Translational Implications of Dysregulated Pathways and microRNA Regulation in Quadruple-Negative Breast Cancer
Abstract
:1. Introduction
2. Definition and Clinical Distinctions of QNBC Compared with TNBC and Other Subtypes of Breast Cancer
3. Significance of Androgen Receptor Expression and Function in Breast Cancer
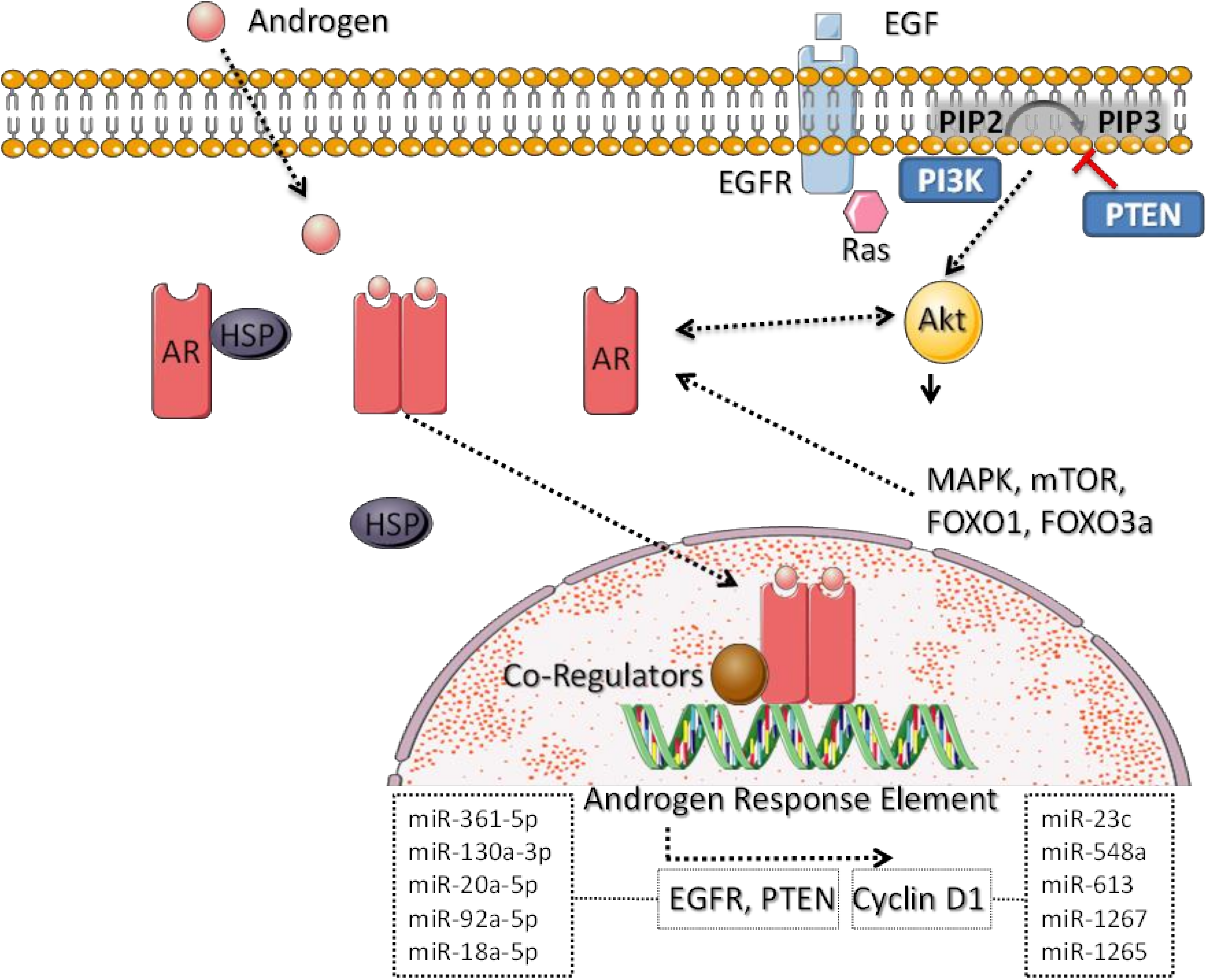
4. Molecular Pathways Dysregulated Specifically in QNBC
5. Molecular Profiles Observed in QNBC
6. Dysregulated miRNAs Related to QNBC and Their Molecular Functions
7. Translational and Clinical Implications of Known Dysregulated miRNAs and Downstream Pathways in QNBC
8. Direction of miRNA Research in QNBC
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brumec, M.; Sobočan, M.; Takač, I.; Arko, D. Clinical Implications of Androgen-Positive Triple-Negative Breast Cancer. Cancers 2021, 13, 1642. [Google Scholar] [CrossRef] [PubMed]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Nikolettos, K.; Dimitroulis, D.; Diamantis, E.; Farmaki, P.; Patsouras, A.; Vouty-Ritsa, E. Molecular Classification and Future Therapeutic Challenges of Triple-negative Breast Cancer. Vivo 2020, 34, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Barton, V.N.; D’Amato, N.C.; Gordon, M.A.; Christenson, J.L.; Elias, A.; Richer, J.K. Androgen Receptor Biology in Triple Negative Breast Cancer: A Case for Classification as AR+ or Quadruple Negative Disease. Horm. Cancer 2015, 6, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayanan, R.; Dalton, J.T. Androgen Receptor: A Complex Therapeutic Target for Breast Cancer. Cancers 2016, 8, 108. [Google Scholar] [CrossRef] [Green Version]
- Gerratana, L.; Basile, D.; Buono, G.; De Placido, S.; Giuliano, M.; Minichillo, S.; Coinu, A.; Martorana, F.; De Santo, I. Androgen receptor in triple negative breast cancer: A potential target for the targetless subtype. Cancer Treat. Rev. 2018, 68, 102–110. [Google Scholar] [CrossRef]
- Qattan, A. Duplicitous Dispositions of Micro-RNAs (miRs) in Breast Cancer. In Genes and Cancer; Lemamy, G.-J., Ed.; IntechOpen: London, UK, 2019; Available online: https://www.intechopen.com/books/genes-and-cancer/duplicitous-dispositions-of-micro-rnas-mirs-in-breast-cancer (accessed on 15 September 2020).
- Shi, P.; Chen, C.; Li, X.; Wei, Z.; Liu, Z.; Liu, Y. microRNA-124 suppresses cell proliferation and invasion of triple negative breast cancer cells by targeting STAT3. Mol. Med. Rep. 2019, 19, 3667–3675. Available online: http://www.spandidos-publications.com/10.3892/mmr.2019.10044 (accessed on 14 September 2020). [CrossRef] [Green Version]
- Nakano, K.; Miki, Y.; Hata, S.; Ebata, A.; Takagi, K.; McNamara, K.M.; Sakurai, M.; Masuda, M.; Hirakawa, H.; Ishida, T. Identification of androgen-responsive microRNAs and androgen-related genes in breast cancer. Anticancer Res. 2013, 33, 4811–4819. [Google Scholar]
- Bandini, E.; Fanini, F. microRNAs and Androgen Receptor: Emerging Players in Breast Cancer. Front. Genet. 2019, 10, 203. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Qattan, A. Novel miRNA Targets and Therapies in the Triple-Negative Breast Cancer Microenvironment: An Emerging Hope for a Challenging Disease. Int. J. Mol. Sci. 2020, 21, 8905. [Google Scholar] [CrossRef]
