Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment
Abstract
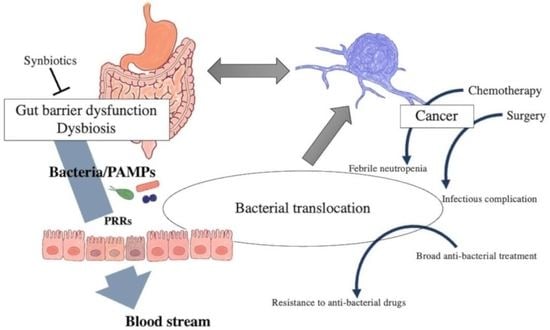
1. Introduction
2. Search Strategy
3. Pathogen-Associated Molecular Patterns
4. Pattern Recognition Receptors
5. Bacterial Translocation and Carcinogenesis
6. Bacterial Translocation and Cancer Surgery, Perioperative Management
7. Febrile Neutropenia
8. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Shi, C.; Yang, Y.; Xia, Y.; Okugawa, Y.; Yang, J.; Liang, Y.; Chen, H.; Zhang, P.; Wang, F.; Han, H.; et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2016, 65, 1470–1481. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Y.; Yu, J.; Chen, T.; Wu, Y.; Shi, L.; Li, Q.; Wu, J.; Fu, X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 2017, 8, 31802–31814. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, H.; Ono, S.; Efron, P.A.; Scumpia, P.O.; Moldawer, L.L.; Mochizuki, H. Role of Toll-like receptors in the development of sepsis. Shock 2008, 29, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Kouzu, K.; Tsujimoto, H.; Kishi, Y.; Ueno, H.; Shinomiya, N. Role of microbial infection-induced inflammation in the development of gastrointestinal cancers. Medicines 2021, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.D.; Owens, W.E. Inhibition of translocation of viable Escherichia coli from the gastrointestinal tract of mice by bacterial antagonism. Infect. Immun. 1979, 25, 820–827. [Google Scholar] [CrossRef]
- Rubartelli, A.; Lotze, M.T. Inside, outside, upside down: Damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007, 28, 429–436. [Google Scholar] [CrossRef]
- De Jong, P.R.; González-Navajas, J.M.; Jansen, N.J. The digestive tract as the origin of systemic inflammation. Crit. Care 2016, 20, 279. [Google Scholar] [CrossRef] [PubMed]
- Assimakopoulos, S.F.; Triantos, C.; Thomopoulos, K.; Fligou, F.; Maroulis, I.; Marangos, M.; Gogos, C.A. Gut-origin sepsis in the critically ill patient: Pathophysiology and treatment. Infection 2018, 46, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Tsujimoto, H.; Yamauchi, A.; Hiraki, S.; Takayama, E.; Mochizuki, H. Detection of microbial DNA in the blood of surgical patients for diagnosing bacterial translocation. World J. Surg. 2005, 29, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, H.; Ono, S.; Mochizuki, H. Role of translocation of pathogen-associated molecular patterns in sepsis. Dig. Surg. 2009, 26, 100–109. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Caballero, S.; Pamer, E.G. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 2015, 33, 227–256. [Google Scholar] [CrossRef]
- Simpson, M.E.; Petri, W.A., Jr. TLR2 as a therapeutic target in bacterial infection. Trends Mol. Med. 2020, 26, 715–717. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- Miao, E.A.; Andersen-Nissen, E.; Warren, S.E.; Aderem, A. TLR5 and Ipaf: Dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 2007, 29, 275–288. [Google Scholar] [CrossRef]
- Cui, B.; Liu, X.; Fang, Y.; Zhou, P.; Zhang, Y.; Wang, Y. Flagellin as a vaccine adjuvant. Expert Rev. Vaccines 2018, 17, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Takeuchi, O.; Akira, S. TLR9 as a key receptor for the recognition of DNA. Adv. Drug Deliv. Rev. 2008, 60, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Echizen, K.; Hirose, O.; Maeda, Y.; Oshima, M. Inflammation in gastric cancer: Interplay of the COX-2/prostaglandin E2 and Toll-like receptor/MyD88 pathways. Cancer Sci. 2016, 107, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.D.; Ji, C.B.; Li, S.B.; Yan, F.; Gu, Q.S.; Balic, J.J.; Yu, L.; Li, J.K. Toll-like receptor 2 stimulation promotes colorectal cancer cell growth via PI3K/Akt and NF-κB signaling pathways. Int. Immunopharmacol. 2018, 59, 375–383. [Google Scholar] [CrossRef]
