Implications of Heterogeneity of Epithelial-Mesenchymal States in Acromegaly Therapeutic Pharmacologic Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA and RNA Isolation
2.3. E-Cadherin Promoter Methylation Assessment
2.4. Reverse Transcription and Quantitative Polymerase Chain Reaction (RT-qPCR)
2.5. RORC Immunohistochemistry
2.6. Biochemical and Hormonal Assays
2.7. Statistical Analysis
3. Results
3.1. E-Cadherin Expression Did Not Correlate with Promoter Methylation
3.2. SRLs Treatment before Surgery Affected the Expression of Some EMT-Related Genes
3.3. Association of EMT Markers with Clinical Variables
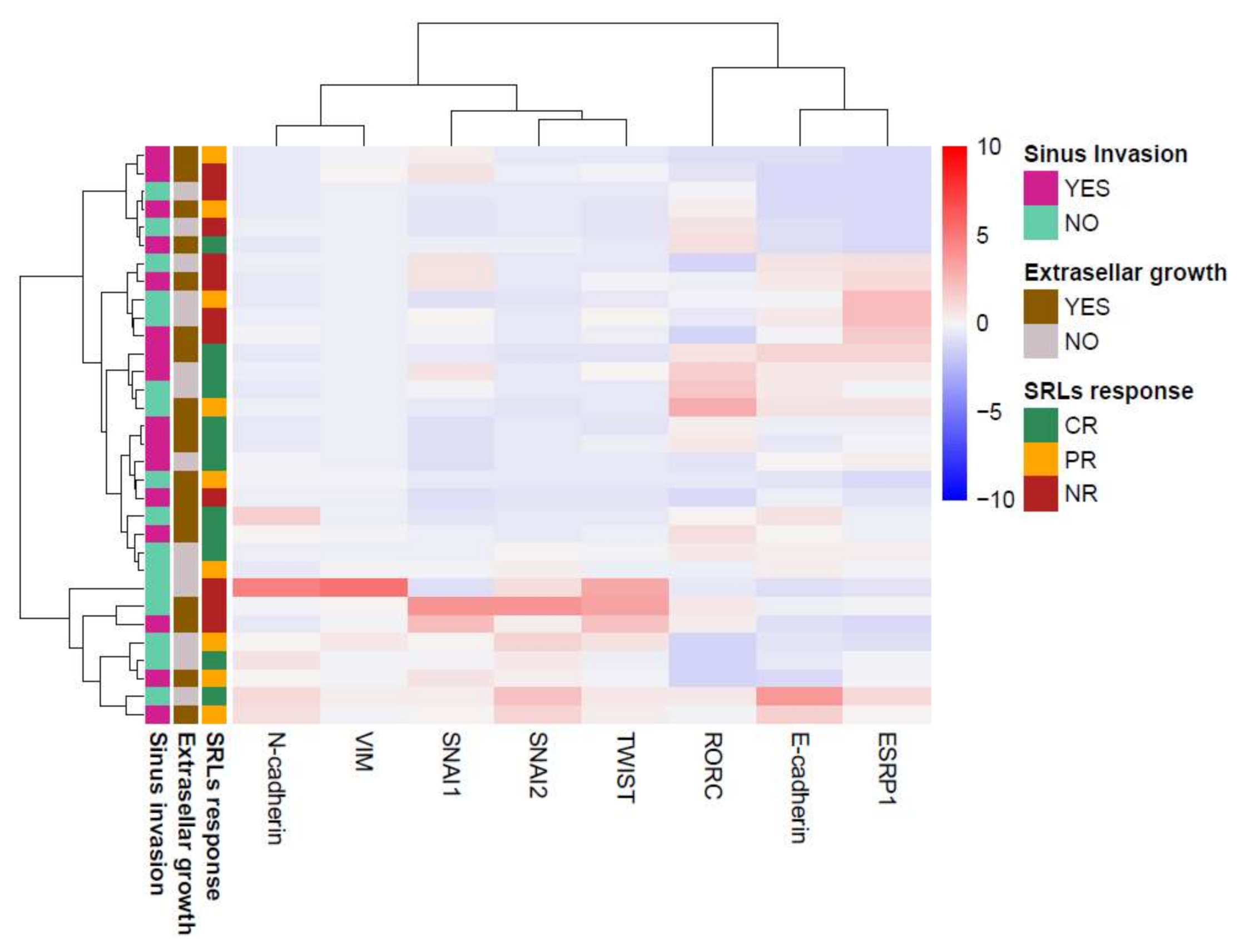
3.4. GH-Producing Tumors Showed Different Intermediate EMT States with No Association with SRL Response
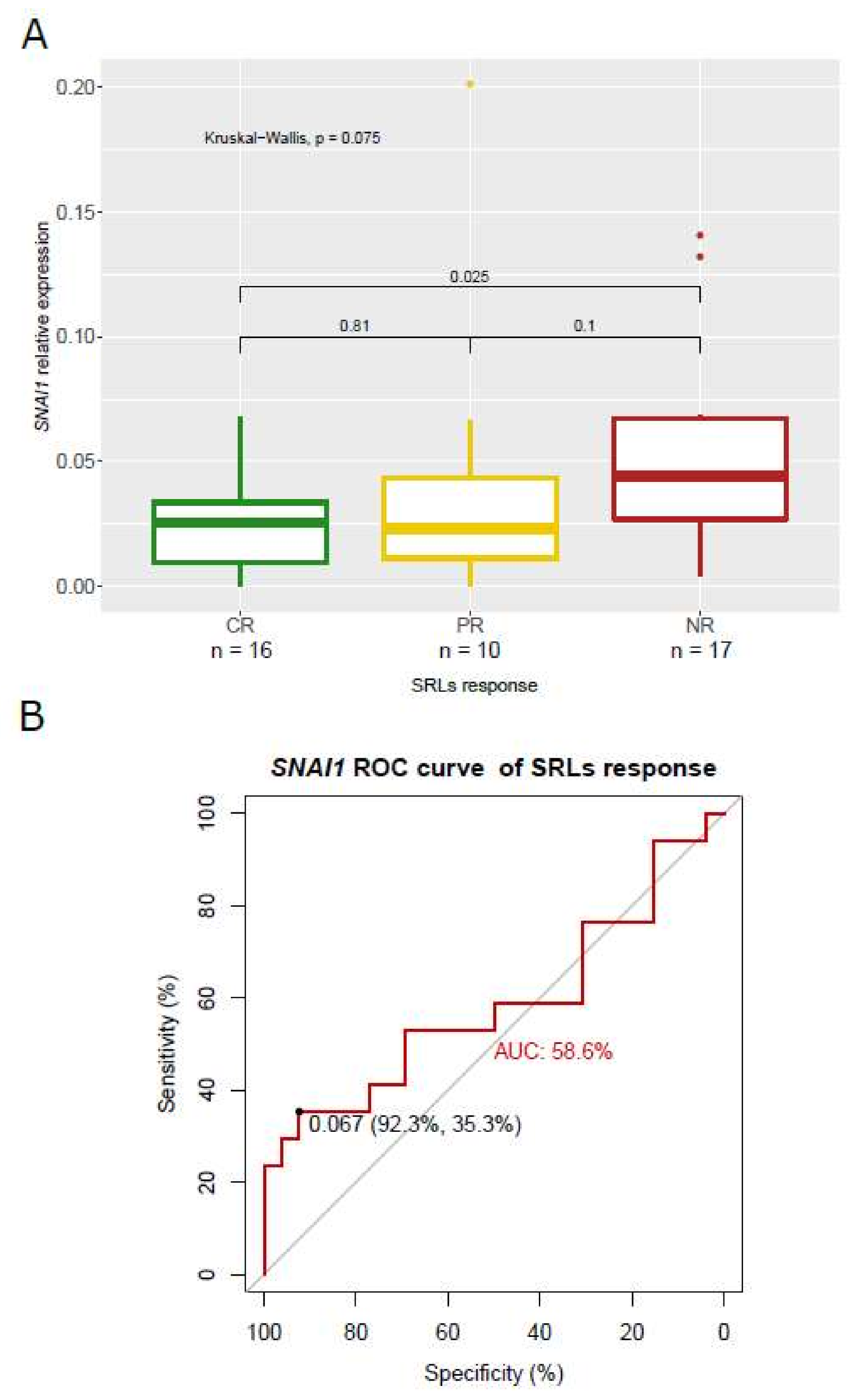
3.5. Association of SNAI1 and RORC Expression with SRL Response
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Colao, A.; Grasso, L.F.S.; Giustina, A.; Melmed, S.; Chanson, P.; Pereira, A.M.; Pivonello, R. Acromegaly. Nat. Rev. Dis. Prim. 2019, 5, 20. [Google Scholar] [CrossRef]
- Katznelson, L.; Laws, E.R.; Melmed, S.; Molitch, M.E.; Murad, M.H.; Utz, A.; Wass, J.A.H. Endocrine Society Acromegaly: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2014, 99, 3933–3951. [Google Scholar] [CrossRef]
- Buchfelder, M.; Schlaffer, S.-M. The surgical treatment of acromegaly. Pituitary 2017, 20, 76–83. [Google Scholar] [CrossRef]
