreaction accelerator. Biosens. Bioelectron. 2018, 114, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Rho, D.; Breaux, C.; Kim, S. Label-Free Optical Resonator-Based Biosensors. Sensors 2020, 20, 5901. [Google Scholar] [CrossRef] [PubMed]
- Gökçe, F.; Ravaynia, P.S.; Modena, M.M.; Hierlemann, A. What is the future of electrical impedance spectroscopy in flow cytometry? Biomicrofluidics 2021, 15, 061302. [Google Scholar] [CrossRef] [PubMed]
- Franks, W.; Schenker, I.; Schmutz, P.; Hierlemann, A. Impedance characterization and modeling of electrodes for biomedical applications. IEEE Trans. Biomed. Eng. 2005, 52, 1295–1302. [Google Scholar] [CrossRef]
- Issakov, V.; Wojnowski, M.; Thiede, A.; Weigel, R. Considerations on the de-embedding of differential devices using two-port techniques. Int. J. Microw. Wirel. Technol. 2010, 2, 349–357. [Google Scholar] [CrossRef]
- Issakov, V.; Heine, C.; Lammert, V.; Stoegmueller, J.; Meindl, M.; Stubenrauch, U.; Geissler, C. Fully Autonomous System-on-Board with Complex Permittivity Sensors and 60 GHz Transmitter for Biomedical Implant Applications. In Proceedings of the IEEE Radio Frequency Integrated Circuits Symposium (RFIC), Los Angeles, CA, USA, 4–6 August 2020; pp. 159–162, ISBN 978-1-7281-6809-8. [Google Scholar]
- Ciocoveanu, R.; Rimmelspacher, J.; Weigel, R.; Hagelauer, A.; Issakov, V. A 1.8-mW low power, PVT-resilient, high linearity, modified Gilbert-cell down-conversion mixer in 28-nm CMOS. In 2018 IEEE 18th Topical Meeting on Silicon Monolithic Integrated Circuits in RF Systems (SiRF); IEEE: Anaheim, CA, USA, 2018; pp. 19–22. ISBN 978-1-5386-1298-9. [Google Scholar]
- Issakov, V.; Thiede, A.; Verweyen, L.; Maurer, L. Wideband Resistive Ring Mixer for Automotive and Industrial Applications in 0.13 μm CMOS. In Proceedings of the 2009 German Microwave Conference (GeMIC 2009), Munich, Germany, 16–18 March 2009; pp. 1–4, ISBN 978-3-9812668-0-1. [Google Scholar]
- Rimmelspacher, J.; Weigel, R.; Hagelauer, A.; Issakov, V. 36% Frequency-tuning-range dual-core 60 GHz push-push VCO in 45 nm RF-SOI CMOS technology. In Proceedings of the IEEE/MTT-S International Microwave Symposium-IMS 2017, Honololu, HI, USA, 4–9 June 2017; pp. 1356–1358, ISBN 978-1-5090-6360-4. [Google Scholar]
- Madou, M.J. Manufacturing Techniques for Microfabrication and Nanotechnology; CRC Press: Boca Raton, FL, USA, 2012; ISBN 978-1420055191. [Google Scholar]
- Gerlach, G.; Dötze, W. Einführung in die Mikrosystemtechnik; Carl Hanser Fachbuchverlag: Berlin, Germany, 2006; ISBN 978-3-446-40523-3. [Google Scholar]
- Schmidt, M.-P.; Oseev, A.; Engel, C.; Brose, A.; Aman, A.; Hirsch, S. A Novel Design and Fabrication of Multichannel Microfluidic Impedance Spectroscopy Sensor for Intensive Electromagnetic Environment Application. Procedia Eng. 2014, 87, 88–91. [Google Scholar] [CrossRef] [Green Version]
- Deal, B.E.; Grove, A.S. General Relationship for the Thermal Oxidation of Silicon. J. Appl. Phys. 1965, 36, 3770–3778. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.R.; Gupta, K.; Wasilik, M. Etch rates for micromachining processing-part II. J. Microelectromech. Syst. 2003, 12, 761–778. [Google Scholar] [CrossRef] [Green Version]
- Stasiak, J.; Richards, S.; Angelos, S. Hewlett Packard’s inkjet MEMS technology: Past, present, and future. In Micro- and Nanotechnology Sensors, Systems, and Applications; George, T., Islam, M.S., Dutta, A.K., Eds.; SPIE Defense, Security, and Sensing: Orlando, FL, USA, 2009; Volume 7318, p. 73180U. [Google Scholar]
- Oseev, A.; Schmidt, M.-P.; Hirsch, S.; Brose, A.; Schmidt, B. Two-component dielectric dispersion impedance biosensor for in-line protein monitoring. Sens. Actuators B Chem. 2017, 239, 1213–1220. [Google Scholar] [CrossRef]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D printed microfluidic devices: Enablers and barriers. Lab. Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Au, A.K.; Huynh, W.; Horowitz, L.F.; Folch, A. 3D-Printed Microfluidics. Angew. Chem. Int. Ed. Engl. 2016, 55, 3862–3881. [Google Scholar] [CrossRef] [PubMed]
- Fastermann, P. 3D-Drucken; Springer: Berlin, Germany, 2016; ISBN 978-3-662-49865-1. [Google Scholar]
- Steimle, A. 3D Micro-Printing: A new Era for Med-Tech Applications. Laser Tech. J. 2018, 15, 32–34. [Google Scholar] [CrossRef] [Green Version]
- Kuhlmann, C.; Blum, J.C.; Schenck, T.L.; Giunta, R.E.; Wiggenhauser, P.S. Evaluation of the Usability of a Low-Cost 3D Printer in a Tissue Engineering Approach for External Ear Reconstruction. Int. J. Mol. Sci. 2021, 22, 11667. [Google Scholar] [CrossRef]
- Domingo-Espin, M.; Puigoriol-Forcada, J.M.; Garcia-Granada, A.-A.; Llumà, J.; Borros, S.; Reyes, G. Mechanical property characterization and simulation of fused deposition modeling Polycarbonate parts. Mater. Des. 2015, 83, 670–677. [Google Scholar] [CrossRef]
- Rinaldi, M.; Ghidini, T.; Cecchini, F.; Brandao, A.; Nanni, F. Additive layer manufacturing of poly (ether ether ketone) via FDM. Compos. Part. B Eng. 2018, 145, 162–172. [Google Scholar] [CrossRef]
- Yang, C.; Tian, X.; Li, D.; Cao, Y.; Zhao, F.; Shi, C. Influence of thermal processing conditions in 3D printing on the crystallinity and mechanical properties of PEEK material. J. Mater. Process. Technol. 2017, 248, 1–7. [Google Scholar] [CrossRef]
- Duran, C.; Subbian, V.; Giovanetti, M.T.; Simkins, J.R.; Beyette, F.R., Jr. Experimental desktop 3D printing using dual extrusion and water-soluble polyvinyl alcohol. Rapid Prototyp. J. 2015, 21, 528–534. [Google Scholar] [CrossRef]
- Quero, R.F.; Da Domingos Silveira, G.; Da Fracassi Silva, J.A.; De Jesus, D.P. Understanding and improving FDM 3D printing to fabricate high-resolution and optically transparent microfluidic devices. Lab. Chip 2021, 21, 3715–3729. [Google Scholar] [CrossRef]
- Bhushan, B.; Caspers, M. An overview of additive manufacturing (3D printing) for microfabrication. Microsyst Technol. 2017, 23, 1117–1124. [Google Scholar] [CrossRef]
- Liberale, C.; Cojoc, G.; Bragheri, F.; Minzioni, P.; Perozziello, G.; La Rocca, R.; Ferrara, L.; Rajamanickam, V.; Di Fabrizio, E.; Cristiani, I. Integrated microfluidic device for single-cell trapping and spectroscopy. Sci. Rep. 2013, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Jonušauskas, L.; Rekštytė, S.; Buividas, R.; Butkus, S.; Gadonas, R.; Juodkazis, S.; Malinauskas, M. Hybrid subtractive-additive-welding microfabrication for lab-on-chip applications via single amplified femtosecond laser source. Opt. Eng. 2017, 56, 094108. [Google Scholar] [CrossRef]
- Aristov, A.I.; Manousidaki, M.; Danilov, A.; Terzaki, K.; Fotakis, C.; Farsari, M.; Kabashin, A.V. 3D plasmonic crystal metamaterials for ultra-sensitive biosensing. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bakhchova, L.; Jonušauskas, L.; Andrijec, D.; Kurachkina, M.; Baravykas, T.; Eremin, A.; Steinmann, U. Femtosecond Laser-Based Integration of Nano-Membranes into Organ-on-a-Chip Systems. Materials 2020, 13, 3076. [Google Scholar] [CrossRef] [PubMed]
- Gissibl, T.; Thiele, S.; Herkommer, A.; Giessen, H. Two-photon direct laser writing of ultracompact multi-lens objectives. Nat. Photonics 2016, 10, 554–560. [Google Scholar] [CrossRef]
- Schnauber, P.; Schall, J.; Bounouar, S.; Höhne, T.; Park, S.-I.; Ryu, G.-H.; Heindel, T.; Burger, S.; Song, J.-D.; Rodt, S.; et al. Deterministic Integration of Quantum Dots into on-Chip Multimode Interference Beamsplitters Using in Situ Electron Beam Lithography. Nano Lett. 2018, 18, 2336–2342. [Google Scholar] [CrossRef]
- Bakhchova, L.; Jantaree, P.; Gupta, A.; Isermann, B.; Steinmann, U.; Naumann, M. On-a-Chip-Based Sensitive Detection of Drug-Induced Apoptosis in Polarized Gastric Epithelial Cells. ACS Biomater. Sci. Eng. 2021, 7, 5474–5483. [Google Scholar] [CrossRef]
- Liu, D.; Zhou, L.; Huang, L.; Zuo, Z.; Ho, V.; Jin, L.; Lu, Y.; Chen, X.; Zhao, J.; Qian, D.; et al. Microfluidic integrated capacitive biosensor for C-reactive protein label-free and real-time detection. Analyst 2021, 146, 5380–5388. [Google Scholar] [CrossRef]
- Schmidt, M.-P.; Oseev, A.; Engel, C.; Brose, A.; Schmidt, B.; Hirsch, S. Flexible free-standing SU-8 microfluidic impedance spectroscopy sensor for 3-D molded interconnect devices application. J. Sens. Sens. Syst. 2016, 5, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Chumbimuni-Torres, K.Y.; Coronado, R.E.; Mfuh, A.M.; Castro-Guerrero, C.; Silva, M.F.; Negrete, G.R.; Bizios, R.; Garcia, C.D. Adsorption of Proteins to Thin-Films of PDMS and Its Effect on the Adhesion of Human Endothelial Cells. RSC Adv. 2011, 1, 706–714. [Google Scholar] [CrossRef]
- Ren, K.; Zhao, Y.; Su, J.; Ryan, D.; Wu, H. Convenient method for modifying poly(dimethylsiloxane) to be airtight and resistive against absorption of small molecules. Anal. Chem. 2010, 82, 5965–5971. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kim, N.; Hong, J.W. Comparison of Surface Modification Techniques on Polydimethylsiloxane to Prevent Protein Adsorption. BioChip J. 2018, 12, 123–127. [Google Scholar] [CrossRef]
- You, J.B.; Lee, B.; Choi, Y.; Lee, C.-S.; Peter, M.; Im, S.G.; Lee, S.S. Nanoadhesive layer to prevent protein absorption in a poly(dimethylsiloxane) microfluidic device. Biotechniques 2020, 69, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Gai, H.; Li, Y.; Yeung, E.S. Optical detection systems on microfluidic chips. Top. Curr. Chem. 2011, 304, 171–201. [Google Scholar] [CrossRef]
- Wu, J.; He, Z.; Chen, Q.; Lin, J.-M. Biochemical analysis on microfluidic chips. TrAC Trends Anal. Chem. 2016, 80, 213–231. [Google Scholar] [CrossRef]
- Mogensen, K.B.; Kutter, J.P. Optical detection in microfluidic systems. Electrophoresis 2009, 30 (Suppl. 1), S92–S100. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, Y.; Li, Y.; Miao, Y.; Gao, S.; Lin, F.; Deng, Y.; Geng, L. Microfluidic chip coupled with optical biosensors for simultaneous detection of multiple analytes: A review. Biosens. Bioelectron. 2019, 126, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Kuswandi, B.; Nuriman; Huskens, J.; Verboom, W. Optical sensing systems for microfluidic devices: A review. Anal. Chim. Acta 2007, 601, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gu, M. Microfluidic sensing: State of the art fabrication and detection techniques. J. Biomed. Opt. 2011, 16, 080901. [Google Scholar] [CrossRef] [Green Version]