- Helminen, O.; Huhta, H.; Lehenkari, P.P.; Saarnio, J.; Karttunen, T.J.; Kauppila, J.H. Nucleic acid-sensing toll-like receptors 3, 7 and 8 in esophageal epithelium, barrett’s esophagus, dysplasia and adenocarcinoma. Oncoimmunology 2016, 5, e1127495. [Google Scholar] [CrossRef][Green Version]
- Sato, Y.; Motoyama, S.; Wakita, A.; Kawakita, Y.; Liu, J.; Nagaki, Y.; Nanjo, H.; Terata, K.; Imai, K.; Saito, H.; et al. TLR3 expression status predicts prognosis in patients with advanced thoracic esophageal squamous cell carcinoma after esophagectomy. Am. J. Surg. 2018, 216, 319–325. [Google Scholar] [CrossRef]
- Kohtz, P.D.; Halpern, A.L.; Eldeiry, M.A.; Hazel, K.; Kalatardi, S.; Ao, L.; Meng, X.; Reece, T.B.; Fullerton, D.A.; Weyant, M.J. Toll-Like Receptor-4 Is a Mediator of Proliferation in Esophageal Adenocarcinoma. Ann. Thorac. Surg. 2019, 107, 233–241. [Google Scholar] [CrossRef]
- Sato, Y.; Motoyama, S.; Wakita, A.; Kawakita, Y.; Liu, J.; Nagaki, Y.; Nanjo, H.; Ito, S.; Terata, K.; Imai, K.; et al. High TLR4 expression predicts a poor prognosis after esophagectomy for advanced thoracic esophageal squamous cell carcinoma. Esophagus 2020, 17, 408–416. [Google Scholar] [CrossRef]
- Ito, N.; Tsujimoto, H.; Ueno, H.; Xie, Q.; Shinomiya, N. Helicobacter pylori-mediated immunity and signaling transduction in gastric cancer. J. Clin. Med. 2020, 9, 3699. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 2017, 152, 851–866. [Google Scholar] [CrossRef]
- Kasurinen, A.; Hagström, J.; Laitinen, A.; Kokkola, A.; Böckelman, C.; Haglund, C. Evaluation of toll-like receptors as prognostic biomarkers in gastric cancer: High tissue TLR5 predicts a better outcome. Sci. Rep. 2019, 9, 12553. [Google Scholar] [CrossRef] [PubMed]
- Beilmann-Lehtonen, I.; Hagström, J.; Mustonen, H.; Koskensalo, S.; Haglund, C.; Böckelman, C. High Tissue TLR5 Expression Predicts Better Outcomes in Colorectal Cancer Patients. Oncology 2021, 99, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kordahi, M.C.; Chac, D.; DePaolo, R.W. Toll-like receptor-6 signaling prevents inflammation and impacts composition of the microbiota during inflammation-induced colorectal cancer. Cancer Prev. Res. 2020, 13, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Sheyhidin, I.; Nabi, G.; Hasim, A.; Zhang, R.P.; Ainiwaer, J.; Ma, H.; Wang, H. Overexpression of TLR3, TLR4, TLR7 and TLR9 in esophageal squamous cell carcinoma. World J. Gastroenterol. 2011, 17, 3745–3751. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Y.; Xiong, Y.; Liu, S.; Qiu, C.; Zhu, Z.; Mao, H.; Yu, M.; Wang, X. TLR 7/8 agonist reverses oxaliplatin resistance in colorectal cancer via directing the myeloid-derived suppressor cells to tumoricidal M1-macrophages. Cancer Lett. 2020, 469, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.R.; Wu, J.; Yao, X.Y.; Huang, J.; Wang, X.Y. Helicobacter pylori DNA promotes cellular proliferation, migration, and invasion of gastric cancer by activating toll-like receptor 9. Saudi J. Gastroenterol. 2019, 25, 181–187. [Google Scholar] [CrossRef]
- Luo, Q.; Zeng, L.; Tang, C.; Zhang, Z.; Chen, Y.; Zeng, C. TLR9 induces colitis-associated colorectal carcinogenesis by regulating NF-κB expression levels. Oncol. Lett. 2020, 20, 110. [Google Scholar] [CrossRef]
- Dowling, J.K.; Mansell, A. Toll-like receptors: The swiss army knife of immunity and vaccine development. Clin. Transl. Immunol. 2016, 5, e85. [Google Scholar] [CrossRef]
