The Role of Mid-Regional Proadrenomedullin in the Differential Diagnosis between Culture-Negative and Culture-Positive Sepsis at Emergency Department Admission
Abstract
1. Introduction
2. Methods
2.1. Definitions
2.2. Biomarkers Assay
2.3. Outcomes
2.4. Statistical Analysis
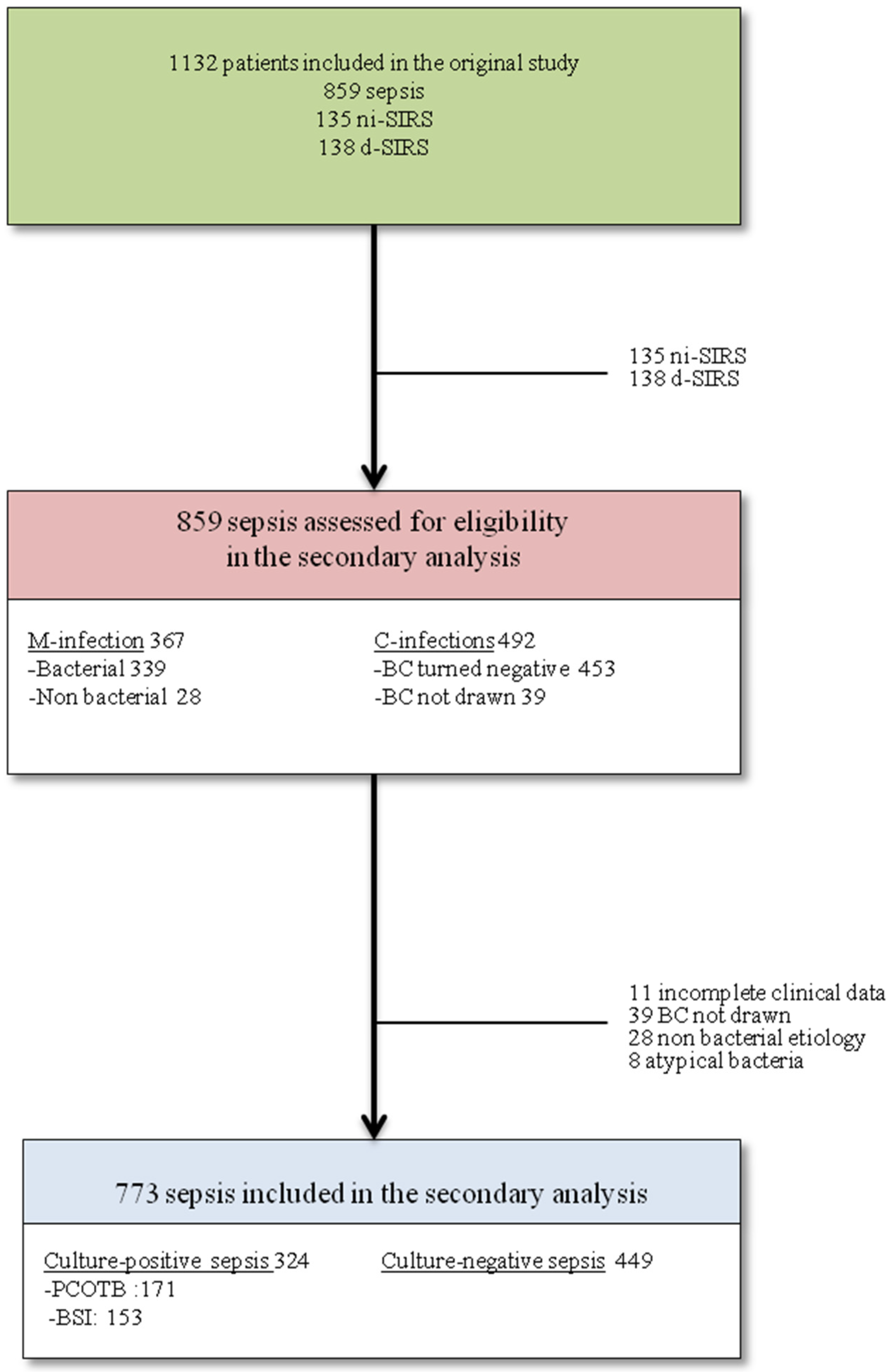
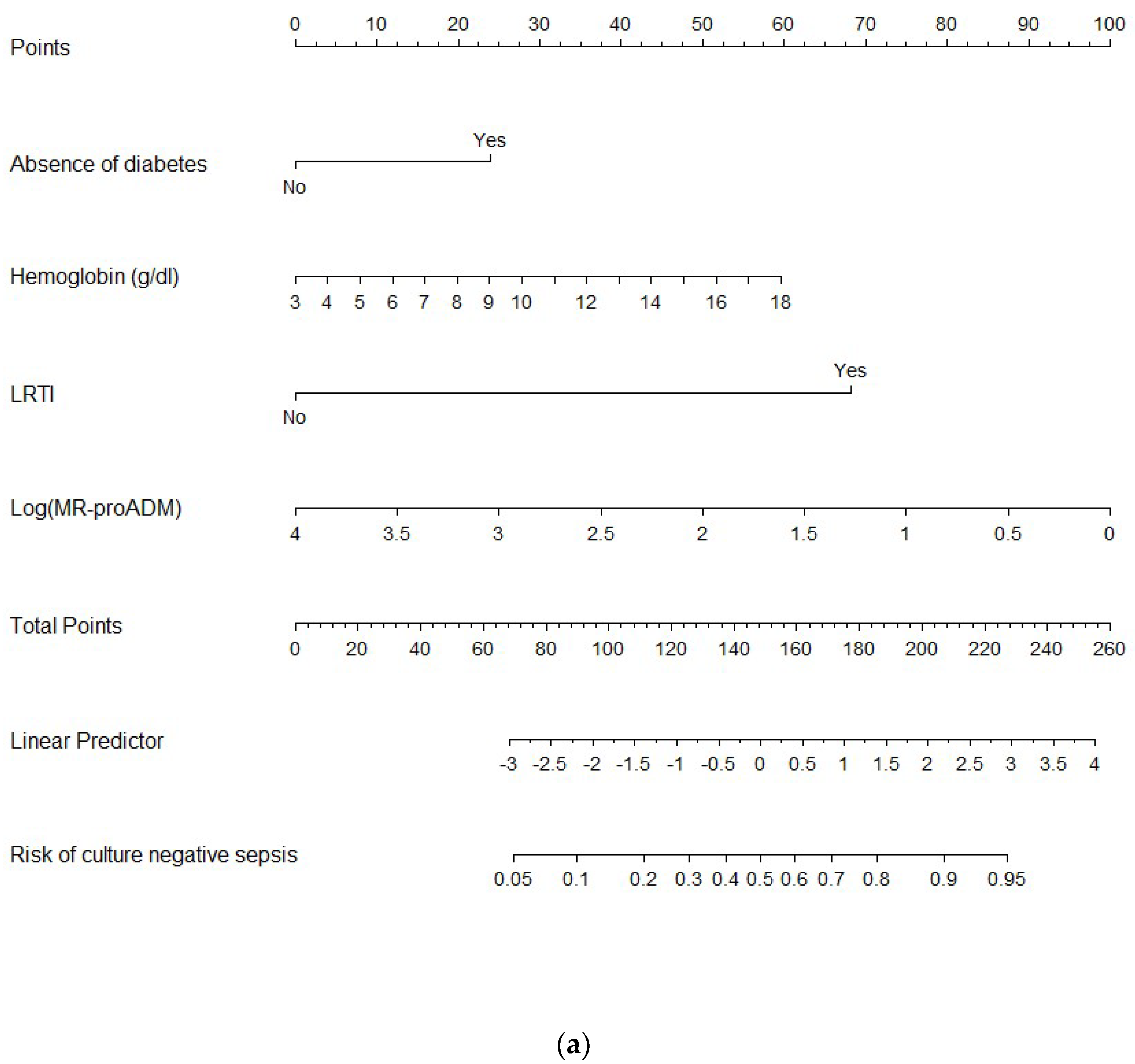
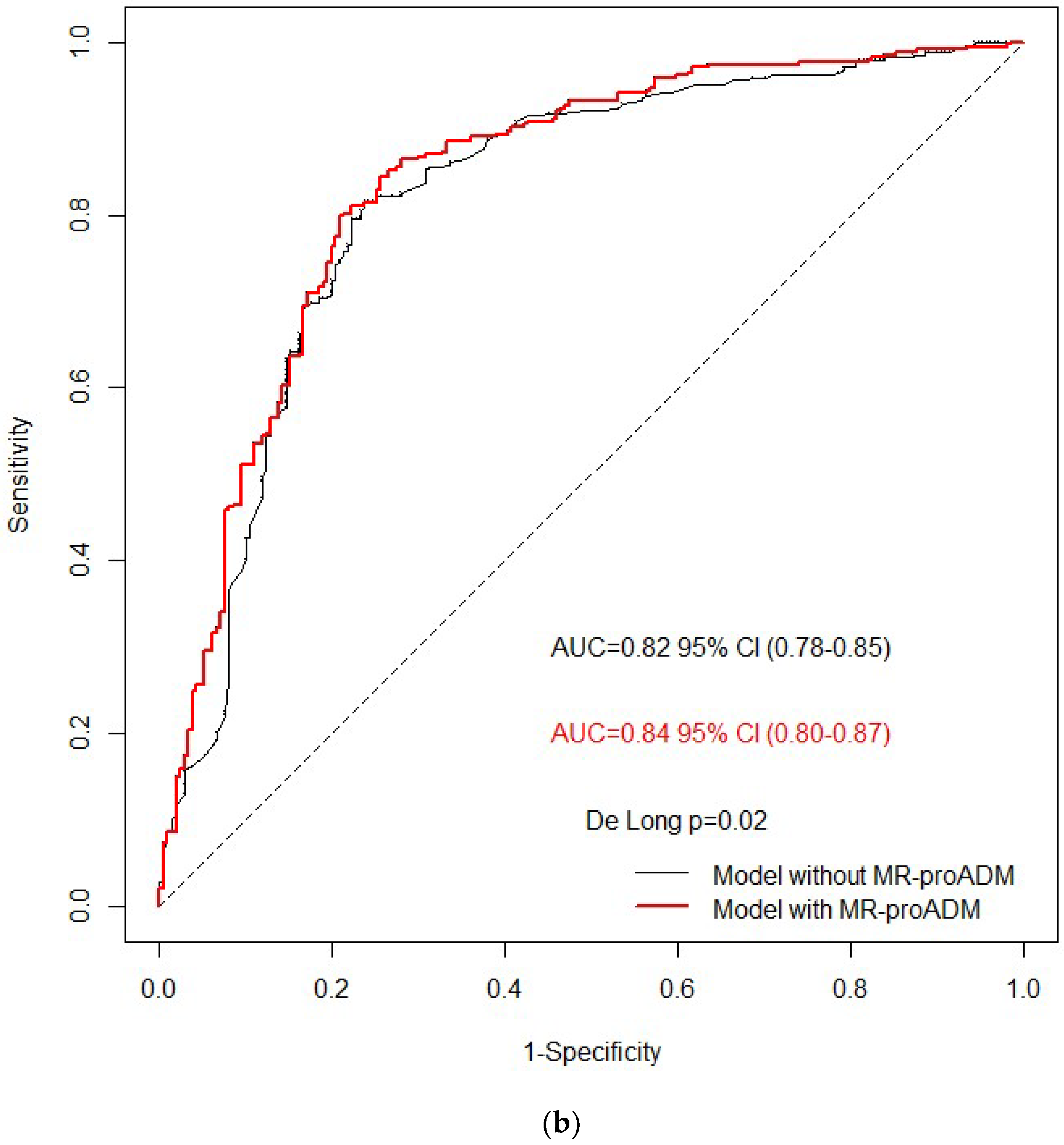
3. Results
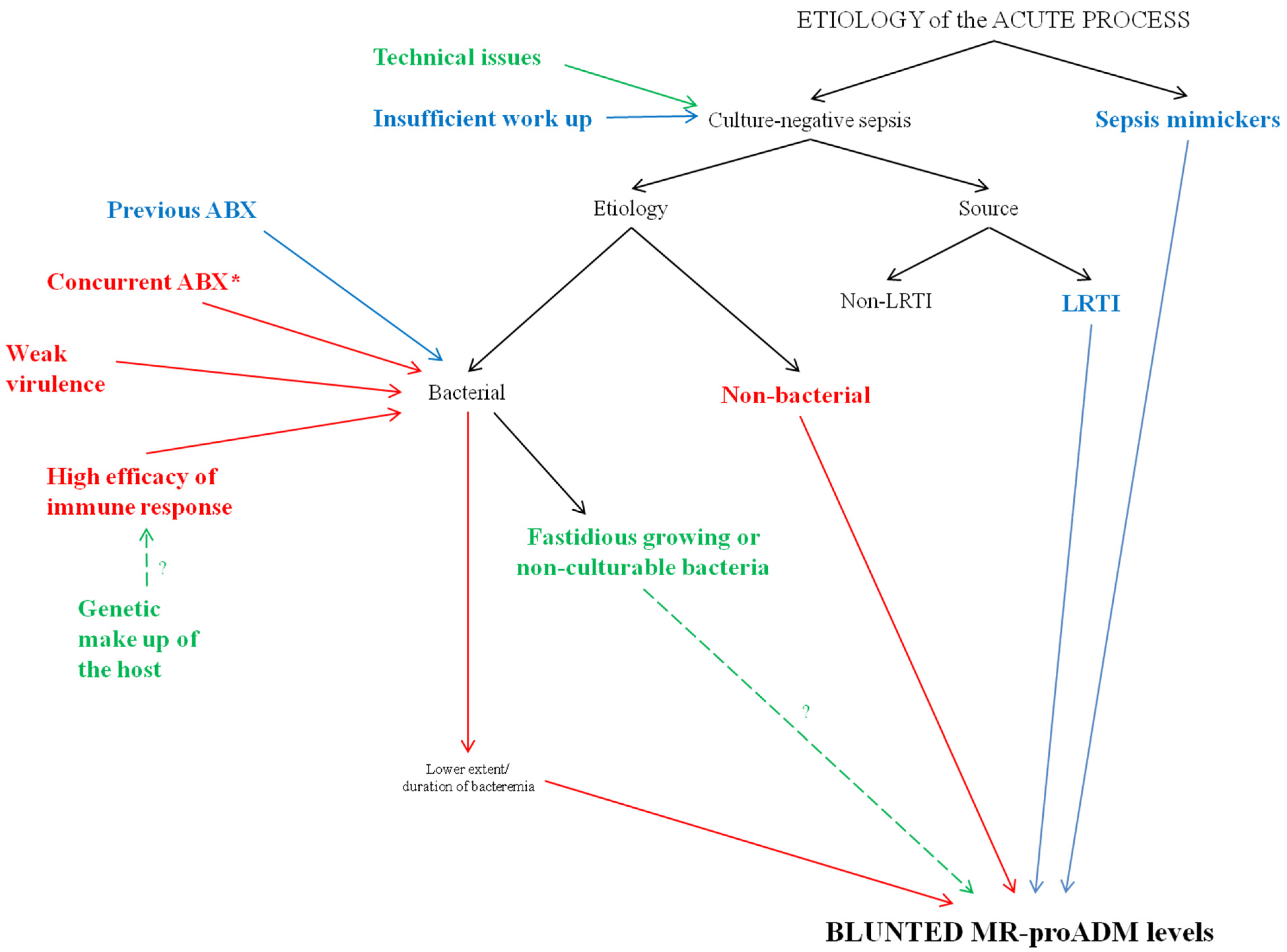
4. Discussion
4.1. Sepsis Mimickers
4.2. Insufficient Work Up and Technical Issues
4.3. Source of Infection
4.4. Bacterial Virulence and Host Response
4.5. Previous or Concurrent Antibacterial Therapy?
4.6. Uncommon Bacteria and Non-Bacterial Etiology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensiv. Care Med. 2018, 44, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Panday, R.S.N.; Lammers, E.M.J.; Alam, N.; Nanayakkara, P.W.B. An overview of positive cultures and clinical outcomes in septic patients: A sub-analysis of the Prehospital Antibiotics Against Sepsis (PHANTASi) trial. Crit. Care 2019, 23, 182. [Google Scholar] [CrossRef]
- Cheng, M.P.; Stenstrom, R.; Paquette, K.; Stabler, S.N.; Akhter, M.; Davidson, A.C.; Gavric, M.; Lawandi, A.; Jinah, R.; Saeed, Z.; et al. Blood Culture Results Before and After Antimicrobial Administration in Patients with Severe Manifestations of Sepsis: A Diagnostic Study. Ann. Intern. Med. 2019, 171, 547–554. [Google Scholar] [CrossRef]
- Scheer, C.; Fuchs, C.; Gründling, M.; Vollmer, M.; Bast, J.; Bohnert, J.; Zimmermann, K.; Hahnenkamp, K.; Rehberg, S.; Kuhn, S.-O. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin. Microbiol. Infect. 2019, 25, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Grace, C.J.; Lieberman, J.; Pierce, K.; Littenberg, B. Usefulness of blood culture for hospitalized patients who are receiving antibi-otic therapy. Clin. Infect. Dis. 2001, 32, 1651–1655. [Google Scholar] [CrossRef] [PubMed]
- Sigakis, M.J.G.; Jewell, E.; Maile, M.D.; Cinti, S.K.; Bateman, B.T.; Engoren, M. Culture-Negative and Culture-Positive Sepsis: A Com-parison of Characteristics and Outcomes. Anesth. Analg. 2019, 129, 1300–1309. [Google Scholar] [CrossRef]
- Lockhart, G.C.; Hanin, J.; Micek, S.T.; Kollef, M.H. Pathogen-Negative Sepsis—An Opportunity for Antimicrobial Stewardship. Open Forum Infect. Dis. 2019, 6, ofz397. [Google Scholar] [CrossRef]