- Manjunath, M.; Choudhary, B. Triple-negative breast cancer: A run-through of features, classification and current therapies. Oncol. Lett. 2021, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Chang-Qing, Y.; Jie, L.; Shi-Qi, Z.; Kun, Z.; Zi-Qian, G.; Ran, X.; Ran, X.; Hui-Meng, L.; Ren-Bin, Z.; Gang, Z.; et al. Recent treatment progress of triple negative breast cancer. Prog. Biophys. Mol. Biol. 2020, 151, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, S.; Saini, G.; Gogineni, K.; Aneja, R. Quadruple-negative breast cancer: Novel implications for a new disease. Breast Cancer Res. 2020, 22, 127. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, J.; Ling, R.; Li, N. Quadruple negative breast cancer. Breast Cancer 2020, 27, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Vera-Badillo, F.E.; Templeton, A.J.; de Gouveia, P.; Diaz-Padilla, I.; Bedard, P.L.; Al-Mubarak, M.; Seruga, B.; Tannock, I.F.; Ocana, A.; Amir, E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, djt319. [Google Scholar] [CrossRef]
- Pistelli, M.; Caramanti, M.; Biscotti, T.; Santinelli, A.; Pagliacci, A.; De Lisa, M.; Ballatore, Z.; Ridolfi, F.; Maccaroni, E. Androgen receptor expression in early triple-negative breast cancer: Clinical significance and prognostic associations. Cancers 2014, 6, 1351–1362. [Google Scholar] [CrossRef] [Green Version]
- Agoff, S.N.; Swanson, P.E.; Linden, H.; Hawes, S.E.; Lawton, T.J. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, and prognostic associations. Am. J. Clin. Pathol. 2003, 120, 725–731. [Google Scholar] [CrossRef]
- Kim, Y.; Jae, E.; Yoon, M. Influence of Androgen Receptor Expression on the Survival Outcomes in Breast Cancer: A Meta-Analysis. J. Breast Cancer 2015, 18, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Qu, Q.; Mao, Y.; Fei, X.; Shen, K. The Impact of Androgen Receptor Expression on Breast Cancer Survival: A Retrospective Study and Meta-Analysis. PLoS ONE 2013, 8, e82650. [Google Scholar] [CrossRef] [Green Version]
- Astvatsaturyan, K.; Yue, Y.; Walts, A.E.; Bose, S. Androgen receptor positive triple negative breast cancer: Clinicopathologic, prognostic, and predictive features. PLoS ONE 2018, 13, e0197827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakaria, Z.; Zulkifle, M.F.; Wan Hasan, W.A.N.; Azhari, A.K.; Abdul Raub, S.H.; Eswaran, J.; Soundararajan, M.; Syed Husain, S.N.A. Epidermal growth factor receptor (EGFR) gene alteration and protein overexpression in Malaysian triple-negative breast cancer (TNBC) cohort. Onco. Targets Ther. 2019, 12, 7749–7756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Doane, A.S.; Danso, M.; Lal, P.; Donaton, M.; Zhang, L.; Hudis, C.; Gerald, W.L. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 2006, 25, 3994–4008. [Google Scholar] [CrossRef] [Green Version]
- Gucalp, A.; Tolaney, S.; Isakoff, S.J.; Ingle, J.N.; Liu, M.C.; Carey, L.A.; Blackwell, K.; Rugo, H.; Nabell, L.; Forero, A. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin. Cancer Res. 2013, 19, 5505–5512. [Google Scholar] [CrossRef] [Green Version]
- Kulshreshtha, P.; Chakraborty, A.; Singh, L.; Mishra, A.K.; Bhatnagar, D.; Saxena, S. Androgen receptor status predicts response to chemotherapy, not risk of breast cancer in Indian women. World J. Surg. Oncol. 2010, 8, 64. [Google Scholar]