- Wang, X.D.; Gao, N.N.; Diao, Y.W.; Liu, Y.; Gao, D.; Li, W.; Wan, Y.Y.; Zhong, J.J.; Jin, G.Y. Conjugation of toll-like receptor-7 agonist to gastric cancer antigen MG7-Ag exerts antitumor effects. World J. Gastroenterol. 2015, 21, 8052–8060. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Man, S.M. Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Lu, Z.; Liu, L.; Li, R.; Liang, Z.; Shen, M.; Xu, H.; Ren, D.; Ji, M.; Yuan, S.; et al. NOD-like receptor signaling in inflammation-associated cancers: From functions to targeted therapies. Phytomedicine 2019, 64, 152925. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, X.; Wu, L.; Wang, X.; Liu, Z. The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl. Res. 2017, 190, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xu, W.Y.; Hu, Z.; Zhang, H.; Shen, Y.; Lu, S.; Wei, C.; Wang, Z.G. RNA virus receptor Rig-I monitors gut microbiota and inhibits colitis-associated colorectal cancer. J. Exp. Clin. Cancer Res. 2017, 36, 2. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Hou, Z.; Han, Q.; Zhang, C.; Tian, Z.; Zhang, J. Poly(I:C) exhibits an anti-cancer effect in human gastric adenocarcinoma cells which is dependent on RLRs. Int. Immunopharmacol. 2013, 17, 814–820. [Google Scholar] [CrossRef]
- Drouin, M.; Saenz, J.; Chiffoleau, E. C-type lectin-like receptors: Head or tail in cell death immunity. Front. Immunol 2020, 11, 251. [Google Scholar] [CrossRef]
- Li, C.; Zhang, J.; Wu, H.; Li, L.; Yang, C.; Song, S.; Peng, P.; Shao, M.; Zhang, M.; Zhao, J.; et al. Lectin-like oxidized low-density lipoprotein receptor-1 facilitates metastasis of gastric cancer through driving epithelial-mesenchymal transition and PI3K/Akt/GSK3β activation. Sci. Rep. 2017, 7, 45275. [Google Scholar] [CrossRef]
- Murdocca, M.; Mango, R.; Pucci, S.; Biocca, S.; Testa, B.; Capuano, R.; Paolesse, R.; Sanchez, M.; Orlandi, A.; di Natale, C.; et al. The lectin-like oxidized LDL receptor-1: A new potential molecular target in colorectal cancer. Oncotarget 2016, 7, 14765–14780. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Uesugi, T.; Froh, M.; Arteel, G.E.; Bradford, B.U.; Thurman, R.G. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology 2001, 34, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, A.T.; Navas, T.A.; Lyons, S.; Godowski, P.J.; Madara, J.L. Cutting edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 2001, 167, 1882–1885. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Yan, H.X.; Liu, Q.; Yang, W.; Wu, H.P.; Dong, W.; Tang, L.; Lin, Y.; He, Y.Q.; Zou, S.S.; et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 2010, 52, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, F.; Yang, J.; Zhong, M.; Zhang, E.; Li, Y.; Zhou, D.; Cao, Y.; Li, W.; Yu, J.; et al. Over-activation of TLR5 signaling by high-dose flagellin induces liver injury in mice. Cell Mol. Immunol. 2015, 12, 729–742. [Google Scholar] [CrossRef]
- Jing, Y.Y.; Han, Z.P.; Sun, K.; Zhang, S.S.; Hou, J.; Liu, Y.; Li, R.; Gao, L.; Zhao, X.; Zhao, Q.D.; et al. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Med. 2012, 10, 98. [Google Scholar] [CrossRef]
- Fedirko, V.; Tran, H.Q.; Gewirtz, A.T.; Stepien, M.; Trichopoulou, A.; Aleksandrova, K.; Olsen, A.; Tjønneland, A.; Overvad, K.; Carbonnel, F.; et al. Exposure to bacterial products lipopolysaccharide and flagellin and hepatocellular carcinoma: A nested case-control study. BMC Med. 2017, 15, 72. [Google Scholar] [CrossRef]
- Yang, B.; Petrick, J.L.; Thistle, J.E.; Pinto, L.A.; Kemp, T.J.; Tran, H.Q.; Gewirtz, A.T.; Waterboer, T.; Fedirko, V.; Jenab, M.; et al. Bacterial translocation and risk of liver cancer in a Finnish cohort. Cancer Epidemiol. Biomark. Prev. 2019, 28, 807–813. [Google Scholar] [CrossRef]