- Momenbeitollahi, N.; Cloet, T.; Li, H. Pushing the detection limits: Strategies towards highly sensitive optical-based protein detection. Anal. Bioanal. Chem. 2021, 413, 5995–6011. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, R. Recent optical sensing technologies for the detection of various biomolecules: Review. Opt. Laser Technol. 2021, 134, 106620. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Y.; Chen, T.; Shen, W.; Tang, S.; Lee, H.K. Application of smartphone-based spectroscopy to biosample analysis: A review. Biosens. Bioelectron. 2021, 172, 112788. [Google Scholar] [CrossRef] [PubMed]
- Pirzada, M.; Altintas, Z. Recent Progress in Optical Sensors for Biomedical Diagnostics. Micromachines 2020, 11, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Fathi, F.; Rashidi, M.-R.; Pakchin, P.S.; Ahmadi-Kandjani, S.; Nikniazi, A. Photonic crystal based biosensors: Emerging inverse opals for biomarker detection. Talanta 2021, 221, 121615. [Google Scholar] [CrossRef]
- Gao, X.-G.; Cheng, L.-X.; Jiang, W.-S.; Li, X.-K.; Xing, F. Graphene and its Derivatives-Based Optical Sensors. Front. Chem. 2021, 9, 5. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, W.; Xing, F. Graphene Optical Biosensors. Int. J. Mol. Sci. 2019, 20, 2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Qiu, J.-G.; Ma, F.; Zhang, C.-Y. Advances in single-molecule fluorescent nanosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1716. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Cao, X.; Yang, S.; Mo, Z.; Wang, W.; Zeng, W. Fluorescent Probes for Detection of Protein: From Bench to Bed. Protein Pept. Lett. 2018, 25, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, L.A.; Wei, Q.; Barui, A.K.; Mohammad, N. Recent Advances in Aptamer-Based Biosensors for Global Health Applications. Annu. Rev. Biomed. Eng. 2021, 23, 433–459. [Google Scholar] [CrossRef]
- Scatena, E.; Baiguera, S.; Del Gaudio, C. Raman Spectroscopy and Aptamers for a Label-Free Approach: Diagnostic and Application Tools. J. Healthc. Eng. 2019, 2019. [Google Scholar] [CrossRef]
- Fan, J.; Qi, L.; Han, H.; Ding, L. Array-Based Discriminative Optical Biosensors for Identifying Multiple Proteins in Aqueous Solution and Biofluids. Front. Chem. 2020, 8, 572234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Qavi, A.J.; Huang, S.H.; Yang, L. Whispering-Gallery Sensors. Matter 2020, 3, 371–392. [Google Scholar] [CrossRef]
- Esfahani Monfared, Y. Overview of Recent Advances in the Design of Plasmonic Fiber-Optic Biosensors. Biosensors 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.; Huertas, C.S.; Lechuga, L.M. Label-free plasmonic biosensors for point-of-care diagnostics: A review. Expert Rev. Mol. Diagn. 2019, 19, 71–81. [Google Scholar] [CrossRef]
- König, K.; Gemeinhardt, A.; Sandoghdar, V. Interferenz von Licht macht einzelne unmarkierte Proteine sichtbar. Biospektrum 2019, 25, 732–736. [Google Scholar] [CrossRef] [Green Version]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards Lateral Flow Quantitative Assays: Detection Approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [Green Version]
- Shan, B.; Pu, Y.; Chen, Y.; Liao, M.; Li, M. Novel SERS labels: Rational design, functional integration and biomedical applications. Coord. Chem. Rev. 2018, 371, 11–37. [Google Scholar] [CrossRef]
- Ciminelli, C.; Campanella, C.M.; Dell’Olio, F.; Campanella, C.E.; Armenise, M.N. Label-free optical resonant sensors for biochemical applications. Prog. Quantum Electron. 2013, 37, 51–107. [Google Scholar] [CrossRef]
- Ma, F.; Li, C.-C.; Zhang, C.-Y. Development of quantum dot-based biosensors: Principles and applications. J. Mater. Chem. B 2018, 6, 6173–6190. [Google Scholar] [CrossRef]
- Piriya, V.S.A.; Joseph, P.; Daniel, S.C.G.K.; Lakshmanan, S.; Kinoshita, T.; Muthusamy, S. Colorimetric sensors for rapid detection of various analytes. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 78, 1231–1245. [Google Scholar] [CrossRef]
- Yang, H.; Gijs, M.A.M. Micro-optics for microfluidic analytical applications. Chem. Soc. Rev. 2018, 47, 1391–1458. [Google Scholar] [CrossRef]
- Wang, P.; Bo, L.; Semenova, Y.; Farrell, G.; Brambilla, G. Optical Microfibre Based Photonic Components and Their Applications in Label-Free Biosensing. Biosensors 2015, 5, 471–499. [Google Scholar] [CrossRef] [Green Version]
- Sinibaldi, A. Cancer Biomarker Detection With Photonic Crystals-Based Biosensors: An Overview. J. Lightwave Technol. 2021, 39, 3871–3881. [Google Scholar] [CrossRef]
- Cabral, A.D.; Radu, T.B.; De Araujo, E.D.; Gunning, P.T. Optical chemosensors for the detection of proximally phosphorylated peptides and proteins. RSC Chem. Biol. 2021, 2, 815–829. [Google Scholar] [CrossRef]
- Li, J.; Si, Y.; Lee, H.J. Recent Research Trend of Biosensors for Colorectal Cancer Specific Protein Biomarkers. Appl. Chem. Eng. 2021, 32, 253–259. [Google Scholar] [CrossRef]
- Das, P.; Sedighi, A.; Krull, U.J. Cancer biomarker determination by resonance energy transfer using functional fluorescent nanoprobes. Anal. Chim. Acta 2018, 1041, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Imas, J.J.; Ruiz Zamarreño, C.; Zubiate, P.; Sanchez-Martín, L.; Campión, J.; Matías, I.R. Optical Biosensors for the Detection of Rheumatoid Arthritis (RA) Biomarkers: A Comprehensive Review. Sensors 2020, 20, 6289. [Google Scholar] [CrossRef] [PubMed]
- Rezabakhsh, A.; Rahbarghazi, R.; Fathi, F. Surface plasmon resonance biosensors for detection of Alzheimer’s biomarkers; an effective step in early and accurate diagnosis. Biosens. Bioelectron. 2020, 167, 112511. [Google Scholar] [CrossRef] [PubMed]
- Ameri, M.; Shabaninejad, Z.; Movahedpour, A.; Sahebkar, A.; Mohammadi, S.; Hosseindoost, S.; Ebrahimi, M.S.; Savardashtaki, A.; Karimipour, M.; Mirzaei, H. Biosensors for detection of Tau protein as an Alzheimer’s disease marker. Int. J. Biol. Macromol. 2020, 162, 1100–1108. [Google Scholar] [CrossRef]
- Sohrabi, H.; kholafazad Kordasht, H.; Pashazadeh-Panahi, P.; Nezhad-Mokhtari, P.; Hashemzaei, M.; Majidi, M.R.; Mosafer, J.; Oroojalian, F.; Mokhtarzadeh, A.; De la Guardia, M. Recent advances of electrochemical and optical biosensors for detection of C-reactive protein as a major inflammatory biomarker. Microchem. J. 2020, 158, 105287. [Google Scholar] [CrossRef]
- Seok, J.S.; Ju, H. Plasmonic Optical Biosensors for Detecting C-Reactive Protein: A Review. Micromachines 2020, 11, 895. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2019, 124–125, 205–215. [Google Scholar] [CrossRef]
- Obermaier, C.; Griebel, A.; Westermeier, R. Principles of protein labeling techniques. Methods Mol. Biol. 2015, 1295, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Gauglitz, G.; Proll, G. Strategies for label-free optical detection. Adv. Biochem. Eng. Biotechnol. 2008, 109, 395–432. [Google Scholar] [CrossRef]
- Gauglitz, G. Critical assessment of relevant methods in the field of biosensors with direct optical detection based on fibers and waveguides using plasmonic, resonance, and interference effects. Anal. Bioanal. Chem. 2020, 412, 3317–3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäntele, W.; Deniz, E. UV-VIS absorption spectroscopy: Lambert-Beer reloaded. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 173, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Beaven, G.H.; Holiday, E.R. Ultraviolet Absorption Spectra of Proteins and Amino Acids. Adv. Protein Chem. 1952, 7, 319–386. [Google Scholar] [CrossRef] [PubMed]
- Stoscheck, C.M. Quantitation of protein. In Guide to Protein Purification; Elsevier: Amsterdam, The Netherlands, 1990; pp. 50–68. ISBN 9780121820831. [Google Scholar]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Demchenko, A.P. Ultraviolet Spectroscopy of Proteins; Springer: Berlin, Germany, 1986; ISBN 978-3-642-70849-7. [Google Scholar]
- Kirschenbaum, D.M. Molar absorptivity and A-1 per cent-1 cm values for proteins at selected wavelengths of the ultraviolet and visible region. VII. Int. J. Pept. Protein Res. 1973, 5, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, A.R.; Saidel, L.; Mosovich, E. The ultraviolet absorption spectra of proteins. J. Biol. Chem. 1951, 193, 397–404. [Google Scholar] [CrossRef]
- Scopes, R.K. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 1974, 59, 277–282. [Google Scholar] [CrossRef]
- Kirschenbaum, D.M. Molar absorptivity and A-1 per cent-1cm values for proteins at selected wavelengths of the ultraviolet and visible region. VI. Int. J. Pept. Protein Res. 1972, 4, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Kirschenbaum, D.M. Molar absorptivity and A 1 1cm values for proteins at selected wavelengths of the ultraviolet and visible region. II. Int. J. Protein Res. 1971, 3, 157–164. [Google Scholar] [CrossRef]
- Kirschenbaum, D.M. Molar absorptivity and A 1 1cm values for proteins at selected wavelengths of the ultraviolet and visible region. III. Int. J. Protein Res. 1971, 3, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Kirschenbaum, D.M. Molar absorptivity and 1 1cm values for proteins at selected wavelengths of the ultraviolet and visible region. IV. Int. J. Protein Res. 1971, 3, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Eftink, M.R. Intrinsic Fluorescence of Proteins. Top. Fluoresc. Spectrosc. 2000, 6, 1–15. [Google Scholar] [CrossRef]
- Longworth, J.W. Intrinsic Fluorescence of Proteins. In Time-Resolved Fluorescence Spectroscopy in Biochemistry and Biology; Cundall, R.B., Dale, R.E., Eds.; Plenum Press: New York, NY, USA, 1983; pp. 651–725. ISBN 978-1-4757-1636-8. [Google Scholar]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Tatulian, S.A. FTIR Analysis of Proteins and Protein-Membrane Interactions. Methods Mol. Biol. 2019, 2003, 281–325. [Google Scholar] [CrossRef] [PubMed]
- Kratz, C.; Furchner, A.; Sun, G.; Rappich, J.; Hinrichs, K. Sensing and structure analysis by in situ IR spectroscopy: From mL flow cells to microfluidic applications. J. Phys. Condens. Matter 2020, 32, 393002. [Google Scholar] [CrossRef] [PubMed]
- Benevides, J.M.; Overman, S.A.; Thomas, G.J. Raman spectroscopy of proteins. Curr. Protoc. Protein Sci. 2004. [Google Scholar] [CrossRef] [PubMed]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Kuhar, N.; Sil, S.; Umapathy, S. Potential of Raman spectroscopic techniques to study proteins. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 258, 119712. [Google Scholar] [CrossRef] [PubMed]
- Tycova, A.; Prikryl, J.; Foret, F. Recent strategies toward microfluidic-based surface-enhanced Raman spectroscopy. Electrophoresis 2017, 38, 1977–1987. [Google Scholar] [CrossRef] [PubMed]