- Loibl, S.; Müller, B.M.; von Minckwitz, G.; Schwabe, M.; Roller, M.; Darb-Esfahani, S.; Ataseven, B.; du Bois, A.; Fissler-Eckhoff, A.; Gerber, B. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2011, 130, 477. [Google Scholar] [CrossRef]
- Gelmann, E.P. Molecular Biology of the Androgen Receptor. J. Clin. Oncol. 2002, 20, 3001–3015. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, F.; Zhang, W.; Song, W.; Liu, Y.; Guan, X. The Androgen Receptor Promotes Cellular Proliferation by Suppression of G-Protein Coupled Estrogen Receptor Signaling in Triple-Negative Breast Cancer. Cell. Physiol. Biochem. 2017, 43, 2047–2061. [Google Scholar] [CrossRef]
- Zhu, A.; Li, Y.; Song, W.; Xu, Y.; Yang, F.; Zhang, W.; Yin, Y.; Guan, X. Antiproliferative Effect of Androgen Receptor Inhibition in Mesenchymal Stem-Like Triple-Negative Breast Cancer. Cell. Physiol. Biochem. 2016, 38, 1003–1014. [Google Scholar] [CrossRef]
- Tang, D.; Xu, S.; Zhang, Q.; Zhao, W. The expression and clinical significance of the androgen receptor and E-cadherin in triple-negative breast cancer. Med. Oncol. 2012, 29, 526–533. [Google Scholar] [CrossRef]
- Liu, Y.-N.; Liu, Y.; Lee, H.-J.; Hsu, Y.-H.; Chen, J.-H. Activated androgen receptor downregulates E-cadherin gene expression and promotes tumor metastasis. Mol. Cell Biol. 2008, 28, 7096–7108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisermann, K.; Fraizer, G. The Androgen Receptor and VEGF: Mechanisms of Androgen-Regulated Angiogenesis in Prostate Cancer. Cancers 2017, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Kwilas, A.R.; Ardiani, A.; Gameiro, S.R.; Richards, J.; Hall, A.B.; Hodge, J.W. Androgen deprivation therapy sensitizes triple negative breast cancer cells to immune-mediated lysis through androgen receptor independent modulation of osteoprotegerin. Oncotarget 2016, 7, 23498–23511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polkinghorn, W.R.; Parker, J.S.; Lee, M.X.; Kass, E.M.; Spratt, D.E.; Iaquinta, P.J.; Arora, V.K.; Yen, W.-F.; Cai, L.; Zheng, D.; et al. Androgen Receptor Signaling Regulates DNA Repair in Prostate Cancers. Cancer Discov. 2013, 3, 1245–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.D.; Yadav, D.K. TNBC: Potential Targeting of Multiple Receptors for a Therapeutic Breakthrough, Nanomedicine, and Immunotherapy. Biomedicines 2021, 9, 876. [Google Scholar] [CrossRef] [PubMed]
- McNamara, K.M.; Moore, N.L.; Hickey, T.E.; Sasano, H.; Tilley, W.D. Complexities of androgen receptor signalling in breast cancer. Endocr.-Relat. Cancer 2014, 21, T161–T181. [Google Scholar] [CrossRef] [PubMed]
- Rangel, N.; Rondon-Lagos, M.; Annaratone, L.; Aristizábal-Pachon, A.F.; Cassoni, P.; Sapino, A.; Castellano, I. AR/ER Ratio Correlates with Expression of Proliferation Markers and with Distinct Subset of Breast Tumors. Cells 2020, 9, E1064. [Google Scholar] [CrossRef]
- Liao, R.S.; Ma, S.; Miao, L.; Li, R.; Yin, Y.; Raj, G.V. Androgen receptor-mediated non-genomic regulation of prostate cancer cell proliferation. Transl. Androl. Urol. 2013, 2, 187. [Google Scholar]