- van Walraven, C.; Wong, J. Independent influence of negative blood cultures and bloodstream infections on in-hospital mor-tality. BMC Infect. Dis. 2014, 14, 36. [Google Scholar] [CrossRef]
- Søgaard, M.; Nørgaard, M.; Pedersen, L.; Sørensen, H.T.; Schønheyder, H.C. Blood culture status and mortality among patients with suspected community-acquired bacteremia: A population-based cohort study. BMC Infect. Dis. 2011, 11, 139. [Google Scholar] [CrossRef]
- Komori, A.; Abe, T.; Kushimoto, S.; Ogura, H.; Shiraishi, A.; Saitoh, D.; Fujishima, S.; Mayumi, T.; Naito, T.; Hifumi, T.; et al. Characteristics and outcomes of bacteremia among ICU-admitted patients with severe sepsis. Sci. Rep. 2020, 10, 2983. [Google Scholar] [CrossRef]
- Cummings, C.J.; Sessler, C.N.; Beall, L.D.; Fisher, B.J.; Best, A.M.; Fowler, A.A., 3rd. Soluble E-selectin levels in sepsis and critical illness. Correlation with infection and hemodynamic dysfunction. Am. J. Respir. Crit. Care Med. 1997, 156, 431–437. [Google Scholar] [CrossRef]
- Caffarini, E.M.; DeMott, J.; Patel, G.; Lat, I. Determining the Clinical Utility of an Absolute Procalcitonin Value for Predicting a Positive Culture Result. Antimicrob. Agents Chemother. 2017, 61, e02007-16. [Google Scholar] [CrossRef] [PubMed]
- Anand, D.; Das, S.; Bhargava, S.; Srivastava, L.M.; Garg, A.; Tyagi, N.; Taneja, S.; Ray, S. Procalcitonin as a rapid diagnostic bi-omarker to differentiate between culture-negative bacterial sepsis and systemic inflammatory response syndrome: A prospec-tive, observational, cohort study. J. Crit. Care 2015, 30, 218.e7-12. [Google Scholar] [CrossRef] [PubMed]
- Fariñas-Alvarez, C.; Fariñas, M.C.; Fernández-Mazarrasa, C.; Llorca, J.; Delgado-Rodríguez, M. Epidemiological differences be-tween sepsis syndrome with bacteremia and culture-negative sepsis. Infect. Control. Hosp. Epidemiol. 2000, 21, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Phua, J.; Ngerng, W.J.; See, K.C.; Tay, C.K.; Kiong, T.; Lim, H.F.; Chew, M.Y.; Yip, H.S.; Tan, A.; Khalizah, H.J.; et al. Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit. Care 2013, 17, R202. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Sakhuja, A.; Kumar, G.; McGrath, E.; Nanchal, R.S.; Kashani, K.B. Culture-Negative Severe Sepsis: Nationwide Trends and Outcomes. Chest 2016, 150, 1251–1259. [Google Scholar] [CrossRef]