- Bates, K.E.; Lu, H. Optics-Integrated Microfluidic Platforms for Biomolecular Analyses. Biophys. J. 2016, 110, 1684–1697. [Google Scholar] [CrossRef] [Green Version]
- Whitaker, J.R.; Granum, P.E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal. Biochem. 1980, 109, 156–159. [Google Scholar] [CrossRef]
- Warburg, O.; Christian, W. Isolation and crystallization of enolase. Biochem. Z. 1942, 310, 384–421. [Google Scholar]
- Anthis, N.J.; Clore, G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013, 22, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Swinney, K.; Bornhop, D.J. Detection in capillary electrophoresis. Electrophoresis 2000, 21, 1239–1250. [Google Scholar] [CrossRef]
- Novo, P.; Janasek, D. Current advances and challenges in microfluidic free-flow electrophoresis-A critical review. Anal. Chim. Acta 2017, 991, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Gomez, F.A. Microfluidics in protein chromatography. Methods Mol. Biol. 2011, 681, 137–150. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, I.; Ackermann, T.N.; Muñoz-Berbel, X.; Llobera, A. Photonic Lab-on-a-Chip: Integration of Optical Spectroscopy in Microfluidic Systems. Anal. Chem. 2016, 88, 6630–6637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minas, G.; Wolffenbuttel, R.F.; Correia, J.H. MCM-based microlaboratory for simultaneous measurement of several biochemical parameters by spectrophotometry. Biomed. Microdevices 2010, 12, 727–736. [Google Scholar] [CrossRef]
- Minas, G.; Wolffenbuttel, R.F.; Correia, J.H. A lab-on-a-chip for spectrophotometric analysis of biological fluids. Lab. Chip 2005, 5, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Minas, G.; Wolffenbuttel, R.F.; Correia, J.H. An array of highly selective Fabry–Perot optical channels for biological fluid analysis by optical absorption using a white light source for illumination. J. Opt. A Pure Appl. Opt. 2006, 8, 272–278. [Google Scholar] [CrossRef] [Green Version]
- Salimi-Moosavi, H.; Jiang, Y.; Lester, L.; McKinnon, G.; Harrison, D.J. A multireflection cell for enhanced absorbance detection in microchip-based capillary electrophoresis devices. Electrophoresis 2000, 21, 1291–1299. [Google Scholar] [CrossRef]
- Blue, R.; Uttamchandani, D. Recent advances in optical fiber devices for microfluidics integration. J. Biophotonics 2016, 9, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Lee, C.S.; DeVoe, D.L. Integrated microfluidic UV absorbance detector with attomol-level sensitivity for BSA. Lab. Chip 2006, 6, 115–120. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Xiao, Y.; Yu, H.; Tong, L. Ultra-sensitive microfibre absorption detection in a microfluidic chip. Lab. Chip 2011, 11, 3720–3724. [Google Scholar] [CrossRef]
- Pereira, F.M.; Bernacka-Wojcik, I.; Ribeiro, R.S.R.; Lobato, M.T.; Fortunato, E.; Martins, R.; Igreja, R.; Jorge, P.A.S.; Águas, H.; Oliva, A.M.G. Hybrid Microfluidic Platform for Multifactorial Analysis Based on Electrical Impedance, Refractometry, Optical Absorption and Fluorescence. Micromachines 2016, 7, 181. [Google Scholar] [CrossRef] [Green Version]
- Libnau, F.O.; Kvalheim, O.M.; Christy, A.A.; Toft, J. Spectra of water in the near- and mid-infrared region. Vib. Spectrosc. 1994, 7, 243–254. [Google Scholar] [CrossRef]
- Ewing, A.V.; Clarke, G.S.; Kazarian, S.G. Attenuated total reflection-Fourier transform infrared spectroscopic imaging of pharmaceuticals in microfluidic devices. Biomicrofluidics 2016, 10, 024125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.L.A.; Gulati, S.; Edel, J.B.; De Mello, A.J.; Kazarian, S.G. Chemical imaging of microfluidic flows using ATR-FTIR spectroscopy. Lab. Chip 2009, 9, 2909–2913. [Google Scholar] [CrossRef] [PubMed]
- Perro, A.; Lebourdon, G.; Henry, S.; Lecomte, S.; Servant, L.; Marre, S. Combining microfluidics and FT-IR spectroscopy: Towards spatially resolved information on chemical processes. React. Chem. Eng. 2016, 1, 577–594. [Google Scholar] [CrossRef]
- Prim, D.; Crelier, S.; Segura, J.-M. Coupling of a microfluidic mixer to a Fourier-transform infrared spectrometer for protein-conformation studies. CHIMIA Int. J. Chem. 2011, 65, 815–816. [Google Scholar] [CrossRef]
- López-Lorente, Á.I.; Mizaikoff, B. Mid-infrared spectroscopy for protein analysis: Potential and challenges. Anal. Bioanal. Chem. 2016, 408, 2875–2889. [Google Scholar] [CrossRef] [PubMed]
- Ataka, K.; Stripp, S.T.; Heberle, J. Surface-enhanced infrared absorption spectroscopy (SEIRAS) to probe monolayers of membrane proteins. Biochim. Biophys. Acta 2013, 1828, 2283–2293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.L.; Wang, L.; Zonderman, J.; Rouse, J.C.; Kim, H.-Y. Automated, High-Throughput Infrared Spectroscopy for Secondary Structure Analysis of Protein Biopharmaceuticals. J. Pharm. Sci. 2020, 109, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Etezadi, D.; Warner, J.B., IV; Ruggeri, F.S.; Dietler, G.; Lashuel, H.A.; Altug, H. Nanoplasmonic mid-infrared biosensor for in vitro protein secondary structure detection. Light Sci. Appl. 2017, 6, e17029. [Google Scholar] [CrossRef] [PubMed]
- Kraiem, H.; Zouari, F.; Abderrazek, R.B.; Manon, Y.; Ayeb, M.E.; Fillaudeau, L.; Bedoui, J.; Bouhaouala-Zahar, B. Two-Dimensional Isoelectric Focusing OFFGEL, Micro-Fluidic Lab-on-Chip Electrophoresis and FTIR for Assessment of Long-Term Stability of rhG-CSF Formulation. IEEE Trans. Nanobioscience 2017, 16, 694–702. [Google Scholar] [CrossRef]
- Choi, S.; Goryll, M.; Sin, L.Y.M.; Wong, P.K.; Chae, J. Microfluidic-based biosensors toward point-of-care detection of nucleic acids and proteins. Microfluid. Nanofluidics 2011, 10, 231–247. [Google Scholar] [CrossRef]
- Wagner, C.; Buchegger, W.; Vellekoop, M.; Kraft, M.; Lendl, B. Time-resolved mid-IR spectroscopy of (bio)chemical reactions in solution utilizing a new generation of continuous-flow micro-mixers. Anal. Bioanal. Chem. 2011, 400, 2487–2497. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, C.B. Microfluidic dissociation and clearance of Alzheimer’s beta-amyloid aggregates. Biomaterials 2010, 31, 6789–6795. [Google Scholar] [CrossRef]
- Schleeger, M.; Wagner, C.; Vellekoop, M.J.; Lendl, B.; Heberle, J. Time-resolved flow-flash FT-IR difference spectroscopy: The kinetics of CO photodissociation from myoglobin revisited. Anal. Bioanal. Chem. 2009, 394, 1869–1877. [Google Scholar] [CrossRef] [Green Version]
- Seiça, A.F.S.; Iqbal, M.H.; Carvalho, A.; Choe, J.-Y.; Boulmedais, F.; Hellwig, P. Study of Membrane Protein Monolayers Using Surface-Enhanced Infrared Absorption Spectroscopy (SEIRAS): Critical Dependence of Nanostructured Gold Surface Morphology. ACS Sens. 2021, 6, 2875–2882. [Google Scholar] [CrossRef]
- Ataka, K.; Heberle, J. Use of surface enhanced infrared absorption spectroscopy (SEIRA) to probe the functionality of a protein monolayer. Biopolymers 2006, 82, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Fallah, M.A.; Hauser, K. Immobilization approaches can affect protein dynamics: A surface-enhanced infrared spectroscopic study on lipid-protein interactions. Biomater. Sci. 2019, 7, 3204–3212. [Google Scholar] [CrossRef] [Green Version]
- Kratz, C.; Furchner, A.; Oates, T.W.H.; Janasek, D.; Hinrichs, K. Nanoliter Sensing for Infrared Bioanalytics. ACS Sens. 2018, 3, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Jameson, D.M.; Croney, J.C.; Moens, P.D. Fluorescence: Basic concepts, practical aspects, and some anecdotes. Biophotonics 2003, 360, 1–43. [Google Scholar] [CrossRef]
- Götz, S.; Karst, U. Recent developments in optical detection methods for microchip separations. Anal. Bioanal. Chem. 2007, 387, 183–192. [Google Scholar] [CrossRef]
- Schulze, P.; Belder, D. Label-free fluorescence detection in capillary and microchip electrophoresis. Anal. Bioanal. Chem. 2009, 393, 515–525. [Google Scholar] [CrossRef]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Baaske, P.; Wienken, C.J.; Reineck, P.; Duhr, S.; Braun, D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed. Engl. 2010, 49, 2238–2241. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.G.; Wanner, R.; Johnson, C.M.; Breitsprecher, D.; Winter, G.; Duhr, S.; Baaske, P.; Ferguson, N. Novel microscale approaches for easy, rapid determination of protein stability in academic and commercial settings. Biochim. Biophys. Acta 2014, 1844, 2241–2250. [Google Scholar] [CrossRef] [Green Version]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, S.A.I.; Dijkman, P.M.; Lea, W.A.; Van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef]
- Seidel, S.A.I.; Wienken, C.J.; Geissler, S.; Jerabek-Willemsen, M.; Duhr, S.; Reiter, A.; Trauner, D.; Braun, D.; Baaske, P. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew. Chem. Int. Ed. Engl. 2012, 51, 10656–10659. [Google Scholar] [CrossRef]
- Tsuji, T.; Matsumoto, Y.; Kugimiya, R.; Doi, K.; Kawano, S. Separation of Nano- and Microparticle Flows Using Thermophoresis in Branched Microfluidic Channels. Micromachines 2019, 10, 321. [Google Scholar] [CrossRef] [Green Version]
- Strutz, W. Exploring Protein Stability by NanoDSF. Biophys. J. 2016, 110, 393a. [Google Scholar] [CrossRef] [Green Version]
- Fohlerova, Z.; Zhu, H.; Hubalek, J.; Ni, S.; Yobas, L.; Podesva, P.; Otahal, A.; Neuzil, P. Rapid Characterization of Biomolecules’ Thermal Stability in a Segmented Flow-Through Optofluidic Microsystem. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Tripathi, A. Measurements of label free protein concentration and conformational changes using a microfluidic UV-LED method. Biotechnol. Prog. 2007, 23, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Challa, P.K.; Peter, Q.; Wright, M.A.; Zhang, Y.; Saar, K.L.; Carozza, J.A.; Benesch, J.L.P.; Knowles, T.P.J. Real-Time Intrinsic Fluorescence Visualization and Sizing of Proteins and Protein Complexes in Microfluidic Devices. Anal. Chem. 2018, 90, 3849–3855. [Google Scholar] [CrossRef] [PubMed]
- Sitkov, N.; Zimina, T.; Kolobov, A.; Karasev, V.; Romanov, A.; Luchinin, V.; Kaplun, D. Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems. Micromachines 2021, 12, 691. [Google Scholar] [CrossRef] [PubMed]
- Schulze, P.; Ludwig, M.; Kohler, F.; Belder, D. Deep UV laser-induced fluorescence detection of unlabeled drugs and proteins in microchip electrophoresis. Anal. Chem. 2005, 77, 1325–1329. [Google Scholar] [CrossRef]