- Chassaing, B.; Etienne-Mesmin, L.; Gewirtz, A.T. Microbiota-liver axis in hepatic disease. Hepatology 2014, 59, 328–339. [Google Scholar] [CrossRef]
- Méndez-Sánchez, N.; Valencia-Rodriguez, A.; Vera-Barajas, A.; Abenavoli, L.; Scarpellini, E.; Ponciano-Rodriguez, G.; Wang, D.Q. The mechanism of dysbiosis in alcoholic liver disease leading to liver cancer. Hepatoma Res. 2020, 6. [Google Scholar] [CrossRef]
- Mitsuhashi, K.; Nosho, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kurihara, H.; Kanno, S.; Igarashi, H.; Naito, T.; Adachi, Y.; et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015, 6, 7209–7220. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019, 178, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Keramidaris, D.; Koronakis, N.; Lagoudianakis, E.E.; Pappas, A.; Koukoutsis, I.; Chrysikos, I.; Karavitis, G.; Toutouzas, K.; Manouras, A. Procalcitonin in patients with colorectal cancer. J. Buon. 2013, 18, 623–628. [Google Scholar] [PubMed]
- Branche, A.; Neeser, O.; Mueller, B.; Schuetz, P. Procalcitonin to guide antibiotic decision making. Curr. Opin. Infect. Dis. 2019, 32, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Matzaraki, V.; Alexandraki, K.I.; Venetsanou, K.; Piperi, C.; Myrianthefs, P.; Malamos, N.; Giannakakis, T.; Karatzas, S.; Diamanti-Kandarakis, E.; Baltopoulos, G. Evaluation of serum procalcitonin and interleukin-6 levels as markers of liver metastasis. Clin. Biochem. 2007, 40, 336–342. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Kobayashi, M.; Sugasawa, H.; Ono, S.; Kishi, Y.; Ueno, H. Potential mechanisms of tumor progression associated with postoperative infectious complications. Cancer Metastasis Rev. 2021, 40, 285–296. [Google Scholar] [CrossRef]
- Woodcock, N.P.; Sudheer, V.; El-Barghouti, N.; Perry, E.P.; MacFie, J. Bacterial translocation in patients undergoing abdominal aortic aneurysm repair. Br. J. Surg. 2000, 87, 439–442. [Google Scholar] [CrossRef]
- Kubo, N.; Sakurai, K.; Tamura, T.; Toyokawa, T.; Tanaka, H.; Muguruma, K.; Yashiro, M.; Maeda, K.; Ohira, M. The duration of systemic inflammatory response syndrome is a reliable indicator of long-term survival after curative esophagectomy for esophageal squamous cell carcinoma. Esophagus 2021, 18, 548–558. [Google Scholar] [CrossRef]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumor Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef]
- Katsurahara, K.; Shiozaki, A.; Fujiwara, H.; Konishi, H.; Kudou, M.; Shoda, K.; Arita, T.; Kosuga, T.; Morimura, R.; Murayama, Y.; et al. Relationship between postoperative crp and prognosis in thoracic esophageal squamous cell carcinoma. Anticancer Res. 2018, 38, 6513–6518. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Ogura, H.; Goto, M.; Asahara, T.; Nomoto, K.; Morotomi, M.; Yoshiya, K.; Matsushima, A.; Sumi, Y.; Kuwagata, Y.; et al. Altered gut flora and environment in patients with severe SIRS. J. Trauma 2006, 60, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.; Windsor, A.C.; Welsh, F.; Barclay, G.R.; Guillou, P.J.; Reynolds, J.V. Lack of correlation between failure of gut barrier function and septic complications after major upper gastrointestinal surgery. Ann. Surg. 2000, 231, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Ohigashi, S.; Sudo, K.; Kobayashi, D.; Takahashi, T.; Nomoto, K.; Onodera, H. Significant changes in the intestinal environment after surgery in patients with colorectal cancer. J. Gastrointest. Surg. 2013, 17, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Yang, Y.; Wang, H.; Wang, H.; Yu, X.; Lu, Y.; Shen, S.; Teng, L. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine 2019, 98, e16626. [Google Scholar] [CrossRef]