- Kethireddy, S.; Bilgili, B.; Sees, A.; Kirchner, H.L.; Ofoma, U.R.; Light, R.B.; Mirzanejad, Y.; Maki, D.; Kumar, A.; Layon, A.J.; et al. Culture-Negative Septic Shock Compared with Culture-Positive Septic Shock: A Retrospective Cohort Study. Crit. Care Med. 2018, 46, 506–512. [Google Scholar] [CrossRef]
- Mearelli, F.; Fiotti, N.; Giansante, C.; Casarsa, C.; Orso, D.; De Helmersen, M.; Altamura, N.; Ruscio, M.; Castello, L.M.; Colonetti, E.; et al. Deriva-tion and Validation of a Biomarker-Based Clinical Algorithm to Rule Out Sepsis from Noninfectious Systemic Inflammatory Response Syndrome at Emergency Department Admission: A Multicenter Prospective Study. Crit. Care Med. 2018, 46, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Variable selection with stepwise and best subset approaches. Ann. Transl. Med. 2016, 4, 136. [Google Scholar] [CrossRef]
- Heffner, A.; Horton, J.M.; Marchick, M.R.; Jones, A.E. Etiology of Illness in Patients with Severe Sepsis Admitted to the Hospital from the Emergency Department. Clin. Infect. Dis. 2010, 50, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, G.; Schønheyder, H.C.; Sørensen, H.T. Source of infection and other factors associated with case fatality in communi-ty-acquired bacteremia--a Danish population-based cohort study from 1992 to 1997. Clin. Microbiol. Infect. 2003, 9, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Teuffel, O.; Ethier, M.-C.; Beyene, J.; Sung, L. Association between tumor necrosis factor-α promoter −308 A/G polymorphism and susceptibility to sepsis and sepsis mortality: A systematic review and meta-analysis. Crit. Care Med. 2010, 38, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Kotsaki, A.; Raftogiannis, M.; Routsi, C.; Baziaka, F.; Kotanidou, A.; Antonopoulou, A.; Orfanos, S.E.; Katsenos, C.; Koutoukas, P.; Pla-chouras, D.; et al. Genetic polymorphisms within tumor necrosis factor gene promoter re-gion: A role for susceptibility to ventilator-associated pneumonia. Cytokine 2012, 59, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Henckaerts, L.; Nielsen, K.R.; Steffensen, R.; Van Steen, K.; Mathieu, C.; Giulietti, A.; Wouters, P.J.; Milants, I.; Vanhorebeek, I.; Langouche, L.; et al. Polymorphisms in innate immunity genes predispose to bacteremia and death in the medical intensive care unit*. Crit. Care Med. 2009, 37, 192–e3. [Google Scholar] [CrossRef]
- Ferwerda, B.; McCall, M.B.; Alonso, S.; Giamarellos-Bourboulis, E.J.; Mouktaroudi, M.; Izagirre, N.; Syafruddin, D.; Kibiki, G.; Cristea, T.; Hijmans, A.; et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of mod-ern humans. Proc. Natl. Acad. Sci. USA 2007, 104, 16645–16650. [Google Scholar] [CrossRef] [PubMed]