- Poehler, E.; Herzog, C.; Lotter, C.; Pfeiffer, S.A.; Aigner, D.; Mayr, T.; Nagl, S. Label-free microfluidic free-flow isoelectric focusing, pH gradient sensing and near real-time isoelectric point determination of biomolecules and blood plasma fractions. Analyst 2015, 140, 7496–7502. [Google Scholar] [CrossRef] [Green Version]
- Saar, K.L.; Peter, Q.; Müller, T.; Challa, P.K.; Herling, T.W.; Knowles, T.P.J. Rapid two-dimensional characterisation of proteins in solution. Microsyst. Nanoeng. 2019, 5, 1–10. [Google Scholar] [CrossRef]
- Schulze, P.; Schüttpelz, M.; Sauer, M.; Belder, D. Two-photon excited fluorescence detection at 420 nm for label-free detection of small aromatics and proteins in microchip electrophoresis. Lab. Chip 2007, 7, 1841–1844. [Google Scholar] [CrossRef]
- Beyreiss, R.; Ohla, S.; Nagl, S.; Belder, D. Label-free analysis in chip electrophoresis applying deep UV fluorescence lifetime detection. Electrophoresis 2011, 32, 3108–3114. [Google Scholar] [CrossRef] [PubMed]
- Barulin, A.; Wenger, J. Ultraviolet Photostability Improvement for Autofluorescence Correlation Spectroscopy on Label-Free Proteins. J. Phys. Chem. Lett. 2020, 11, 2027–2035. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, Y.; He, C.; Yang, X.; Xie, Y.; Hu, X.; Chen, C.; Wang, L.; Pu, J.; Liao, F. Selective and sensitive homogenous assay of serum albumin with 1-anilinonaphthalene-8-sulphonate as a biosensor. Anal. Chim. Acta 2014, 829, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Barer, R.; Joseph, S. Refractometry of Living Cells. Quart. J. Microscop. Sci. 1954, 95, 399–423. [Google Scholar]
- Briend-Marshal, A.; Medaille, C.; Braun, J.P. Comparison of total protein measurement by biuret method and refractometry in canine and feline plasma. Rev. Méd. Vét. 2005, 156, 615–619. [Google Scholar]
- Anderle, H.; Weber, A. Rediscovery and Revival of Analytical Refractometry for Protein Determination: Recombining Simplicity With Accuracy in the Digital Era. J. Pharm. Sci. 2016, 105, 1097–1103. [Google Scholar] [CrossRef]
- Deelen, S.M.; Ollivett, T.L.; Haines, D.M.; Leslie, K.E. Evaluation of a Brix refractometer to estimate serum immunoglobulin G concentration in neonatal dairy calves. J. Dairy Sci. 2014, 97, 3838–3844. [Google Scholar] [CrossRef]
- Chigerwe, M.; Hagey, J.V. Refractometer assessment of colostral and serum IgG and milk total solids concentrations in dairy cattle. BMC Vet. Res. 2014, 10, 178. [Google Scholar] [CrossRef] [Green Version]
- Ziska, S.M.; Schumacher, J.; Duran, S.H.; Brock, K.V. Effects of serial harvest of plasma on total plasma protein and total immunoglobulin G concentrations in donor horses involved in a plasmapheresis program. Am. J. Vet. Res. 2012, 73, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Elsohaby, I.; McClure, J.T.; Keefe, G.P. Evaluation of digital and optical refractometers for assessing failure of transfer of passive immunity in dairy calves. J. Vet. Intern. Med. 2015, 29, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Tvedten, H.W.; Norén, A. Comparison of a Schmidt and Haensch refractometer and an Atago PAL-USG Cat refractometer for determination of urine specific gravity in dogs and cats. Vet. Clin. Pathol. 2014, 43, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Hunsaker, J.J.H.; Wyness, S.P.; Snow, T.M.; Genzen, J.R. Clinical performance evaluation of total protein measurement by digital refractometry and characterization of non-protein solute interferences. Pract. Lab. Med. 2016, 6, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsoulos, P.D.; Athanasiou, L.V.; Karatzia, M.A.; Giadinis, N.; Karatzias, H.; Boscos, C.; Polizopoulou, Z.S. Comparison of biuret and refractometry methods for the serum total proteins measurement in ruminants. Vet. Clin. Pathol. 2017, 46, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Krioukov, E.; Greve, J.; Otto, C. Performance of integrated optical microcavities for refractive index and fluorescence sensing. Sens. Actuators B Chem. 2003, 90, 58–67. [Google Scholar] [CrossRef]
- Niehusmann, J.; Vörckel, A.; Bolivar, P.H.; Wahlbrink, T.; Henschel, W.; Kurz, H. Ultrahigh-quality-factor silicon-on-insulator microring resonator. Opt. Lett. 2004, 29, 2861–2863. [Google Scholar] [CrossRef] [PubMed]
- Kippenberg, T.J.; Spillane, S.M.; Armani, D.K.; Vahala, K.J. Fabrication and coupling to planar high-Q silica disk microcavities. Appl. Phys. Lett. 2003, 83, 797–799. [Google Scholar] [CrossRef] [Green Version]
- Armani, D.K.; Kippenberg, T.J.; Spillane, S.M.; Vahala, K.J. Ultra-high-Q toroid microcavity on a chip. Nature 2003, 421, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Krioukov, E.; Klunder, D.J.W.; Driessen, A.; Greve, J.; Otto, C. Sensor based on an integrated optical microcavity. Opt. Lett. 2002, 27, 512–514. [Google Scholar] [CrossRef]
- Chao, C.Y.; Fung, W.; Guo, L.J. Polymer microring resonators for biochemical sensing applications. IEEE J. Sel. Top. Quant. Electron. 2006, 12, 134–142. [Google Scholar] [CrossRef]
- Heideman, R.G.; Lambeck, P.V. Remote opto-chemical sensing with extreme sensitivity: Design, fabrication and performance of a pigtailed integrated optical phase-modulated Mach–Zehnder interferometer system. Sens. Actuators B Chem. 1999, 61, 100–127. [Google Scholar] [CrossRef]
- Dumais, P.; Callender, C.L.; Noad, J.P.; Ledderhof, C.J. Integrated optical sensor using a liquid-core waveguide in a Mach-Zehnder interferometer. Opt. Express 2008, 16, 18164–18172. [Google Scholar] [CrossRef]
- Cusano, A.; Iadicicco, A.; Campopiano, S.; Giordano, M.; Cutolo, A. Thinned and micro-structured fibre Bragg gratings: Towards new all-fibre high-sensitivity chemical sensors. J. Opt. A Pure Appl. Opt. 2005, 7, 734–741. [Google Scholar] [CrossRef]
- Liang, W.; Huang, Y.; Xu, Y.; Lee, R.K.; Yariv, A. Highly sensitive fiber Bragg grating refractive index sensors. Appl. Phys. Lett. 2005, 86, 151122. [Google Scholar] [CrossRef]
- Domachuk, P.; Littler, I.C.M.; Cronin-Golomb, M.; Eggleton, B.J. Compact resonant integrated microfluidic refractometer. Appl. Phys. Lett. 2006, 88, 093513. [Google Scholar] [CrossRef]
- Duveneck, G.L.; Abel, A.P.; Bopp, M.A.; Kresbach, G.M.; Ehrat, M. Planar waveguides for ultra-high sensitivity of the analysis of nucleic acids. Anal. Chim. Acta 2002, 469, 49–61. [Google Scholar] [CrossRef]
- Yuen, P.K.; Fontaine, N.H.; Quesada, M.A.; Mazumder, P.; Bergman, R.; Mozdy, E.J. Self-referencing a single waveguide grating sensor in a micron-sized deep flow chamber for label-free biomolecular binding assays. Lab. Chip 2005, 5, 959–965. [Google Scholar] [CrossRef]
- Choi, C.J.; Cunningham, B.T. Single-step fabrication and characterization of photonic crystal biosensors with polymer microfluidic channels. Lab. Chip 2006, 6, 1373–1380. [Google Scholar] [CrossRef]
- Sidorov, A.I.; Ignatieva, L.A. 1D photonic crystal with defect for microfluidic applications in near IR and THz spectral ranges. Optik 2021, 245, 167685. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Spackova, B.; Wrobel, P.; Bockova, M.; Homola, J. Optical Biosensors Based on Plasmonic Nanostructures: A Review. Proc. IEEE 2016, 104, 2380–2408. [Google Scholar] [CrossRef]
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A comprehensive review on plasmonic-based biosensors used in viral diagnostics. Commun. Biol. 2021, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kretschmann, E. Die Bestimmung optischer Konstanten von Metallen durch Anregung von Oberflächenplasmaschwingungen. Z. Phys. 1971, 241, 313–324. [Google Scholar] [CrossRef]
- Kretschmann, E.; Raether, H. Notizen: Radiative Decay of Non Radiative Surface Plasmons Excited by Light. Z. Für Nat. A 1968, 23, 2135–2136. [Google Scholar] [CrossRef]
- Kashyap, R.; Nemova, G. Surface Plasmon Resonance-Based Fiber and Planar Waveguide Sensors. J. Sens. 2009, 2009, 1–9. [Google Scholar] [CrossRef]
- Reather, H. Surface Plasmons on Smooth and Rough Surfaces and on Gratings; Springer: Berlin, Germany, 1988; Volume 111, pp. 1–3. [Google Scholar]
- Hooper, I.R.; Sambles, J.R. Surface plasmon polaritons on narrow-ridged short-pitch metal gratings in the conical mount. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2003, 20, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Nemova, G.; Kashyap, R. A Compact Integrated Planar-Waveguide Refractive-Index Sensor Based on a Corrugated Metal Grating. J. Lightwave Technol. 2007, 25, 2244–2250. [Google Scholar] [CrossRef]
- Jun, L.; Nianqiang, W. Biosensors Based on Nanomaterials and Nanodevices; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781315216317. [Google Scholar]
- Boken, J.; Khurana, P.; Thatai, S.; Kumar, D.; Prasad, S. Plasmonic nanoparticles and their analytical applications: A review. Appl. Spectrosc. Rev. 2017, 52, 774–820. [Google Scholar] [CrossRef]
- Petryayeva, E.; Krull, U.J. Localized surface plasmon resonance: Nanostructures, bioassays and biosensing—A review. Anal. Chim. Acta 2011, 706, 8–24. [Google Scholar] [CrossRef]
- Nehl, C.L.; Hafner, J.H. Shape-dependent plasmon resonances of gold nanoparticles. J. Mater. Chem. 2008, 18, 2415–2419. [Google Scholar] [CrossRef] [Green Version]
- Jain, P.K.; Lee, K.S.; El-Sayed, I.H.; El-Sayed, M.A. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: Applications in biological imaging and biomedicine. J. Phys. Chem. B 2006, 110, 7238–7248. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Sun, W. Mie theory for light scattering by a spherical particle in an absorbing medium. Appl. Opt. 2001, 40, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Link, S.; El-Sayed, M.A. Optical properties and ultrafast dynamics of metallic nanocrystals. Annu. Rev. Phys. Chem. 2003, 54, 331–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Cushing, S.K.; Wu, N. Plasmon-enhanced optical sensors: A review. Analyst 2015, 140, 386–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byun, K.-M. Development of Nanostructured Plasmonic Substrates for Enhanced Optical Biosensing. J. Opt. Soc. Korea 2010, 14, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Cappi, G.; Spiga, F.M.; Moncada, Y.; Ferretti, A.; Beyeler, M.; Bianchessi, M.; Decosterd, L.; Buclin, T.; Guiducci, C. Label-free detection of tobramycin in serum by transmission-localized surface plasmon resonance. Anal. Chem. 2015, 87, 5278–5285. [Google Scholar] [CrossRef]
- Rothenhäusler, B.; Knoll, W. Surface–plasmon microscopy. Nature 1988, 332, 615–617. [Google Scholar] [CrossRef]