- Pang, T.P.S.; Clarke, M.V.; Ghasem-Zadeh, A.; Lee, N.K.L.; Davey, R.A.; MacLean, H.E. A physiological role for androgen actions in the absence of androgen receptor DNA binding activity. Mol. Cell Endocrinol. 2012, 348, 189–197. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar] [PubMed]
- Coussy, F.; Lavigne, M.; de Koning, L.; Botty, R.E.; Nemati, F.; Naguez, A.; Bataillon, G.; Ouine, B.; Dahmani, A.; Montaudon, E. Response to mTOR and PI3K inhibitors in enzalutamide-resistant luminal androgen receptor triple-negative breast cancer patient-derived xenografts. Theranostics 2020, 10, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.K.; Sadar, M.D. Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front. Endocrinol. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; He, X.; Ngeow, J.; Eng, C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum. Mol. Genet. 2012, 21, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Hon, J.D.C.; Singh, B.; Sahin, A.; Du, G.; Wang, J.; Wang, V.Y.; Wang, V.Y.; Deng, F.-M.; Zhang, D.Y.; Monaco, M.E. Breast cancer molecular subtypes: From TNBC to, Q.N.B.C. Am. J. Cancer Res. 2016, 6, 1864–1872. [Google Scholar] [PubMed]
- Saini, G.; Bhattarai, S.; Gogineni, K.; Aneja, R. Quadruple-Negative Breast Cancer: An Uneven Playing Field. JCO Glob. Oncol. 2020, 6, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Tripathi, S.; Hughley, R.; He, Q.; Bae, S.; Karanam, B.; Martini, R.; Newman, L.; Colomb, W. AR negative triple negative or “quadruple negative” breast cancers in African American women have an enriched basal and immune signature. PLoS ONE 2018, 13, e0196909. [Google Scholar] [CrossRef] [Green Version]
- Angajala, A.; Mothershed, E.; Davis, M.B.; Tripathi, S.; He, Q.; Bedi, D.; Dean-Colomb, W.; Yates, C. Quadruple Negative Breast Cancers (QNBC) Demonstrate Subtype Consistency among Primary and Recurrent or Metastatic Breast Cancer. Transl. Oncol. 2019, 12, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Wang, J.; Wen, X.; Marcus, M.T.; Daniels, G.; Zhang, D.Y.; Ye, F.; Wang, L.H.; Du, X.; et al. Long chain fatty Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone resistance in human breast cancer. PLoS ONE 2013, 8, e77060. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.E.; Creighton, C.J.; Lee, P.; Zou, X.; Topham, M.K.; Stafforini, D.M. Expression of Long-chain Fatty Acyl-CoA Synthetase 4 in Breast and Prostate Cancers Is Associated with Sex Steroid Hormone Receptor Negativity. Transl. Oncol. 2010, 3, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Beltran, A.S.; Graves, L.M.; Blancafort, P. Novel role of Engrailed 1 as a prosurvival transcription factor in basal-like breast cancer and engineering of interference peptides block its oncogenic function. Oncogene 2014, 33, 4767–4777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peluffo, G.; Subedee, A.; Harper, N.W.; Kingston, N.; Jovanović, B.; Flores, F.; Stevens, L.E.; Beca, F.; Trinh, A. EN1 Is a Transcriptional Dependency in Triple-Negative Breast Cancer Associated with Brain Metastasis. Cancer Res. 2019, 79, 4173–4183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorolla, A.; Wang, E.; Clemons, T.D.; Evans, C.W.; Plani-Lam, J.H.; Golden, E.; Dessauvagie, B.; Redfern, A.D.; Swaminathan-Iyer, K.; Blancafort, P. Triple-hit therapeutic approach for triple negative breast cancers using docetaxel nanoparticles, EN1-iPeps and RGD peptides. Nanomed. Nanotechnol. Biol. Med. 2019, 20, 102003. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, J.; Lin, Y.; Sun, X.; Zhang, J.; Cheng, Z. Identification of MiR-211-5p as a tumor suppressor by targeting ACSL4 in Hepatocellular Carcinoma. J. Transl. Med. 2020, 18, 326. [Google Scholar] [CrossRef]