- Khor, C.C.; Chapman, S.J.; Vannberg, F.O.; Dunne, A.; Murphy, C.; Ling, E.Y.; Frodsham, A.J.; Walley, A.J.; Kyrieleis, O.; Khan, A.; et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat. Genet. 2007, 39, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Ye, Q.; Ma, Y.; Zhou, J. Genetic Association of Interleukin-1β (−511C/T) and Its Receptor Antagonist (86-bpVNTR) Gene Polymorphism with Susceptibility to Bacteremia in Kidney Transplant Recipients. Transplant. Proc. 2012, 44, 3026–3028. [Google Scholar] [CrossRef]
- Casqueiro, J.; Casqueiro, J.; Alves, C. Infections in patients with diabetes mellitus: A review of pathogenesis. Indian J. Endocrinol. Metab. 2012, 16 (Suppl. 1), S27–S36. [Google Scholar] [PubMed]
- Önal, U.; Valenzuela-Sánchez, F.; Vandana, K.E.; Rello, J. Mid-Regional Pro-Adrenomedullin (MR-proADM) as a Biomarker for Sepsis and Septic Shock: Narrative Review. Healthcare 2018, 6, 110. [Google Scholar] [CrossRef]
- Mearelli, F.; Barbati, G.; Casarsa, C.; Giansante, C.; Breglia, A.; Spica, A.; Moras, C.; Olivieri, G.; Occhipinti, A.A.; De Nardo, M.; et al. The Integration of qSOFA with Clinical Variables and Serum Biomarkers Improves the Prognostic Value of qSOFA Alone in Patients with Suspected or Confirmed Sepsis at ED Admission. J. Clin. Med. 2020, 9, 1205. [Google Scholar] [CrossRef]
- Schermer, C.R.; Sanchez, D.P.; Qualls, C.R.; Demarest, G.B.; Albrecht, R.M.; Fry, D.E. Blood Culturing Practices in a Trauma Intensive Care Unit: Does Concurrent Antibiotic Use Make a Difference? J. Trauma: Inj. Infect. Crit. Care 2002, 52, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, R.; Curtis, S.; Schilling, W.; James, P.R. Blood culture negative endocarditis in the modern era of 16S rRNA sequencing. Clin. Med. 2020, 20, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Cesario, T.C. Viruses Associated with Pneumonia in Adults. Clin. Infect. Dis. 2012, 55, 107–113. [Google Scholar] [CrossRef]
- Sakr, Y.; Jaschinski, U.; Wittebole, X.; Szakmany, T.; Lipman, J.; Ñamendys-Silva, S.A.; Martin-Loeches, I.; Leone, M.; Lupu, M.-N.; Vincent, J.-L.; et al. Sepsis in Intensive Care Unit Patients: Worldwide Data from the Intensive Care over Nations Audit. Open Forum Infect. Dis. 2018, 5, ofy313. [Google Scholar] [CrossRef]