- Bocková, M.; Slabý, J.; Špringer, T.; Homola, J. Advances in Surface Plasmon Resonance Imaging and Microscopy and Their Biological Applications. Annu. Rev. Anal. Chem. 2019, 12, 151–176. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-enhanced Raman spectroscopy: Benefits, trade-offs and future developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.; Oh, S.; Kang, D.; Choi, Y. Protein Quantification and Imaging by Surface-Enhanced Raman Spectroscopy and Similarity Analysis. Adv. Sci. 2020, 7, 1903638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almehmadi, L.M.; Curley, S.M.; Tokranova, N.A.; Tenenbaum, S.A.; Lednev, I.K. Surface Enhanced Raman Spectroscopy for Single Molecule Protein Detection. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Li, Y.; Ling, L.; Xiang, X.; Han, X.; Zhao, B.; Guo, X. Label-Free and Highly Sensitive Detection of Native Proteins by Ag IANPs via Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 14325–14329. [Google Scholar] [CrossRef]
- Zheng, X.-S.; Jahn, I.J.; Weber, K.; Cialla-May, D.; Popp, J. Label-free SERS in biological and biomedical applications: Recent progress, current challenges and opportunities. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 197, 56–77. [Google Scholar] [CrossRef]
- Oliverio, M.; Perotto, S.; Messina, G.C.; Lovato, L.; De Angelis, F. Chemical Functionalization of Plasmonic Surface Biosensors: A Tutorial Review on Issues, Strategies, and Costs. ACS Appl. Mater. Interfaces 2017, 9, 29394–29411. [Google Scholar] [CrossRef] [Green Version]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, F.; Galvan, D.D.; Jain, P.; Yu, Q. Multi-functional, thiophenol-based surface chemistry for surface-enhanced Raman spectroscopy. Chem. Commun. 2017, 53, 4550–4561. [Google Scholar] [CrossRef] [PubMed]
- Veerabaghu, P.P.; Ramasamy, P.; Sathe, V.; Ramasamy, A.; Mahalingam, U. Plasmonic silver nanospheres embedded ε-caprolactone/reduced graphite oxide nanolayers as active SERS substrates. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 101, 431–437. [Google Scholar] [CrossRef]
- Kamińska, A.; Winkler, K.; Kowalska, A.; Witkowska, E.; Szymborski, T.; Janeczek, A.; Waluk, J. SERS-based Immunoassay in a Microfluidic System for the Multiplexed Recognition of Interleukins from Blood Plasma: Towards Picogram Detection. Sci. Rep. 2017, 7, 10656. [Google Scholar] [CrossRef] [PubMed]
- Gallo, V.; Lai, A.; Pasquo, A.; Almaviva, S.; Iacobelli, S.; Persichetti, L.; Capellini, G.; Antonini, G. Surface-enhanced Raman scattering (SERS)-based immunosystem for ultrasensitive detection of the 90K biomarker. Anal. Bioanal. Chem. 2020, 412, 7659–7667. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, A.R.; Cannistraro, S. SERS detection of thrombin by protein recognition using functionalized gold nanoparticles. Nanomedicine 2007, 3, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Lin, Y.; Zheng, M.; Lin, Y.; Lin, K.; Xie, S.; Yu, Y.; Lin, J. Label-free determination of liver cancer stages using surface-enhanced Raman scattering coupled with preferential adsorption of hydroxyapatite microspheres. Anal. Methods 2021, 13, 3885–3893. [Google Scholar] [CrossRef] [PubMed]
- Hassanain, W.A.; Theiss, F.L.; Izake, E.L. Label-free identification of Erythropoietin isoforms by surface enhanced Raman spectroscopy. Talanta 2022, 236, 122879. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, J.; Tang, S.; Zhao, X.; Zheng, M.; Gong, W.; Xie, S.; Gao, S.; Yu, Y.; Lin, J. Label-free diagnosis of breast cancer based on serum protein purification assisted surface-enhanced Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 263, 120234. [Google Scholar] [CrossRef]
- Singh, S.; Agarwal, A.; Avni, A.; Mukhopadhyay, S. Ultrasensitive Characterization of the Prion Protein by Surface-Enhanced Raman Scattering: Selective Enhancement via Electrostatic Tethering of the Intrinsically Disordered Domain with Functionalized Silver Nanoparticles. J. Phys. Chem. Lett. 2021, 12, 3187–3194. [Google Scholar] [CrossRef]
- Ma, H.; Liu, S.; Liu, Y.; Zhu, J.; Han, X.X.; Ozaki, Y.; Zhao, B. In-situ fingerprinting phosphorylated proteins via surface-enhanced Raman spectroscopy: Single-site discrimination of Tau biomarkers in Alzheimer’s disease. Biosens. Bioelectron. 2021, 171, 112748. [Google Scholar] [CrossRef] [PubMed]
- Fălămaș, A.; Rotaru, H.; Hedeșiu, M. Surface-enhanced Raman spectroscopy (SERS) investigations of saliva for oral cancer diagnosis. Lasers Med. Sci. 2020, 35, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Hassanain, W.A.; Izake, E.L. Toward Label-Free SERS Detection of Proteins through Their Disulfide Bond Structure. SLAS Discov. 2020, 25, 87–94. [Google Scholar] [CrossRef]
- Reza, K.K.; Sina, A.A.I.; Wuethrich, A.; Grewal, Y.S.; Howard, C.B.; Korbie, D.; Trau, M. A SERS microfluidic platform for targeting multiple soluble immune checkpoints. Biosens. Bioelectron. 2019, 126, 178–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Cao, F.; Tian, Y.; Li, A.; Xu, W.; Chen, Q.; Shi, W.; Xu, S. Label-Free Detection of Multiplexed Metabolites at Single-Cell Level via a SERS-Microfluidic Droplet Platform. Anal. Chem. 2019, 91, 15484–15490. [Google Scholar] [CrossRef]
- TunÇ, İ.; Susapto, H.H. Label-Free Detection of Ovarian Cancer Antigen CA125 by Surface Enhanced Raman Scattering. J. Nanosci. Nanotechnol. 2020, 20, 1358–1365. [Google Scholar] [CrossRef]
- Muhammad, M.; Shao, C.; Huang, Q. Label-free SERS diagnostics of radiation-induced injury via detecting the biomarker Raman signal in the serum and urine bio-samples based on Au-NPs array substrates. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 223, 117282. [Google Scholar] [CrossRef]
- Wang, J.; Lin, D.; Lin, J.; Yu, Y.; Huang, Z.; Chen, Y.; Lin, J.; Feng, S.; Li, B.; Liu, N.; et al. Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening. J. Biomed. Opt. 2014, 19, 087003. [Google Scholar] [CrossRef] [PubMed]
- Pecora, R. Doppler Shifts in Light Scattering from Pure Liquids and Polymer Solutions. J. Chem. Phys. 1964, 40, 1604–1614. [Google Scholar] [CrossRef]
- Foord, R.; Jakeman, E.; Oliver, C.J.; Pike, E.R.; Blagrove, R.J.; Wood, E.; Peacocke, A.R. Determination of diffusion coefficients of haemocyanin at low concentration by intensity fluctuation spectroscopy of scattered laser light. Nature 1970, 227, 242–245. [Google Scholar] [CrossRef]
- Pecora, R. (Ed.) Dynamic Light Scattering; Springer: Boston, MA, USA, 1985. [Google Scholar]
- Chastek, T.Q.; Beers, K.L.; Amis, E.J. Miniaturized dynamic light scattering instrumentation for use in microfluidic applications. Rev. Sci. Instrum. 2007, 78, 072201. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, P.; Li, W.; Xia, M.; Guo, W. Multi-angle Fiber DLS system Based on Microfluidics Technology. In Proceedings of the International Applied Computational Electromagnetics Society Symposium-China (ACES), Nanjing, China, 8–11 August 2019; pp. 1–2, ISBN 978-0-9960078-9-4. [Google Scholar]
- Gross, J.; Sayle, S.; Karow, A.R.; Bakowsky, U.; Garidel, P. Nanoparticle tracking analysis of particle size and concentration detection in suspensions of polymer and protein samples: Influence of experimental and data evaluation parameters. Eur. J. Pharm. Biopharm. 2016, 104, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Hole, P.; Sillence, K.; Hannell, C.; Maguire, C.M.; Roesslein, M.; Suarez, G.; Capracotta, S.; Magdolenova, Z.; Horev-Azaria, L.; Dybowska, A.; et al. Interlaboratory comparison of size measurements on nanoparticles using nanoparticle tracking analysis (NTA). J. Nanopart. Res. 2013, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Faez, S.; Carattino, A.; Mosk, A. PyNTA: An Open Source Software Application for Live Particle Tracking. Preprints 2019. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, X.; Zuo, Y.; Hu, X.; Shi, Y.; Liang, L.; Yang, Y. Optofluidics: The interaction between light and flowing liquids in integrated devices. Opto-Electron. Adv. 2019, 2, 11190007. [Google Scholar] [CrossRef]
- Yao, B.; Luo, G.; Wang, L.; Gao, Y.; Lei, G.; Ren, K.; Chen, L.; Wang, Y.; Hu, Y.; Qiu, Y. A microfluidic device using a green organic light emitting diode as an integrated excitation source. Lab. Chip 2005, 5, 1041–1047. [Google Scholar] [CrossRef]
- Novo, P.; Prazeres, D.M.F.; Chu, V.; Conde, J.P. Microspot-based ELISA in microfluidics: Chemiluminescence and colorimetry detection using integrated thin-film hydrogenated amorphous silicon photodiodes. Lab. Chip 2011, 11, 4063–4071. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shu, W.; Wang, Y.; Gong, Y.; Gong, C.; Chen, Q.; Tan, X.; Peng, G.-D.; Fan, X.; Rao, Y.-J. Turbidimetric inhibition immunoassay revisited to enhance its sensitivity via an optofluidic laser. Biosens. Bioelectron. 2019, 131, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, I.; Conejero-Muriel, M.; Ackermann, T.N.; Gavira, J.A.; Llobera, A. A multiple path photonic lab on a chip for parallel protein concentration measurements. Lab. Chip 2015, 15, 1133–1139. [Google Scholar] [CrossRef]
- Yang, H.; Cornaglia, M.; Gijs, M.A.M. Photonic nanojet array for fast detection of single nanoparticles in a flow. Nano Lett. 2015, 15, 1730–1735. [Google Scholar] [CrossRef] [PubMed]
- Ghenuche, P.; De Torres, J.; Ferrand, P.; Wenger, J. Multi-focus parallel detection of fluorescent molecules at picomolar concentration with photonic nanojets arrays. Appl. Phys. Lett. 2014, 105, 131102. [Google Scholar] [CrossRef] [Green Version]
- Mittal, V.; Devitt, G.; Nedeljkovic, M.; Carpenter, L.G.; Chong, H.M.H.; Wilkinson, J.S.; Mahajan, S.; Mashanovich, G.Z. Ge on Si waveguide mid-infrared absorption spectroscopy of proteins and their aggregates. Biomed. Opt. Express 2020, 11, 4714–4722. [Google Scholar] [CrossRef]
- Wang, C.; Xu, Y.; Wang, R.; Zhao, H.; Xiang, S.; Chen, L.; Qi, X. Fabrication and effect study of microfluidic SERS chip with integrated surface liquid core optical waveguide modified with nano gold. Microsyst. Technol. 2017, 23, 3059–3068. [Google Scholar] [CrossRef]
- Liang, L.; Jin, L.; Ran, Y.; Sun, L.-P.; Guan, B.-O. Fiber Light-Coupled Optofluidic Waveguide (FLOW) Immunosensor for Highly Sensitive Detection of p53 Protein. Anal. Chem. 2018, 90, 10851–10857. [Google Scholar] [CrossRef]
- Sulabh; Kumar, M. Slot Waveguide with Grating Based Cladding for Protein Detection. In ICOL-2019; Singh, K., Gupta, A.K., Khare, S., Dixit, N., Pant, K., Eds.; Springer: Singapore, 2021; pp. 495–498. ISBN 978-981-15-9258-4. [Google Scholar]