- Melmed, S.; Bronstein, M.D.; Chanson, P.; Klibanski, A.; Casanueva, F.F.; Wass, J.A.H.; Strasburger, C.J.; Luger, A.; Clemmons, D.R.; Giustina, A. A Consensus Statement on acromegaly therapeutic outcomes. Nat. Rev. Endocrinol. 2018, 14, 552–561. [Google Scholar] [CrossRef] [Green Version]
- Gadelha, M.R.; Wildemberg, L.E.; Bronstein, M.D.; Gatto, F.; Ferone, D. Somatostatin receptor ligands in the treatment of acromegaly. Pituitary 2017, 20, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Colao, A.; Auriemma, R.S.; Lombardi, G.; Pivonello, R. Resistance to Somatostatin Analogs in Acromegaly. Endocr. Rev. 2011, 32, 247–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paragliola, R.M.; Corsello, S.M.; Salvatori, R. Somatostatin receptor ligands in acromegaly: Clinical response and factors predicting resistance. Pituitary 2017, 20, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Geraedts, V.; Andela, C.D.; Stalla, G.K.; Pereira, A.M.; Van Furth, W.R.; Sievers, C.; Biermasz, N.R. Predictors of Quality of Life in Acromegaly: No Consensus on Biochemical Parameters. Front. Endocrinol. 2017, 8, 40. [Google Scholar] [CrossRef] [Green Version]
- Crespo, I.; Valassi, E.; Webb, S.M. Update on quality of life in patients with acromegaly. Pituitary 2017, 20, 185–188. [Google Scholar] [CrossRef]
- Gadelha, M.R.; Kasuki, L.; Korbonits, M. Novel pathway for somatostatin analogs in patients with acromegaly. Trends Endocrinol. Metab. 2013, 24, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Domingo, M.P. Treatment of acromegaly in the era of personalized and predictive medicine. Clin. Endocrinol. 