- Tabah, A.; Bassetti, M.; Kollef, M.H.; Zahar, J.R.; Paiva, J.A.; Timsit, J.F.; Roberts, J.A.; Schouten, J.; Giamarellou, H.; Rello, J.; et al. Antimicrobial de-escalation in critically ill patients: A posi-tion statement from a task force of the European Society of Intensive Care Medicine (ESICM) and European Society of Clini-cal Microbiology and Infectious Diseases (ESCMID) Critically Ill Patients Study Group (ESGCIP). Intensive Care Med. 2020, 46, 245–265. [Google Scholar] [PubMed]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Shehadeh, F.; Zacharioudakis, I.M.; Zervou, F.N.; Mylonakis, E. Cost-effectiveness of rapid diagnostic assays that perform directly on blood samples for the diagnosis of septic shock. Diagn. Microbiol. Infect. Dis. 2019, 94, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Shiraishi, A.; Gando, S.; Abe, T.; Kushimoto, S.; Mayumi, T.; Fujishima, S.; Hagiwara, A.; Shiino, Y.; Shiraishi, S.-I.; Hifumi, T.; et al. Quick sequential organ failure assessment versus systemic inflammatory response syndrome criteria for emergency department patients with suspected infection. Sci. Rep. 2021, 11, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International guidelines for the management of severe sepsis and septic shock, 2012. Intensiv. Care Med. 2013, 39, 165–228. [Google Scholar] [CrossRef] [PubMed]
- Baron, E.J.; Miller, J.M.; Weinstein, M.P.; Richter, S.S.; Gilligan, P.H.; Thomson Jr, R.B.; Bourbeau, P.; Carroll, K.C.; Kehl, S.C.; Dunne, W.M.; et al. Executive summary: A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. 2013, 57, 485–488. [Google Scholar] [CrossRef]
- Woodhead, M.; Blasi, F.; Ewig, S.; Garau, J.; Huchon, G.; Ieven, M.; Ortqvist, A.; Schaberg, T.; Torres, A.; Van Der Heijden, G.; et al. Guidelines for the management of adult lower respiratory tract infections. Clin. Microbiol. Infect. 2011, 17 (Suppl. S6), E1–E59. [Google Scholar] [CrossRef] [PubMed]
- Mandell, L.A.; Wunderink, R.G.; Anzueto, A.; Bartlett, J.G.; Campbell, G.D.; Dean, N.C.; Dowell, S.F.; File Jr, T.M.; Musher, D.M.; Niederman, M.S.; et al. Infectious Diseases Society of America/American Thoracic Society cosensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007, 44 (Suppl. s2), S27–S72. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Hooton, T.M.; Naber, K.G.; Wullt, B.; Colgan, R.; Miller, L.G.; Moran, G.J.; Nicolle, L.E.; Raz, R.; Schaeffer, A.J.; et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 2011, 52, e103–e120. [Google Scholar] [CrossRef]