- Li, K.; Zhou, W.; Zeng, S. Optical Micro/Nanofiber-Based Localized Surface Plasmon Resonance Biosensors: Fiber Diameter Dependence. Sensors 2018, 18, 3295. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Goddard, N.J. A study of diffraction-based chitosan leaky waveguide (LW) biosensors. Analyst 2021, 146, 4964–4971. [Google Scholar] [CrossRef] [PubMed]
- Shivananju, B.N.; Prasad, V.; Chenrayan, G.A.; Misra, A.; Varma, M.M.; Asokan, S. Nanomaterials coated multiplexed fiber Bragg grating for multiparameter sensing. In Quantum Sensing and Nanophotonic Devices XII; Razeghi, M., Tournié, E., Brown, G.J., Eds.; SPIE OPTO: San Francisco, CA, USA, 2015; p. 937036. [Google Scholar]
- Guo, T.; Liu, F.; Liang, X.; Qiu, X.; Huang, Y.; Xie, C.; Xu, P.; Mao, W.; Guan, B.-O.; Albert, J. Highly sensitive detection of urinary protein variations using tilted fiber grating sensors with plasmonic nanocoatings. Biosens. Bioelectron. 2016, 78, 221–228. [Google Scholar] [CrossRef] [Green Version]
- Esposito, F.; Sansone, L.; Srivastava, A.; Baldini, F.; Campopiano, S.; Chiavaioli, F.; Giordano, M.; Giannetti, A.; Iadicicco, A. Long period grating in double cladding fiber coated with graphene oxide as high-performance optical platform for biosensing. Biosens. Bioelectron. 2021, 172, 112747. [Google Scholar] [CrossRef]
- Yang, C.-J.; Yan, H.; Tang, N.; Zou, Y.; Al-Hadeethi, Y.; Xu, X.; Dalir, H.; Chen, R.T. Ultra Sensitivity Silicon-Based Photonic Crystal Microcavity Biosensors for Plasma Protein Detection in Patients with Pancreatic Cancer. Micromachines 2020, 11, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Hou, X.; Si, J. Protein analysis by Mach-Zehnder interferometers with a hybrid plasmonic waveguide with nano-slots. Opt. Express 2017, 25, 31294–31308. [Google Scholar] [CrossRef]
- Tu, L.; Huang, L.; Wang, W. A novel micromachined Fabry-Perot interferometer integrating nano-holes and dielectrophoresis for enhanced biochemical sensing. Biosens. Bioelectron. 2019, 127, 19–24. [Google Scholar] [CrossRef]
- Chatzipetrou, M.; Gounaridis, L.; Tsekenis, G.; Dimadi, M.; Vestering-Stenger, R.; F Schreuder, E.; Trilling, A.; Besselink, G.; Scheres, L.; Van der Meer, A.; et al. A Miniature Bio-Photonics Companion Diagnostics Platform for Reliable Cancer Treatment Monitoring in Blood Fluids. Sensors 2021, 21, 2230. [Google Scholar] [CrossRef] [PubMed]
- Martens, D.; Ramirez-Priego, P.; Murib, M.S.; Elamin, A.A.; Gonzalez-Guerrero, A.B.; Stehr, M.; Jonas, F.; Anton, B.; Hlawatsch, N.; Soetaert, P.; et al. A low-cost integrated biosensing platform based on SiN nanophotonics for biomarker detection in urine. Anal. Methods 2018, 10, 3066–3073. [Google Scholar] [CrossRef] [Green Version]
- Stambaugh, A.; Parks, J.W.; Stott, M.A.; Meena, G.G.; Hawkins, A.R.; Schmidt, H. Optofluidic multiplex detection of single SARS-CoV-2 and influenza A antigens using a novel bright fluorescent probe assay. Proc. Natl. Acad. Sci. USA 2021, 118, e2103480118. [Google Scholar] [CrossRef]
- Meena, G.G.; Stambaugh, A.M.; Ganjalizadeh, V.; Stott, M.A.; Hawkins, A.R.; Schmidt, H. Ultrasensitive detection of SARS-CoV-2 RNA and antigen using single-molecule optofluidic chip. APL Photonics 2021, 6, 066101. [Google Scholar] [CrossRef]
- Gao, R.; Lu, D.; Guo, D.; Xin, X. Dual-optofluidic waveguide in-line fiber biosensor for real-time label-free detection of interferon-gamma with temperature compensation. Opt. Express 2020, 28, 10491–10504. [Google Scholar] [CrossRef]
- Yang, J.-M.; Yang, N.-Z.; Chen, C.-H.; Huang, C.-S. Gradient Waveguide Thickness Guided-Mode Resonance Biosensor. Sensors 2021, 21, 376. [Google Scholar] [CrossRef] [PubMed]
- Soni, V.; Chang, C.-W.; Xu, X.; Wang, C.; Yan, H.; D’Agati, M.; Tu, L.-W.; Chen, Q.Y.; Tian, H.; Chen, R.T. Portable Automatic Microring Resonator System Using a Subwavelength Grating Metamaterial Waveguide for High-Sensitivity Real-Time Optical-Biosensing Applications. IEEE Trans. Biomed. Eng. 2021, 68, 1894–1902. [Google Scholar] [CrossRef]
- Chang, N.; Zhai, J.; Liu, B.; Zhou, J.; Zeng, Z.; Zhao, X. Low cost 3D microfluidic chips for multiplex protein detection based on photonic crystal beads. Lab. Chip 2018, 18, 3638–3644. [Google Scholar] [CrossRef]
- Mohammed, M.-I.; Desmulliez, M.P.Y. Planar lens integrated capillary action microfluidic immunoassay device for the optical detection of troponin I. Biomicrofluidics 2013, 7, 064112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Li, T.; Li, Z.; Long, J.; Zhang, X. Optofluidic Tunable Lenses for In-Plane Light Manipulation. Micromachines 2018, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Chao, K.-S.; Lin, M.-S.; Yang, R.-J. An in-plane optofluidic microchip for focal point control. Lab. Chip 2013, 13, 3886–3892. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Qiu, G.; Cao, X.; Yue, Y.; Zhang, X.; Schmitt, J.; Wang, J. Self-aligned 3D microlenses in a chip fabricated with two-photon stereolithography for highly sensitive absorbance measurement. Lab. Chip 2020, 20, 2334–2342. [Google Scholar] [CrossRef] [PubMed]
- Dietvorst, J.; Goyvaerts, J.; Ackermann, T.N.; Alvarez, E.; Muñoz-Berbel, X.; Llobera, A. Microfluidic-controlled optical router for lab on a chip. Lab. Chip 2019, 19, 2081–2088. [Google Scholar] [CrossRef] [Green Version]
- Mishra, K.; Van den Ende, D.; Mugele, F. Recent Developments in Optofluidic Lens Technology. Micromachines 2016, 7, 102. [Google Scholar] [CrossRef]
- Darafsheh, A. Photonic nanojets and their applications. J. Phys. Photonics 2021, 3, 022001. [Google Scholar] [CrossRef]
- Patel, H.S.; Kushwaha, P.K.; Swami, M.K. Photonic nanojet assisted enhancement of Raman signal: Effect of refractive index contrast. J. Appl. Phys. 2018, 123, 023102. [Google Scholar] [CrossRef]
- Minin, I.V.; Geints, Y.E.; Zemlyanov, A.A.; Minin, O.V. Specular-reflection photonic nanojet: Physical basis and optical trapping application. Opt. Express 2020, 28, 22690–22704. [Google Scholar] [CrossRef] [PubMed]
- Surdo, S.; Duocastella, M.; Diaspro, A. Nanopatterning with Photonic Nanojets: Review and Perspectives in Biomedical Research. Micromachines 2021, 12, 256. [Google Scholar] [CrossRef] [PubMed]
- Ge, A.; Hu, L.; Fan, J.; Ge, M.; Wang, X.; Wang, S.; Feng, X.; Du, W.; Liu, B.-F. A low-cost microfluidic platform coupled with light emitting diode for optogenetic analysis of neuronal response in C. elegans. Talanta 2021, 223, 121646. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.-G.; Hu, S.; Peng, Y. Applications of fiber-optic biochemical sensor in microfluidic chips: A review. Biosens. Bioelectron. 2020, 166, 112447. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J. An Introduction to Optical Waveguides; John Wiley: Chichester, UK, 1981; ISBN 978-0471279693. [Google Scholar]
- Li, Z.; Xu, Y.; Fang, W.; Tong, L.; Zhang, L. Ultra-sensitive nanofiber fluorescence detection in a microfluidic chip. Sensors 2015, 15, 4890–4898. [Google Scholar] [CrossRef] [Green Version]
- Yadav, T.K.; Narayanaswamy, R.; Abu Bakar, M.H.; Kamil, Y.M.; Mahdi, M.A. Single mode tapered fiber-optic interferometer based refractive index sensor and its application to protein sensing. Opt. Express 2014, 22, 22802–22807. [Google Scholar] [CrossRef]
- Mukundan, H.; Kubicek, J.Z.; Holt, A.; Shively, J.E.; Martinez, J.S.; Grace, K.; Grace, W.K.; Swanson, B.I. Planar optical waveguide-based biosensor for the quantitative detection of tumor markers. Sens. Actuators B Chem. 2009, 138, 453–460. [Google Scholar] [CrossRef]
- Tong, X.; Tian, H.; Guo, Y.; Zhang, M.; Zhang, L. Dynamic Nanoparticle Trapping By Cascaded Nanophotonic Traps in a Silicon Slot Waveguide. IEEE J. Sel. Top. Quantum Electron. 2021, 27, 1–8. [Google Scholar] [CrossRef]
- Tong, L.; Gattass, R.R.; Ashcom, J.B.; He, S.; Lou, J.; Shen, M.; Maxwell, I.; Mazur, E. Subwavelength-diameter silica wires for low-loss optical wave guiding. Nature 2003, 426, 816–819. [Google Scholar] [CrossRef]
- Wongkaew, N. Nanofiber-integrated miniaturized systems: An intelligent platform for cancer diagnosis. Anal. Bioanal. Chem. 2019, 411, 4251–4264. [Google Scholar] [CrossRef]
- Gupta, R.; Goddard, N.J. Leaky waveguides (LWs) for chemical and biological sensing−A review and future perspective. Sens. Actuators B Chem. 2020, 322, 128628. [Google Scholar] [CrossRef]
- Zourob, M.; Goddard, N.J. Metal clad leaky waveguides for chemical and biosensing applications. Biosens. Bioelectron. 2005, 20, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Sypabekova, M.; Korganbayev, S.; González-Vila, Á.; Caucheteur, C.; Shaimerdenova, M.; Ayupova, T.; Bekmurzayeva, A.; Vangelista, L.; Tosi, D. Functionalized etched tilted fiber Bragg grating aptasensor for label-free protein detection. Biosens. Bioelectron. 2019, 146, 111765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Zhao, Y.; Zhou, T.; Wu, Q. Applications and developments of on-chip biochemical sensors based on optofluidic photonic crystal cavities. Lab. Chip 2017, 18, 57–74. [Google Scholar] [CrossRef]
- Maia, J.M.; Amorim, V.A.; Viveiros, D.; Marques, P.V.S. Femtosecond laser micromachining of an optofluidics-based monolithic whispering-gallery mode resonator coupled to a suspended waveguide. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Singh, R.; Singh, R.R.; Priye, V. Optical Biosensor Based on Guided Mode Grating Coupler. Front. Opt./Laser Sci. 2018, JTu2A.114. [Google Scholar] [CrossRef]
- Persichetti, G.; Testa, G.; Bernini, R. Optofluidic jet waveguide for laser-induced fluorescence spectroscopy. Opt. Lett. 2012, 37, 5115–5117. [Google Scholar] [CrossRef]
- Testa, G.; Persichetti, G.; Bernini, R. Optofluidic approaches for enhanced microsensor performances. Sensors 2014, 15, 465–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozcelik, D.; Parks, J.W.; Wall, T.A.; Stott, M.A.; Cai, H.; Parks, J.W.; Hawkins, A.R.; Schmidt, H. Optofluidic wavelength division multiplexing for single-virus detection. Proc. Natl. Acad. Sci. USA 2015, 112, 12933–12937. [Google Scholar] [CrossRef] [Green Version]
- Testa, G.; Huang, Y.; Zeni, L.; Sarro, P.M.; Bernini, R. Liquid Core ARROW Waveguides by Atomic Layer Deposition. IEEE Photonics Technol. Lett. 2010, 22, 616–618. [Google Scholar] [CrossRef]
- Testa, G.; Huang, Y.; Zeni, L.; Sarro, P.M.; Bernini, R. Hybrid Silicon-PDMS Optofluidic ARROW Waveguide. IEEE Photonics Technol. Lett. 2012, 24, 1307–1309. [Google Scholar] [CrossRef]
- Schmidt, H.; Hawkins, A.R. The photonic integration of non-solid media using optofluidics. Nat. Photonics 2011, 5, 598–604. [Google Scholar] [CrossRef]