- Reddy, B.S.; Gatt, M.; Sowdi, R.; MacFie, J. Surgical manipulation of the large intestine increases bacterial translocation in patients undergoing elective colorectal surgery. Colorectal. Dis. 2006, 8, 596–600. [Google Scholar] [CrossRef]
- Nishigaki, E.; Abe, T.; Yokoyama, Y.; Fukaya, M.; Asahara, T.; Nomoto, K.; Nagino, M. The detection of intraoperative bacterial translocation in the mesenteric lymph nodes is useful in predicting patients at high risk for postoperative infectious complications after esophagectomy. Ann. Surg. 2014, 259, 477–484. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Fukaya, M.; Mizuno, T.; Ebata, T.; Asahara, T.; Nagino, M. Clinical importance of "occult-bacterial translocation" in patients undergoing highly invasive gastrointestinal surgery: A review. Surg. Today 2021, 51, 485–492. [Google Scholar] [CrossRef]
- Schietroma, M.; Pessia, B.; Carlei, F.; Cecilia, E.M.; De Santis, G.; Amicucci, G. Laparoscopic versus open colorectal surgery for colon cancer: The effect of surgical trauma on the bacterial translocation. A prospective randomized study. Am. J. Surg. 2015, 210, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, E.; Kotzampassi, K.; Papanotas, K.; Heliadis, N.; Sarris, K. Gut ischemia, oxidative stress, and bacterial translocation in elevated abdominal pressure in rats. World J. Surg. 1996, 20, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Han, T.; Han, C.; Sun, H.; Yang, X.; Zhang, D.; Ni, X. Propofol Regulates the TLR4/NF-κB Pathway Through miRNA-155 to Protect Colorectal Cancer Intestinal Barrier. Inflammation 2021, 44, 2078–2090. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, G.; Nagino, M.; Nishio, H.; Ebata, T.; Takagi, K.; Asahara, T.; Nomoto, K.; Nimura, Y. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery: A randomized controlled trial. Ann. Surg. 2006, 244, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, S.; Takatsuki, M.; Hidaka, M.; Soyama, A.; Ichikawa, T.; Kanematsu, T. Perioperative synbiotic treatment to prevent infectious complications in patients after elective living donor liver transplantation: A prospective randomized study. Am. J. Surg. 2011, 201, 498–502. [Google Scholar] [CrossRef]
- Usami, M.; Miyoshi, M.; Kanbara, Y.; Aoyama, M.; Sakaki, H.; Shuno, K.; Hirata, K.; Takahashi, M.; Ueno, K.; Tabata, S.; et al. Effects of perioperative synbiotic treatment on infectious complications, intestinal integrity, and fecal flora and organic acids in hepatic surgery with or without cirrhosis. JPEN J. Parenter. Enteral Nutr. 2011, 35, 317–328. [Google Scholar] [CrossRef]
- Tanaka, K.; Yano, M.; Motoori, M.; Kishi, K.; Miyashiro, I.; Ohue, M.; Ohigashi, H.; Asahara, T.; Nomoto, K.; Ishikawa, O. Impact of perioperative administration of synbiotics in patients with esophageal cancer undergoing esophagectomy: A prospective randomized controlled trial. Surgery 2012, 152, 832–842. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Nishigaki, E.; Abe, T.; Fukaya, M.; Asahara, T.; Nomoto, K.; Nagino, M. Randomized clinical trial of the effect of perioperative synbiotics versus no synbiotics on bacterial translocation after oesophagectomy. Br. J. Surg. 2014, 101, 189–199. [Google Scholar] [CrossRef]
- Okazaki, M.; Matsukuma, S.; Suto, R.; Miyazaki, K.; Hidaka, M.; Matsuo, M.; Noshima, S.; Zempo, N.; Asahara, T.; Nomoto, K. Perioperative synbiotic therapy in elderly patients undergoing gastroenterological surgery: A prospective, randomized control trial. Nutrition 2013, 29, 1224–1230. [Google Scholar] [CrossRef]