- Stevens, D.L.; Bisno, A.L.; Chambers, H.F.; Dellinger, E.P.; Goldstein, E.J.; Gorbach, S.L.; Hirschmann, J.V.; Kaplan, S.L.; Montoya, J.G.; Wade, J.C. Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the infectious diseases society of America. Clin. Infect. Dis. 2014, 59, 147–159. [Google Scholar] [CrossRef]
- Solomkin, J.S.; Mazuski, J.E.; Bradley, J.S.; Rodvold, K.A.; Goldstein, E.J.; Baron, E.J.; O’Neill, P.J.; Chow, A.W.; Dellinger, E.P.; Eachempati, S.R.; et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: Guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Surg. Infect. (Larchmt) 2010, 11, 79–109. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | CnS (n = 449) | CpS (n = 324) | Un p | Un OR |
|---|---|---|---|---|
| Male, n (%) | 246 (54) | 172 (53) | 0.639 | |
| Median age (IQR) | 82 (73–88) | 79 (72–85) | 0.156 | |
| Median Charlson comorbidity index (IQR) | 3 (1–4) | 3 (1–4) | 0.736 | |
| Diabetes, n (%) | 63 (14) | 70 (21) | 0.006 | 0.592 (0.407–0.862) |
| Chronic heart failure, n (%) | 121 (27) | 63 (19) | 0.016 | 1.528 (1.082–2.158) |
| Previous acute myocardial infarction, n (%) | 114 (25) | 59 (18) | 0.019 | 1.528 (1.074–2.176) |
| Solid cancer, n (%) | 33 (7) | 30 (9) | 0.339 | |
| Haematologic cancer, n (%) | 12 (3) | 10 (3) | 0.733 | |
| Chronic liver disease, n (%) | 33 (7) | 28 (9) | 0.511 | |
| Chronic pulmonary disease, n (%) | 140 (31) | 62 (19) | <0.001 | 1.915 (1.361–2.693) |
| Chronic kidney disease, n (%) | 82 (18) | 64 (19) | 0.602 | |
| Dementia, n (%) | 136 (30) | 96 (30) | 0.843 | |
| Chronic rheumatologic disease, n (%) | 16 (3) | 14 (4) | 0.591 | |
| AIDS, n (%) | 2 (0) | 0 (0) | 0.999 | |
| Antibacterials within 30 days from ED admission ^, n (%) | 129 (29) | 108 (33) | 0.171 | |
| Patients untreated with antibacterials during their stay in ED *, n (%) | 26 (6) | 6 (2) | 0.009 | 3.257 (1.325–8.009) |
| Prosthetic devices, n (%) | 52 (11) | 74 (23) | <0.001 | 0.443 (0.300–0.653) |
| Median body temperature (°C), (IQR) | 37.5 (36.6–38.2) | 37.8 (36.7–38.2) | 0.331 | |
| Meadian mean arterial pressure (mmHg), (IQR) | 86 (76–95) | 85 (75–93) | <0.001 | 1.019 (1.009–1.029) |
| Median heart rate (beats/min), (IQR) | 100 (90–110) | 104 (78–120) | 0.912 | |
| Median respiratory rate (breaths/min), (IQR) | 24 (20–28) | 24 (21–32) | 0.361 | |
| Median Glasgow Coma Scale (IQR) | 15 (15–15) | 15 (15–15) | 0.183 | |