- Cui, W.; Zhang, S.; Shan, C.; Zhou, L.; Zhou, Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013, 280, 3962–3974. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Yu, J.; Dai, Y.; Li, J.; Guo, M.; Song, J.; Zhou, X. Overexpression of miR-361-5p in triple-negative breast cancer (TNBC) inhibits migration and invasion by targeting RQCD1 and inhibiting the EGFR/PI3K/Akt pathway. Bosn. J. Basic Med. Sci. 2019, 19, 52–59. [Google Scholar] [CrossRef]
- Uva, P.; Cossu-Rocca, P.; Loi, F.; Pira, G.; Murgia, L.; Orrù, S.; Floris, M.; Muroni, M.R.; Sanges, F.; Carru, C. miRNA-135b Contributes to Triple Negative Breast Cancer Molecular Heterogeneity: Different Expression Profile in Basal-like Versus non-Basal-like Phenotypes. Int. J. Med. Sci. 2018, 15, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Angajala, A.; Hughley, R.; DeanColomb, W.; Tripathi, S.; Tan, M.; Yates, C. Abstract 4769: Identification of differentially expressed microRNAs in African American women with quadruple-negative breast cancer. Cancer Res. 2018, 78 (Suppl. 13), 4769. [Google Scholar]
- Bhattarai, S.; Sugita, B.M.; Bortoletto, S.M.; Fonseca, A.S.; Cavalli, L.R.; Aneja, R. QNBC Is Associated with High Genomic Instability Characterized by Copy Number Alterations and miRNA Deregulation. Int. J. Mol. Sci. 2021, 22, 11548. [Google Scholar] [CrossRef]
- Becuwe, P.; Ennen, M.; Klotz, R.; Barbieux, C.; Grandemange, S. Manganese superoxide dismutase in breast cancer: From molecular mechanisms of gene regulation to biological and clinical significance. Free Radic. Biol. Med. 2014, 77, 139–151. [Google Scholar] [CrossRef]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Lehmann, B.D.; Bauer, J.A.; Schafer, J.M.; Pendleton, C.S.; Tang, L.; Johnson, K.C.; Chen, X.; Balko, J.M.; Gómez, H.; Arteaga, C.L. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vora, S.R.; Juric, D.; Kim, N.; Mino-Kenudson, M.; Huynh, T.; Costa, C.; Lockerman, E.L.; Pollack, S.F.; Liu, M.; Li, X. CDK 4/6 Inhibitors Sensitize PIK3CA Mutant Breast Cancer to PI3K Inhibitors. Cancer Cell 2014, 26, 136–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef] [Green Version]
- Omarini, C.; Guaitoli, G.; Pipitone, S.; Moscetti, L.; Cortesi, L.; Cascinu, S.; Piacentini, F. Neoadjuvant treatments in triple-negative breast cancer patients: Where we are now and where we are going. Cancer Manag. Res. 2018, 10, 91–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burguin, A.; Diorio, C.; Durocher, F. Breast Cancer Treatments: Updates and New Challenges. J. Pers. Med. 2021, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Angulo, A.M.; Akcakanat, A.; Liu, S.; Green, M.C.; Murray, J.L.; Chen, H.; Palla, S.L.; Koenig, K.B.; Brewster, A.M.; Valero, V. Open-label randomized clinical trial of standard neoadjuvant chemotherapy with paclitaxel followed by FEC versus the combination of paclitaxel and everolimus followed by FEC in women with triple receptor-negative breast cancer†. Ann. Oncol. 