- Komatsu, S.; Sakamoto, E.; Norimizu, S.; Shingu, Y.; Asahara, T.; Nomoto, K.; Nagino, M. Efficacy of perioperative synbiotics treatment for the prevention of surgical site infection after laparoscopic colorectal surgery: A randomized controlled trial. Surg. Today 2016, 46, 479–490. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; O’Toole, P.W.; Shanahan, F. The gut microbiota in causation, detection, and treatment of cancer. Am. J. Gastroenterol. 2019, 114, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Hughes, W.T.; Armstrong, D.; Bodey, G.P.; Bow, E.J.; Brown, A.E.; Calandra, T.; Feld, R.; Pizzo, P.A.; Rolston, K.V.I.; Shenep, J.L.; et al. 2002 Guidelines for the Use of Antimicrobial Agents in Neutropenic Patients with Cancer. Clin. Infect. Dis. 2002, 34, 730–751. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Matsuoka, H.; Tsukada, J.; Masuda, M.; Ikeda, S.; Matsuishi, E.; Kawano, F.; Izumi, Y.; Uike, N.; Utsunomiya, A.; et al. Cefepime or carbapenem treatment for febrile neutropenia as a single agent is as effective as a combination of 4th-generation cephalosporin + aminoglycosides: Comparative study. Am. J. Hematol. 2002, 71, 248–255. [Google Scholar] [CrossRef]
- van der Velden, W.J.; Herbers, A.H.; Netea, M.G.; Blijlevens, N.M. Mucosal barrier injury, fever and infection in neutropenic patients with cancer: Introducing the paradigm febrile mucositis. Br. J. Haematol. 2014, 167, 441–452. [Google Scholar] [CrossRef]
- Ota, A.; Morita, S.; Matsuoka, A.; Shimokata, T.; Maeda, O.; Mitsuma, A.; Yagi, T.; Asahara, T.; Ando, Y. Detection of bacteria in blood circulation in patients receiving cancer chemotherapy. Int. J. Clin. Oncol. 2020, 25, 210–215. [Google Scholar] [CrossRef]
- Wong, M.; Barqasho, B.; Ohrmalm, L.; Tolfvenstam, T.; Nowak, P. Microbial translocation contribute to febrile episodes in adults with chemotherapy-induced neutropenia. PLoS ONE 2013, 8, e68056. [Google Scholar] [CrossRef]
- Papanicolas, L.E.; Gordon, D.L.; Wesselingh, S.L.; Rogers, G.B. Not just antibiotics: Is cancer chemotherapy driving antimicrobial resistance? Trends Microbiol. 2018, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; Ebringer, L.; Drgona, L.; Mardiak, J.; Trupl, J.; Greksak, R.; Nemova, I.; Oravcova, E.; Zajac, V.; Koza, I. Prevention of febrile neutropenia in cancer patients by probiotic strain Enterococcus faecium M-74. Pilot study phase I. Neoplasma 2005, 52, 159–164. [Google Scholar] [PubMed]
- Mego, M.; Koncekova, R.; Mikuskova, E.; Drgona, L.; Ebringer, L.; Demitrovicova, L.; Nemova, I.; Trupl, J.; Mardiak, J.; Koza, I.; et al. Prevention of febrile neutropenia in cancer patients by probiotic strain Enterococcus faecium M-74. Phase II study. Support. Care Cancer 2006, 14, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.; Clarke, S.; Vitetta, L. The role of adjuvant probiotics to attenuate intestinal inflammatory responses due to cancer treatments. Benef. Microbes 2018, 9, 899–916. [Google Scholar] [CrossRef]
- Motoori, M.; Yano, M.; Miyata, H.; Sugimura, K.; Saito, T.; Omori, T.; Fujiwara, Y.; Miyoshi, N.; Akita, H.; Gotoh, K.; et al. Randomized study of the effect of synbiotics during neoadjuvant chemotherapy on adverse events in esophageal cancer patients. Clin. Nutr. 2017, 36, 93–99. [Google Scholar] [CrossRef]
- Kanazawa, H.; Nagino, M.; Kamiya, S.; Komatsu, S.; Mayumi, T.; Takagi, K.; Asahara, T.; Nomoto, K.; Tanaka, R.; Nimura, Y. Synbiotics reduce postoperative infectious complications: A randomized controlled trial in biliary cancer patients undergoing hepatectomy. Langenbeck’s Arch. Surg. 2005, 390, 104–113. [Google Scholar] [CrossRef]