| Median white blood cell count x1000/mm3 (IQR) | 12.5 (9.0–16.4) | 13.4 (9.8–18.5) | 0.025 | 0.806 (0.629–1.001) * |
| Median hemoglobin (g/L) (IQR) | 12.6 (11.0–13.8) | 11.6 (10.6–13.0) | <0.001 | 1.249 (1.159–1.346) |
| Median platelets count x1000/mm3 (IQR) | 222 (160–298) | 215 (151–300) | 0.330 | |
| Median serum urea (mg/dL) (IQR) | 43 (30–63) | 46 (30–74) | 0.033 | 0.996 (0.993–1.000) |
| Median creatinine/mg/dL) (IQR) | 1.0 (0.8–1.6) | 1.1 (0.9–1.7) | 0.039 | 0.873 (0.768–0.993) |
| Median sodium (mEq/L) (IQR) | 137 (134–140) | 135 (133–138) | <0.001 | 1.052 (1.025–1.079) |
| Median potassium (mEq/L) (IQR) | 3.9 (3.5–4.4) | 3.9 (3.5–4.4) | 0.631 | |
| Median AST (U/L) (IQR) | 24 (17–36) | 27 (19–59) | 0.910 | |
| Median ALT (U/L) (IQR) | 17 (11–29) | 23 (12–41) | 0.456 | |
| Median total bilirubin (mg/dL) (IQR) | 0.9 (0.6–1.4) | 0.9 (0.7–1.4) | 0.152 | |
| Median INR (IQR) | 1.1 (1.0–1.3) | 1.37 (1.17–3.2) | 0.223 | |
| Median fibrinogen (mg/dL) (IQR) | 478 (382–637) | 495 (378–668) | 0.340 | |
| Biomarkers | ||||
| Median C-reactive protein (mg/dL) (IQR) | 81 (31–170) | 127 (53–215) | <0.001 | 0.769 (0.678–0.872) |
| Median lactate (mg/dL) (IQR) | 14 (9–19) | 14 (10–21) | 0.103 | |
| Median procalcitonin (ng/mL) (IQR) | 0.51 (0.16–2.43) | 1.12 (0.29–9.51) | <0.001 | 0.698 (0.619–0.787) |
| Median sIL2Rα (pg/mL) (IQR) | 13367 (9050–16817) | 17510 (10635–29856) | <0.001 | 0.588 (0.481–0.718) |
| Median sTREM-1 (pg/mL) (IQR) | 398 (269–634) | 456 (292–742) | 0.101 | |
| Median sPLA2GIIA (ng/mL) (IQR) | 30.8 (24.1–35.5) | 32.4 (28.0–36.3) | 0.002 | 0.600 (0.434–0.830) |
| Median presepsin (pg/mL) (IQR) | 525 (321–918) | 675 (359–1328) | <0.001 | 0.645 (0.547–0.762) |
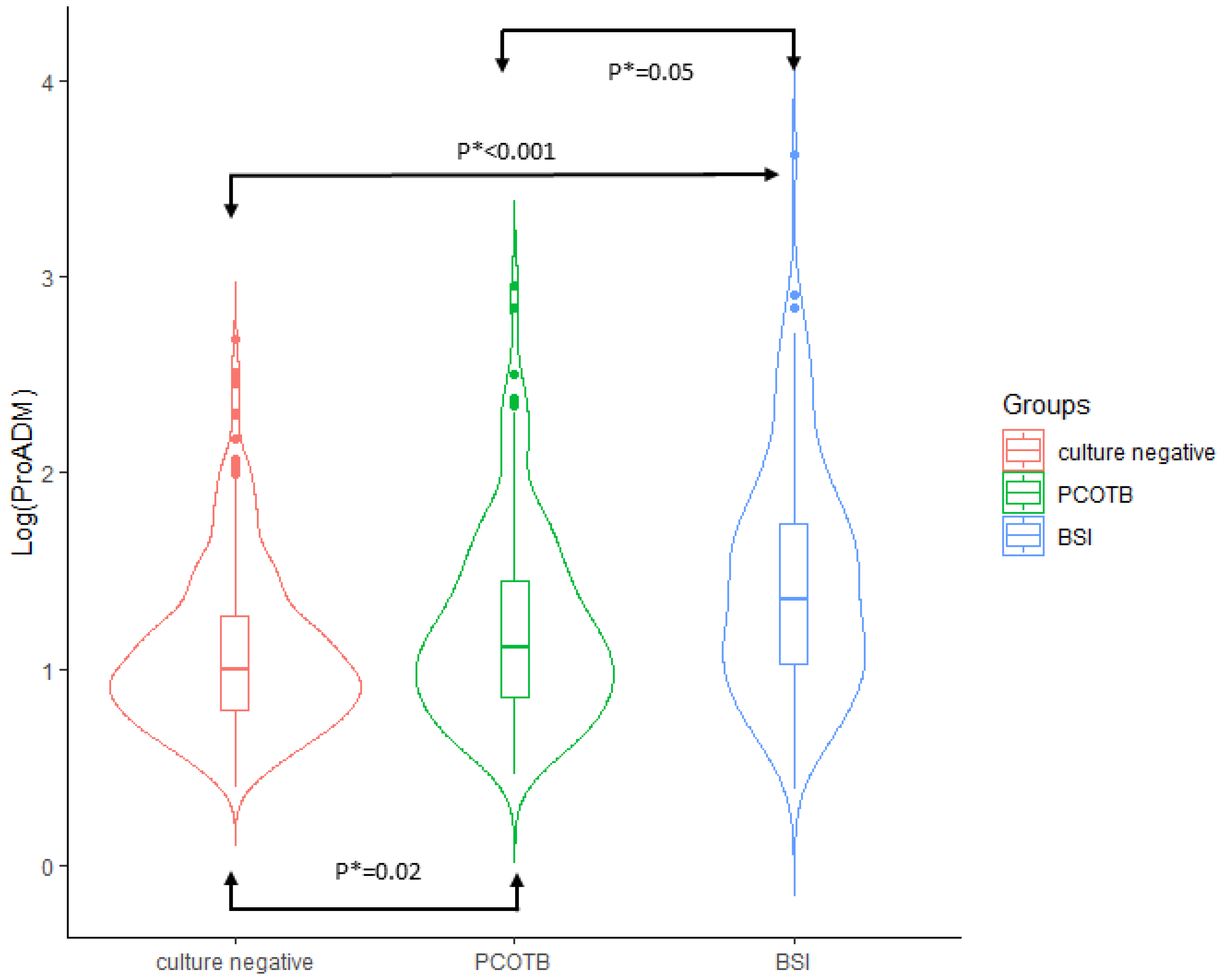
| Median MR-proADM (nmol/L) (IQR) | 1.93 (1.29–3.06) | 2.31 (1.45–4.13) | <0.001 | 0.349 (0.248–0.490) |
| Source of infection | ||||
| Single Source, n (%) | 413 (92) | 266 (82) | <0.001 | 2.375 (1.501–3.757) |
| -LRTI, n (%) | 339 (82) | 71 (27) | <0.001 | 11.394 (7.974–16.281) |
| -Non LRTI, n (%) | 74 (18) | 195 (73) | ||
| Multiple Source, n (%) | 36 (8) | 58 (18) | ||
| Microbiological work up | ||||
| At least one biological sample other than blood, n (%) | 441 (98) | 322 (99) | 0.205 | |
| Median SOFA score (IQR) | 3 (2–4) | 3 (1–5) | 0.092 |
| Characteristics | CnS (n = 449) | CpS (n = 324) | p |
|---|---|---|---|
| Organ dysfunction * | |||
| Renal | 116 (26) | 113 (35) | 0.007 |