- Yin, D.; Lunt, E.J.; Barman, A.; Hawkins, A.R.; Schmidt, H. Microphotonic control of single molecule fluorescence correlation spectroscopy using planar optofluidics. Opt. Express 2007, 15, 7290–7295. [Google Scholar] [CrossRef]
- Soldano, L.B.; Pennings, E. Optical multi-mode interference devices based on self-imaging: Principles and applications. J. Lightwave Technol. 1995, 13, 615–627. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, D.; Yue, H.; Spiering, M.M.; Zhao, C.; Benkovic, S.J.; Huang, T.J. Dark-field illumination on zero-mode waveguide/microfluidic hybrid chip reveals T4 replisomal protein interactions. Nano Lett. 2014, 14, 1952–1960. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Baibakov, M.; Claude, J.-B.; Wenger, J. Surface passivation of zero-mode waveguide nanostructures: Benchmarking protocols and fluorescent labels. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Barulin, A.; Claude, J.-B.; Patra, S.; Bonod, N.; Wenger, J. Deep Ultraviolet Plasmonic Enhancement of Single Protein Autofluorescence in Zero-Mode Waveguides. Nano Lett. 2019, 19, 7434–7442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunte, A.A.; Gaikwad, A.N. Dielectric constant measurement of low loss liquids using stacked multi ring resonator. Sādhanā 2018, 43, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hengcharoen, T.; Eaiprasertsak, K.; Fuangfoong, M. Microwave Dielectric Measurement of Liquids by using Waveguide Plunger Technique. Procedia Eng. 2011, 8, 270–274. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Luzio, S.; Beutler, J.; Porch, A. Microwave noninvasive blood glucose monitoring sensor: Human clinical trial results. In Proceedings of the IEEE/MTT-S International Microwave Symposium-IMS 2017, Honololu, HI, USA, 4–9 June 2017; pp. 876–879, ISBN 978-1-5090-6360-4. [Google Scholar]
- Kordzadeh, A.; De Zanche, N. Permittivity measurement of liquids, powders, and suspensions using a parallel-plate cell. Concepts Magn. Reson. 2016, 46, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Murphy, E.K.; Mahara, A.; Khan, S.; Hyams, E.S.; Schned, A.R.; Pettus, J.; Halter, R.J. Comparative study of separation between ex vivo prostatic malignant and benign tissue using electrical impedance spectroscopy and electrical impedance tomography. Physiol. Meas. 2017, 38, 1242–1261. [Google Scholar] [CrossRef] [PubMed]
- Tobon Vasquez, J.A.; Scapaticci, R.; Turvani, G.; Bellizzi, G.; Rodriguez-Duarte, D.O.; Joachimowicz, N.; Duchêne, B.; Tedeschi, E.; Casu, M.R.; Crocco, L.; et al. A Prototype Microwave System for 3D Brain Stroke Imaging. Sensors 2020, 20, 2607. [Google Scholar] [CrossRef]
- Tang, L.; Chang, S.J.; Chen, C.-J.; Liu, J.-T. Non-Invasive Blood Glucose Monitoring Technology: A Review. Sensors 2020, 20, 6925. [Google Scholar] [CrossRef]
- Dean, D.A.; Ramanathan, T.; Machado, D.; Sundararajan, R. Electrical Impedance Spectroscopy Study of Biological Tissues. J. Electrostat. 2008, 66, 165–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, K.S.; Cole, R.H. Dispersion and Absorption in Dielectrics I. Alternating Current Characteristics. J. Chem. Phys. 1941, 9, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Wolf, M.; Gulich, R.; Lunkenheimer, P.; Loidl, A. Relaxation dynamics of a protein solution investigated by dielectric spectroscopy. Biochim. Biophys. Acta 2012, 1824, 723–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibi, F.; Villain, M.; Guillaume, C.; Sorli, B.; Gontard, N. A Review: Origins of the Dielectric Properties of Proteins and Potential Development as Bio-Sensors. Sensors 2016, 16, 1232. [Google Scholar] [CrossRef] [Green Version]
- Maji, D.; De La Fuente, M.; Kucukal, E.; Sekhon, U.D.S.; Schmaier, A.H.; Sen Gupta, A.; Gurkan, U.A.; Nieman, M.T.; Stavrou, E.X.; Mohseni, P.; et al. Assessment of whole blood coagulation with a microfluidic dielectric sensor. J. Thromb. Haemost. 2018, 16, 2050–2056. [Google Scholar] [CrossRef] [Green Version]
- Moazeni, M.; Karimzadeh, F.; Kermanpur, A. Peptide modified paper based impedimetric immunoassay with nanocomposite electrodes as a point-of-care testing of Alpha-fetoprotein in human serum. Biosens. Bioelectron. 2018, 117, 748–757. [Google Scholar] [CrossRef]
- Basey-Fisher, T.H.; Guerra, N.; Triulzi, C.; Gregory, A.; Hanham, S.M.; Stevens, M.M.; Maier, S.A.; Klein, N. Microwaving blood as a non-destructive technique for haemoglobin measurements on microlitre samples. Adv. Healthc. Mater. 2014, 3, 536–542. [Google Scholar] [CrossRef]
- Alahnomi, R.A.; Zakaria, Z.; Ruslan, E.; Ab Rashid, S.R.; Mohd Bahar, A.A.; Shaaban, A. Microwave bio-sensor based on symmetrical split ring resonator with spurline filters for therapeutic goods detection. PLoS ONE 2017, 12, e0185122. [Google Scholar] [CrossRef] [Green Version]
- Fok, M.; Bashir, M.; Fraser, H.; Strouther, N.; Mason, A. A novel microwave sensor to detect specific biomarkers in human cerebrospinal fluid and their relationship to cellular ischemia during thoracoabdominal aortic aneurysm repair. J. Med. Syst. 2015, 39, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Lisdat, F.; Schäfer, D. The use of electrochemical impedance spectroscopy for biosensing. Anal. Bioanal. Chem. 2008, 391, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-B.; Zhang, H.-D.; Xu, S.-P.; Gan, T.; Huang, K.-J.; Liu, Y.-M. A sensitive and label-free electrochemical impedance biosensor for protein detection based on terminal protection of small molecule-linked DNA. Sens. Actuators B Chem. 2014, 194, 478–483. [Google Scholar] [CrossRef]
- Sharma, R.; Deacon, S.E.; Nowak, D.; George, S.E.; Szymonik, M.P.; Tang, A.A.S.; Tomlinson, D.C.; Davies, A.G.; McPherson, M.J.; Wälti, C. Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity. Biosens. Bioelectron. 2016, 80, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, J.; Mindrinos, M.N.; Davis, R.W.; Javanmard, M. Digital microfluidic assay for protein detection. Proc. Natl. Acad. Sci. USA 2014, 111, 2110–2115. [Google Scholar] [CrossRef] [Green Version]
- Santos, D.R.; Soares, R.R.G.; Pinto, I.F.; Caneira, C.R.F.; Pinto, R.M.R.; Chu, V.; Conde, J.P. Label-Free Detection of Biomolecules in Microfluidic Systems Using On-Chip UV and Impedimetric Sensors. IEEE Sens. J. 2019, 19, 7803–7812. [Google Scholar] [CrossRef]
- Stelson, A.C.; Liu, M.; Little, C.A.E.; Long, C.J.; Orloff, N.D.; Stephanopoulos, N.; Booth, J.C. Label-free detection of conformational changes in switchable DNA nanostructures with microwave microfluidics. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Jantaree, P.; Bakhchova, L.; Steinmann, U.; Naumann, M. From 3D Back to 2D Monolayer Stomach Organoids-on-a-Chip. Trends Biotechnol. 2021, 39, 745–748. [Google Scholar] [CrossRef]
- De Oliveira, G.C.M.; De Souza Carvalho, J.H.; Brazaca, L.C.; Vieira, N.C.S.; Janegitz, B.C. Flexible platinum electrodes as electrochemical sensor and immunosensor for Parkinson’s disease biomarkers. Biosens. Bioelectron. 2020, 152, 112016. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Primiceri, E.; Montanaro, A.; De Feo, F.; Leone, L.; Rinaldi, R.; Maruccio, G. On-chip screening for prostate cancer: An EIS microfluidic platform for contemporary detection of free and total PSA. Analyst 2013, 138, 5404–5410. [Google Scholar] [CrossRef] [PubMed]
- Mihailescu, C.M.; Stan, D.; Savin, M.; Moldovan, C.A.; Dinulescu, S.; Radulescu, C.H.; Firtat, B.; Muscalu, G.; Brasoveanu, C.; Ion, M.; et al. Platform with biomimetic electrochemical sensors for adiponectin and leptin detection in human serum. Talanta 2020, 210, 120643. [Google Scholar] [CrossRef] [PubMed]
- Vasantham, S.; Alhans, R.; Singhal, C.; Nagabooshanam, S.; Nissar, S.; Basu, T.; Ray, S.C.; Wadhwa, S.; Narang, J.; Mathur, A. Paper based point of care immunosensor for the impedimetric detection of cardiac troponin I biomarker. Biomed. Microdevices 2019, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, H.-G.; Krinke, D.; Seidel, D.; Lilienthal, K.; Schmidt, S.; Azendorf, R.; Fischer, M.; Mack, T.; Striggow, F.; Althaus, H.; et al. A novel 384-multiwell microelectrode array for the impedimetric monitoring of Tau protein induced neurodegenerative processes. Biosens. Bioelectron. 2017, 88, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.; Ozhukil Kollath, V.; Kundra, V.; Nguyen, M.D.; Debert, C.; Sen, A.; Karan, K.; Sanati-Nezhad, A. Polyethylenimine Modified Graphene-Oxide Electrochemical Immunosensor for the Detection of Glial Fibrillary Acidic Protein in Central Nervous System Injury. ACS Sens. 2018, 3, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.-M.; Kim, D.-M.; Kim, M.H.; Han, J.-Y.; Jung, D.-K.; Shim, Y.-B. A disposable amperometric dual-sensor for the detection of hemoglobin and glycated hemoglobin in a finger prick blood sample. Biosens. Bioelectron. 2017, 91, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Mondal, K.; Jiao, Y.; Oren, S.; Xu, Z.; Sharma, A.; Dong, L. Microfluidic Immuno-Biochip for Detection of Breast Cancer Biomarkers Using Hierarchical Composite of Porous Graphene and Titanium Dioxide Nanofibers. ACS Appl. Mater. Interfaces 2016, 8, 20570–20582. [Google Scholar] [CrossRef]
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-Based Impedimetric Sensor for Detecting Dengue Fever Biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
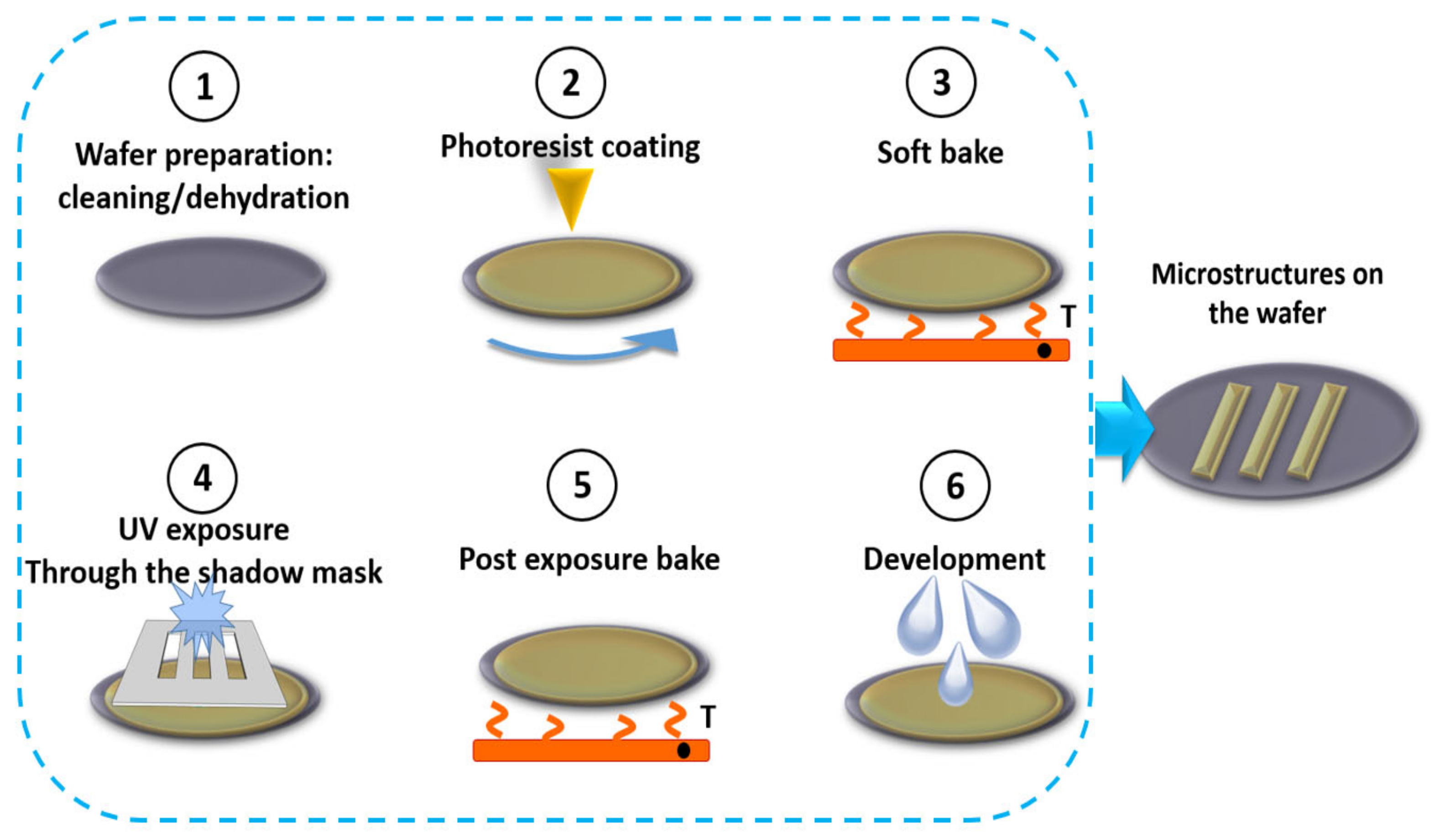
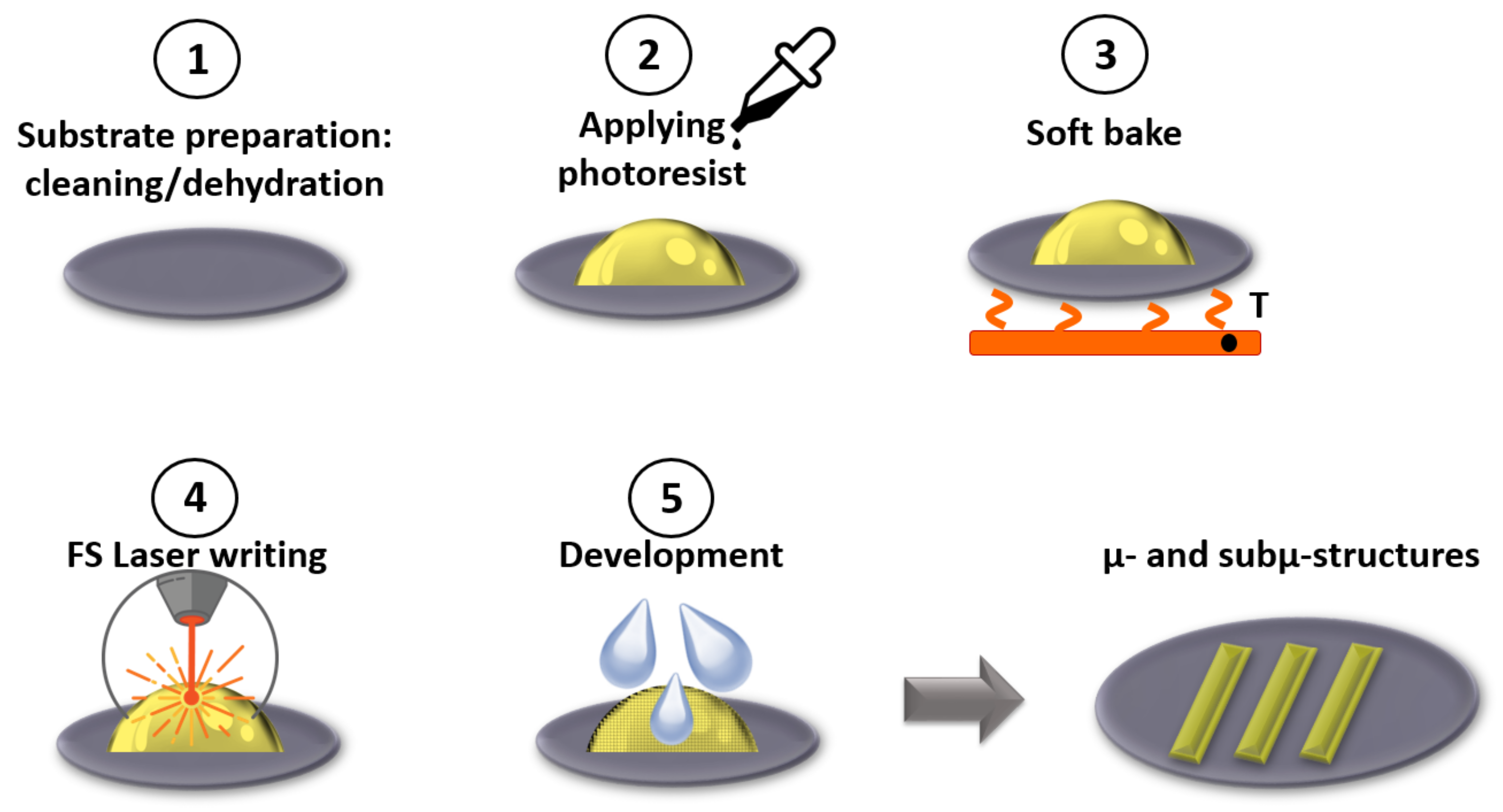




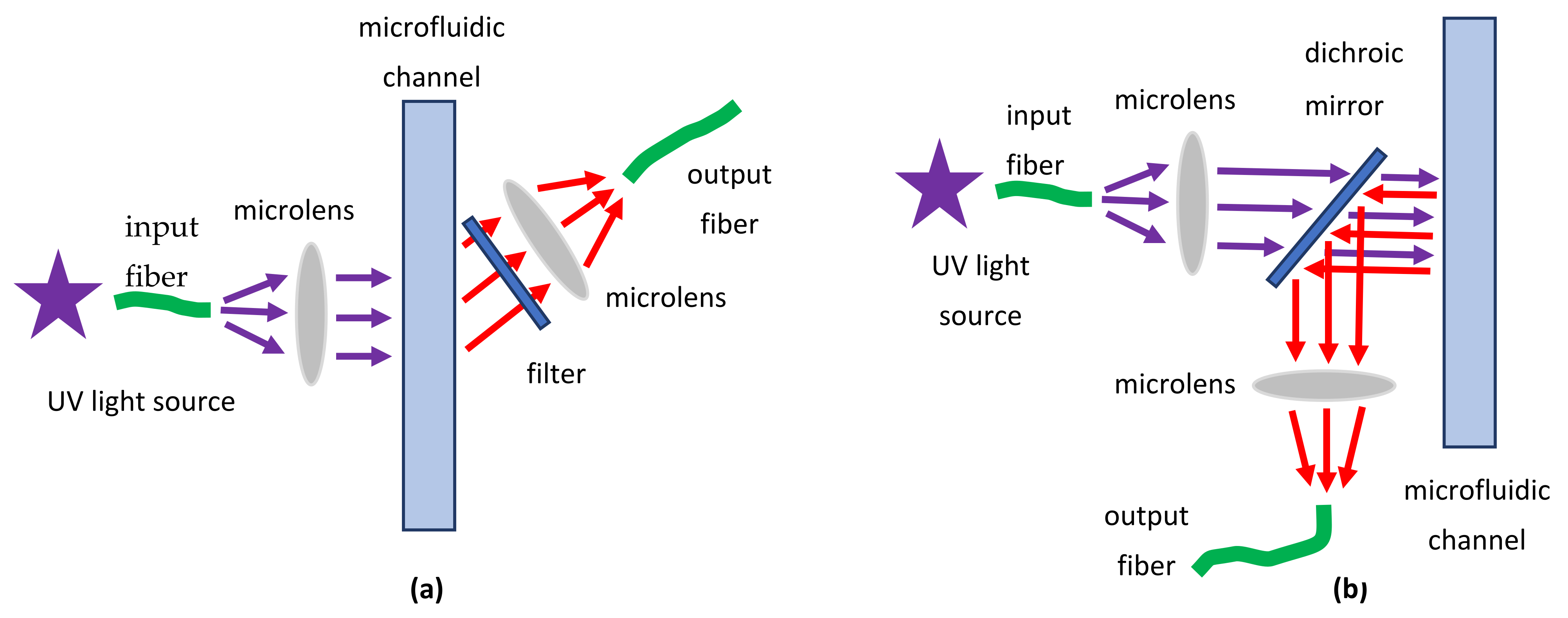







| Protein | Absorption Coefficient, 1/cm | |
|---|---|---|
| 280 nm | 205 nm | |
| Human serum albumin | 0.59 [153] | 32.7/30.5 (fraction II/IV) [154] |
| Bovine serum albumin | 0.66 [155] | 29.6 [153] |
| Human IgG (serum) | 1.33 [156] | |
| Human IgA (serum) | 1.34 [156] | |
| Human IgM | 1.33 [156] | |
| Human myoglobin | 1.74 [157] | |
| Insulin | 1.05 [158] | 38.1 [154] |
| Fibrinogen | 1.56 [159] | |
| β2-microglobuline (human urine) | 1.70 [159] | |
| Human α-lactoalbumine | 1.53 [156] | |
| Chicken lysozyme | 2.65 [159] | 35.4 [155] |
| Amino Acid | Optical Properties | ||||
|---|---|---|---|---|---|
| Absorption | Fluorescence | ||||
| λmax, nm | Fluorescence Lifetime, ns | λmax, nm | Quantum Yield | ||
| Tryptophane | 280 | 5600 | 3.1 | 348 | 0.2 |
| Tyrosine | 274 | 1400 | 3.6 | 303 | 0.14 |
| Phenylalanine | 257 | 200 | 3.4 | 272 | 0.04 |
| Absorption Band | Wavenumber, cm−1 | Description |
|---|---|---|
| Amide I | 1600–1690 [163] | C=O stretching |
| Amide II | 1480–1575 [163] | CN stretching, NH bending |
| Amide III | 1229–1301 [163] | CN stretching, NH bending |
| Arginine | 1652–1695 [163] | νas(CN3H5+ ) |