2014, 25, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Basho, R.K.; Gilcrease, M.; Murthy, R.K.; Helgason, T.; Karp, D.D.; Meric-Bernstam, F.; Hess, K.R.; Herbrich, S.M.; Valero, V.; Albarracin, C. Targeting the PI3K/AKT/mTOR Pathway for the Treatment of Mesenchymal Triple-Negative Breast Cancer: Evidence from a Phase 1 Trial of mTOR Inhibition in Combination With Liposomal Doxorubicin and Bevacizumab. JAMA Oncol. 2017, 3, 509–515. [Google Scholar] [CrossRef]
- Wawruszak, A.; Halasa, M.; Okon, E.; Kukula-Koch, W.; Stepulak, A. Valproic Acid and Breast Cancer: State of the Art in 2021. Cancers 2021, 13, 3409. [Google Scholar] [CrossRef]


| Expression-Based Molecular Classification | ||||||
|---|---|---|---|---|---|---|
| BL1 (17.9%) | BL2 (11.1%) | IM (21.1%) | M (20.8%) | MSL (6.5%) | LAR (9.2%) | Unstable (13.5%) |
| Molecular Targets | ||||||
| PARP1,CHEK1, RAD51, PLK1, TTK, AURKA/B | EGFR, mTOR, MET,EPHA2 | JAK1/2, STAT,BTK, NFκB,LYN,IRF1 | PI3K,IGF1R mTOR, SRC, FGFR PDGFR | PI3K, mTOR, MEK1/2, SRC, IGF1R, FGFR, PDGFR, NFkB | AR, HSP90, PI3K, FGFR4 | PARP1, RAD51, PLK1, AURKA/BTTK,CHEK1 |
| Treatment | ||||||
| Antimitotic agents (platinum, PARPi) | TKI,mTORi, eribulin mesylate | Anti-androgen | ||||
| Genetic Mutation-Based Profiling | ||||||
| BRCA1/2, PARPi | PIK3CA (PI3Ki) | PD-L1 (Immunotherapy) | CDK4/6 (CDK4/6i) | TP53 | PTEN | EGFR |
| Expression-Based Cellular Classification | ||||||
| Basal-like | Claudin-high | Claudin-low | LAR | |||
| Androgen Receptor (IHC)-Based Profiling | ||||||
| Androgen receptor-positive (Androgen antagonists, e.g., bicalutamide, anzalutamide) | Androgen receptor-negative (Restricted to chemotherapy) | |||||
| Molecular Targets in QNBC | ||||||
| Cell metabolism | acyl-CoA synthetase4 (ASCL4) | |||||
| Tumor immune microenvironment | Tumor-infiltrating lymphocytes (TIL), Tumor necrosis factor superfamily member 10 (TNFSF10), Programmed death ligand 1 (PD-L1) | |||||
| Cell growth and proliferation | EGFR,HER4, CK5/6,CDK6,PTEN,ki-67 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qattan, A.; Al-Tweigeri, T.; Suleman, K. Translational Implications of Dysregulated Pathways and microRNA Regulation in Quadruple-Negative Breast Cancer. Biomedicines 2022, 10, 366. https://doi.org/10.3390/biomedicines10020366
Qattan A, Al-Tweigeri T, Suleman K. Translational Implications of Dysregulated Pathways and microRNA Regulation in Quadruple-Negative Breast Cancer. Biomedicines. 2022; 10(2):366. https://doi.org/10.3390/biomedicines10020366
Chicago/Turabian StyleQattan, Amal, Taher Al-Tweigeri, and Kausar Suleman. 2022. "Translational Implications of Dysregulated Pathways and microRNA Regulation in Quadruple-Negative Breast Cancer" Biomedicines 10, no. 2: 366. https://doi.org/10.3390/biomedicines10020366
APA StyleQattan, A., Al-Tweigeri, T., & Suleman, K. (2022). Translational Implications of Dysregulated Pathways and microRNA Regulation in Quadruple-Negative Breast Cancer. Biomedicines, 10(2), 366. https://doi.org/10.3390/biomedicines10020366