| Cardiovascular | 190 (42) | 93 (29) | <0.001 |
| -Narrow/broad complex tachycardia | 38 (8) | 25 (8) | 0.790 |
| -Atrial fibrillation | 29 (6) | 13 (4) | 0.092 |
| -Newly detected | 10 (2) | 6 (2) | 0.718 |
| -Acute decompensated heart failure | 116 (26) | 54 (17) | 0.002 |
| -Acute coronary syndrome | 68 (15) | 25 (8) | 0.002 |
| -Shock | 26 (6) | 16 (5) | 0.606 |
| Respiratory | 271 (60) | 133 (41) | <0.001 |
| Haemostasis | 77 (17) | 73 (22) | 0.062 |
| Haematologic | 29 (6) | 30 (9) | 0.148 |
| Neurologic | 122 (27) | 114 (35) | 0.002 |
| Liver | 12 (3) | 13 (4) | 0.299 |
| Median number of organ | 2 (1–3) | 2 (1–3) | 0.053 |
| -Multiorgan failure | 4 (1) | 11 (3) | 0.013 |
| Median length of hospital stay | 9 (7–16) | 12 (7–19) | <0.001 |
| Mortality at 30 days | 88 (19) | 66 (20) | 0.791 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mearelli, F.; Barbati, G.; Spagnol, F.; Nunnari, A.; Castello, L.M.; Lupia, E.; Muiesan, M.L.; Di Somma, S.; Avanzi, G.C.; Biolo, G. The Role of Mid-Regional Proadrenomedullin in the Differential Diagnosis between Culture-Negative and Culture-Positive Sepsis at Emergency Department Admission. Biomedicines 2022, 10, 357. https://doi.org/10.3390/biomedicines10020357
Mearelli F, Barbati G, Spagnol F, Nunnari A, Castello LM, Lupia E, Muiesan ML, Di Somma S, Avanzi GC, Biolo G. The Role of Mid-Regional Proadrenomedullin in the Differential Diagnosis between Culture-Negative and Culture-Positive Sepsis at Emergency Department Admission. Biomedicines. 2022; 10(2):357. https://doi.org/10.3390/biomedicines10020357
Chicago/Turabian StyleMearelli, Filippo, Giulia Barbati, Francesca Spagnol, Alessio Nunnari, Luigi Mario Castello, Enrico Lupia, Maria Lorenza Muiesan, Salvatore Di Somma, Gian Carlo Avanzi, and Gianni Biolo. 2022. "The Role of Mid-Regional Proadrenomedullin in the Differential Diagnosis between Culture-Negative and Culture-Positive Sepsis at Emergency Department Admission" Biomedicines 10, no. 2: 357. https://doi.org/10.3390/biomedicines10020357
APA StyleMearelli, F., Barbati, G., Spagnol, F., Nunnari, A., Castello, L. M., Lupia, E., Muiesan, M. L., Di Somma, S., Avanzi, G. C., & Biolo, G. (2022). The Role of Mid-Regional Proadrenomedullin in the Differential Diagnosis between Culture-Negative and Culture-Positive Sepsis at Emergency Department Admission. Biomedicines, 10(2), 357. https://doi.org/10.3390/biomedicines10020357