- McNaught, C.E.; Woodcock, N.P.; Anderson, A.D.G.; MacFie, J. A prospective randomised trial of probiotics in critically ill patients. Clin. Nutr. 2005, 24, 211–219. [Google Scholar] [CrossRef]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Luo, Y.; Ray Chaudhuri, K.; Reynolds, R.; Tan, E.K.; Pettersson, S. The role of gut dysbiosis in Parkinson’s disease: Mechanistic insights and therapeutic options. Brain 2021, 144, 2571–2593. [Google Scholar] [CrossRef]
- Elfil, M.; Kamel, S.; Kandil, M.; Koo, B.B.; Schaefer, S.M. Implications of the Gut Microbiome in Parkinson’s Disease. Mov. Disord. 2020, 35, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Tamtaji, O.R.; Taghizadeh, M.; Daneshvar Kakhaki, R.; Kouchaki, E.; Bahmani, F.; Borzabadi, S.; Oryan, S.; Mafi, A.; Asemi, Z. Clinical and metabolic response to probiotic administration in people with Parkinson’s disease: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. 2019, 38, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Akkasheh, G.; Kashani-Poor, Z.; Tajabadi-Ebrahimi, M.; Jafari, P.; Akbari, H.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z.; Esmaillzadeh, A. Clinical and metabolic response to probiotic administration in patients with major depressive disorder: A randomized, double-blind, placebo-controlled trial. Nutrition 2016, 32, 315–320. [Google Scholar] [CrossRef]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Dong, T.S.; Gupta, A. Influence of early life, diet, and the environment on the microbiome. Clin. Gastroenterol. Hepatol. 2019, 17, 231–242. [Google Scholar] [CrossRef]
- Lofgren, J.L.; Whary, M.T.; Ge, Z.; Muthupalani, S.; Taylor, N.S.; Mobley, M.; Potter, A.; Varro, A.; Eibach, D.; Suerbaum, S.; et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 2011, 140, 210–220. [Google Scholar] [CrossRef]



| TLRs | Exogenous Ligands | Endogenous Ligands | Cancer | Citation |
|---|---|---|---|---|
| TLR1 | Triacyl lipopeptide, LPS, Peptidoglycan | HSP, HMGB1, and Proteoglycans | ||
| TLR 2 | LPS, Peptidoglycan | HSP, HMGB1, Proteoglycans | GC, CRC | [23,24] |
| TLR 3 | Double-stranded RNA | mRNA and tRNA | EAC, ESCC | [25,26] |
| TLR 4 | LPS | Fibronectin, Polysaccharide fragments of heparan sulfate, HSP, Surfactant protein A in the lung epithelium 1, Neutrophil elastase, HMGB1, Biglycan | EAC, ESCC, GC, CRC | [27,28,29,30] |
| TLR 5 | Flagellin | GC, CRC | [31,32] | |
| TLR 6 | Diacyl lipopeptide, Zymosan | CRC | [33] | |
| TLR 7 | Single-stranded RNA | Single-stranded RNA complex | EAC, ESCC, CRC | [25,34,35] |
| TLR 8 | Single-stranded RNA, imidazoquinolines, guanosine analogs | Single-stranded RNA complex | EAC, CRC | [25,35] |
| TLR 9 | Unmethylated CpG DNA | Chromatin–IgG complex | ESCC, GC, CRC | [34,36,37] |
| TLR 10 | HIV-1 proteins | |||
| TLR 11 | Uropathogenic Escherichia coli |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouzu, K.; Tsujimoto, H.; Kishi, Y.; Ueno, H.; Shinomiya, N. Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines 2022, 10, 380. https://doi.org/10.3390/biomedicines10020380
Kouzu K, Tsujimoto H, Kishi Y, Ueno H, Shinomiya N. Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines. 2022; 10(2):380. https://doi.org/10.3390/biomedicines10020380
Chicago/Turabian StyleKouzu, Keita, Hironori Tsujimoto, Yoji Kishi, Hideki Ueno, and Nariyoshi Shinomiya. 2022. "Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment" Biomedicines 10, no. 2: 380. https://doi.org/10.3390/biomedicines10020380
APA StyleKouzu, K., Tsujimoto, H., Kishi, Y., Ueno, H., & Shinomiya, N. (2022). Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines, 10(2), 380. https://doi.org/10.3390/biomedicines10020380