| Glutamine | 1556–1560 [163] | νas(COO−) |
| Tyrosine | 1498–1500 [162] | ν(CC), δ(CH) |
| Tyrosine | 1269–1273 [162] | ν(C–O), ν(CC) |
| Spectroscopic Technique | Target Proteins | Characterization | Reference |
|---|---|---|---|
| MMS | Therapeutic monoclonal antibodies (mAb) | Secondary Structure Analysis | [193] |
| SEIRAS with plasmonic nanoantennas | Alpha-synuclein | Secondary structure | [194] |
| Derivative ATR FTIR spectroscopy | Granulocyte colony-stimulating factor (rhG-CSF) | Secondary Structure | [195] |
| TM-FTIR | Hemoglobin (H2500), poly-l-lysine (P2636) | Protein-Conformation Studies | [196] |
| Time-resolved FTIR | Ubiquitin | Protein folding | [197] |
| ATR FTIR | Alzheimer’s b-amyloid | Secondary structure | [198] |
| FTIR difference spectroscopy | Myoglobin | CO photodissociation | [199] |
| SEIRAS | Cytochrome c | Monolayer structure morphology | [200] |
| SEIRAS | Cytochrome c | Functionality of a Protein Monolayer | [201] |
| SEIRAS | α-synuclein | Lipid–protein interactions | [202] |
| SEIRAS | Tripeptide glutathione (GSH) | Ultrasensitive detection | [203] |
| Spectrosocopic Technique/Notes | Target Protein | LOD | Reference |
|---|---|---|---|
| Intrinsic protein fluorescence, conformational changes UV LED excitation at 295 nm, detection at 330 nm | tryptophan, bovine serum albumin (BSA), bovine carbonic anhydrase (BCA) | 72 nM 128 nM 250 nM | [216] |
| Intrinsic protein fluorescence, visualization UV LED excitation at 280 nm | BSA | 500 nM | [217] |
| Intrinsic protein fluorescence, qualitative determination UV LED excitation at 280 nm | troponin T | 6.5 ng/mL | [218] |
| Intrinsic protein fluorescence, detection for microchip electrophoresis Laser excitation at 266 nm detection with PMT based spectrometer | lysozyme, trypsinogen, chymotrypsinogen conalbumin, ovalbumin | 12.5 μg/mL | [219] |
| Intrinsic protein fluorescence, continuous electrophoretic separation via free flow isoelectric focusing (FFIEF) Laser excitation at 266 nm | α-Lactalbumin, β-Lactoglobulin B, Albumin, Globulins | 300 μmol/L | [220] |
| Intrinsic protein fluorescence, electrophoresis visualization, two-dimensional fingerprinting UV LED excitation at 280 nm | BSA, human lysozyme | 100 nM | [221] |
| Two photon excited (TPE) fluorescence laser excitation at 420 nmdetection at 320 nm | tryptophan lysozyme, trypsinogen and chymotrypsinogen | 12.5 ug/mL | [222] |
| Fluorescence lifetime detection, microchip electrophoresis laser excitation at 266 nm | lysozyme, trypsinogen and chymotrypsinogen | 2.5 mg/L | [223] |
| Fluorescence Correlation Spectroscopy laser excitation at 266 nm detection in 310−410 nm | β-galactosidase streptavidin penicillin amidase | - | [224] |
| Förster resonance energy transfer (FRET) Excitation at 280 nm, detection at 350 nm | Albumin | 0.15 nM | [225] |
| Target Protein | Pathological Condition | Reference |
|---|---|---|
| Human serum albumin | different stage liver cancer | [284] |
| Erythropoietin isophorms | anemia in cancer patients, athletes | [285] |
| Serum proteins | breast cancer | [286] |
| Prion proteins | Creutzfeldt-Jakob disease, kuru, fatal familial insomnia, and Gerstmann−Sträussler−Scheinker (GSS) | [287] |
| Phosphorylated proteins (Tau Biomarkers) | Alzheimer’s disease | [288] |
| Saliva proteins | oral cancer | [289] |
| Insulin | diabetes, hyperinsulinemia | [290] |
| Immune checkpoints proteins | cancer | [291] |
| Single cell metabolites | cancer | [292] |
| Ovarian Cancer Antigen CA125 | ovarian Cancer | [293] |
| Myoglobin | radiation-induced injury | [294] |
| Serum proteins | colorectal cancer | [295] |
| Interleukins | immunological disoderes | [281] |
| 90 K biomarker | cancer | [282] |
| Thrombin | blood coagulation | [263] |
| Optical Component or Structure | Detection Principle/Notes | Target Analyte | Label-Free | Limit of Detection (LOD) // Sensitivity | [Ref.], Year |
|---|---|---|---|---|---|
| Optofluidic laser TIIA (OFL-TIIA) | Dependence of the laser emission intensity on the IgG concentration in the RhB solution inside a Fabry-Pérot cavity/Wide dynamic range (1.8 × 10−10–1.8 × 10−5 g/L) | Rabbit IgG | No | 1.8 × 10−10 g/L // - | [307] 2019 |
| 2D microlenses, mirrors and optical fibers | Absorbance in six channels/Parallel measurements at different optical lengths (MPHIL concept) | Proteins: HEWL, GI, BLL, FASE, DHP | Yes | From 1.28 ± 0.04 μM to 8 ± 2 μM for different channel for the GI // - | [308] 2015 |
| Array of dielectric microspheres | Fluorescence of functionalized Au NPs enhanced by PNjs from microspheres | Biotin and mouse IgG | No | Fluorescence intensity was enhanced by a factor ∼40 | [309] 2015 |
| Array of dielectric microspheres | Fluorescence (FCS method)/Increasing detection volumes up to several tens of femtoliters | Protein Annexin A5b | No | Concentrations in the picomolar range | [310] 2014 |
| Ge on Si (GOS) WG | MIR spectroscopy/Measurement of aqueous protein | BSA protein | Yes | Chip was tested with 900 µM BSA solution | [311] 2020 |
| Liquid-core WG modified with gold NPs | SERS with enhancement factor of 2.7 × 108 for R6G | R6G, BSA protein | Yes | 10−11 mol/L (R6G) // - Chip was tested with 10−5 mol/L of BSA | [312] 2017 |
| FLOW | Wavelength shift of the transmission spectrum in the optical fiber/Log–linear response at concentrations ranging from 10 fg/mL up to 10 ng/mL | Protein p53 | Yes | 10 fg/mL // 22.2 pm/(fg/mL) | [313] 2018 |
| Slot WG with grating | Wavelength shift of resonance in grating for different analyte n | hemoglobin, globulin and BSA protein | Yes | - // 600 nm/RIU (300 FOM) | [314] 2019–2021 |
| OMNFs | LSPR (gold NPs on the fiber surface) | Streptavidin | Yes | 1 pg/mL // - | [315] 2018 |
| Diffraction-based leaky waveguides (LWGs) | RI sensing (Defining resonances in reflectivity curves)/Chitosan WGs | BSA protein | Yes | 1.9 × 10−6 ± 1.3 × 10−6 RIU // 125.5 ± 3.8 deg/RIU | [316] 2021 |
| Array of nanomaterials coated FBGs | Bragg wavelength shift/Multiparameter sensing (pH, temperature, humidity, gas concentration, light intensity and protein concentration) | BSA protein | Yes | - // 5 pm/μg/mL | [317] 2015 |
| Plasmonic TFBG sensor | RI sensing; differential amplitude measurement between the plasmonic and cut-off resonances/Minimal temperature cross-sensitivity | Rat urinary protein | Yes | 10−5 RIU; 1.5 × 10−3 mg/mL // 8000 dB/RIU; 5.5 dB/(mg/mL) | [318] 2016 |
| LPG coated with graphene oxide | Wavelength shift of the resonance in a transmission spectrum/Wide dynamic range (1 ng/mL–100 μg/mL) | C-reactive protein | Yes | 0.15 ng/mL // - | [319] 2021 |
| PC cavity and WG | Resonance wavelength shift/Q = (1.2–2.2) × 104 | Protein markers: fasligand (f), chemokine ligand 4 (MIP1) and hepatic growth factor (HGF) | Yes | 9.813 pg/mL (HGF); 15.437 pg/mL (MIP1); 0.3346 pg/mL (f) // 68–112 nm/RIU | [320] 2020 |
| MZI + hybrid plasmonic waveguide (HPWG) + nano-slots | Change in the dip (resonance) depth in the transmission spectrum at fixed wavelengths—amplitude measurements/Bulk RI measurement; biosensor are designed and theoretically investigated | HepV | Yes | Output transmission spectra and methodology for calculating parameters were reported | [321] 2017 |
| µFPI with sub-wavelength nano-hole arrays | Change in EOT-modulated SPR patterns/Q factor up to 128.4 | BSA protein | Yes | 1 pM // 593 nm/RIU | [322] 2019 |
| Array of six asymmetric MZI (aMZI) | Measuring the phase shift of the output signal (RI sensing) | Periostin (POSTN) and transforming growth factor beta-induced protein (TGFBI) | Yes | 16 × 10−8 RIU; 10 ng/mL // ≤5000 nm/RIU | [323] 2021 |
| MZI and AWG | Spectral shift (RI sensing)/Low-cost instrumentation | C-reactive protein | Yes | 6 × 10−6 RIU; 19.478 ng/mL // - | [324] 2018 |
| MMI-ARROW | Fluorescence excitation via MMI waveguides orthogonal to the microfluidic channel aligned with the ARROW, which traps the luminescence emission | SARS-CoV-2 and influenza A antigens | No | 30 ng/mL // - | [325] 2021 |
| multi-channel MMI-ARROW chip | The same/Dual detection of nucleic acid and antigen biomarkers with single molecule sensitivity | SARS-CoV-2 nucleic acids and proteins | No | 0.7 ng/mL (SARS-CoV-2 antigens) // - | [326] 2021 |
| 2 ARROWs in HCF | Wavelength shift of two resonance/Temperature compensation | Interferon-gamma (IFN-γ) | Yes | 0.5 ng/mL // 1413 nm/ RIU | [327] 2020 |
| GWT-GMR sensor | Sensor converts spectral information to spatial information on a CCD (resonance band shift when n (biomolecule concentration) changes) | Albumin, creatinine | Yes | 2.92 µg/mL (albumin); 12.05 µg/mL (creatinine) // 24,497 µm/RIU | [328] 2021 |
| Subwavelength grating metamaterial (SGM) ring resonator | Wavelength shift of resonance in transmission spectra/Q~2180 at 1548.81 nm. | Streptavidin | Yes | 10 μg/mL or ∼4.2 × 10−4 RIU // 310–423 nm/RIU | [329] 2021 |
| Photonic crystal beads (PCBs) | Fluorescence immunoassays/Multiplex bioassays | Human IgG, AFP and CEA | No | 25.33–105.45 ng/mL (IgG); 18.92 ng/mL (AFP) // - | [330] 2018 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konoplev, G.; Agafonova, D.; Bakhchova, L.; Mukhin, N.; Kurachkina, M.; Schmidt, M.-P.; Verlov, N.; Sidorov, A.; Oseev, A.; Stepanova, O.; et al. Label-Free Physical Techniques and Methodologies for Proteins Detection in Microfluidic Biosensor Structures. Biomedicines 2022, 10, 207. https://doi.org/10.3390/biomedicines10020207
Konoplev G, Agafonova D, Bakhchova L, Mukhin N, Kurachkina M, Schmidt M-P, Verlov N, Sidorov A, Oseev A, Stepanova O, et al. Label-Free Physical Techniques and Methodologies for Proteins Detection in Microfluidic Biosensor Structures. Biomedicines. 2022; 10(2):207. https://doi.org/10.3390/biomedicines10020207
Chicago/Turabian StyleKonoplev, Georgii, Darina Agafonova, Liubov Bakhchova, Nikolay Mukhin, Marharyta Kurachkina, Marc-Peter Schmidt, Nikolay Verlov, Alexander Sidorov, Aleksandr Oseev, Oksana Stepanova, and et al. 2022. "Label-Free Physical Techniques and Methodologies for Proteins Detection in Microfluidic Biosensor Structures" Biomedicines 10, no. 2: 207. https://doi.org/10.3390/biomedicines10020207
APA StyleKonoplev, G., Agafonova, D., Bakhchova, L., Mukhin, N., Kurachkina, M., Schmidt, M.-P., Verlov, N., Sidorov, A., Oseev, A., Stepanova, O., Kozyrev, A., Dmitriev, A., & Hirsch, S. (2022). Label-Free Physical Techniques and Methodologies for Proteins Detection in Microfluidic Biosensor Structures. Biomedicines, 10(2), 207. https://doi.org/10.3390/biomedicines10020207