2015, 83, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Puig-Domingo, M.; Gil, J.; Nuñez, M.S.; Jordà, M.; Webb, S.M.; Serra, G.; Pons, L.; Salinas, I.; Blanco, A.; Marques-Pamies, M.; et al. Molecular profiling for acromegaly treatment: A validation study. Endocr.-Relat. Cancer 2020, 27, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, A.M.; Na, T.-Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Lekva, T.; Berg, J.P.; Fougner, S.L.; Olstad, O.K.; Ueland, T.; Bollerslev, J. Gene Expression Profiling Identifies ESRP1 as a Potential Regulator of Epithelial Mesenchymal Transition in Somatotroph Adenomas from a Large Cohort of Patients with Acromegaly. J. Clin. Endocrinol. Metab. 2012, 97, E1506–E1514. [Google Scholar] [CrossRef] [Green Version]
- Lekva, T.; Berg, J.P.; Lyle, R.; Heck, A.; Ringstad, G.; Olstad, O.K.; Michelsen, A.E.; Casar-Borota, O.; Bollerslev, J.; Ueland, T. Epithelial Splicing Regulator Protein 1 and Alternative Splicing in Somatotroph Adenomas. Endocrinology 2013, 154, 3331–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fougner, S.L.; Lekva, T.; Casar-Borota, O.; Hald, J.K.; Bollerslev, J.; Berg, J.P. The Expression of E-Cadherin in Somatotroph Pituitary Adenomas Is Related to Tumor Size, Invasiveness, and Somatostatin Analog Response. J. Clin. Endocrinol. Metab. 2010, 95, 2334–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venegas-Moreno, E.; Flores-Martinez, A.; Dios, E.; Vázquez-Borrego, M.C.; Ibáñez-Costa, A.; Madrazo-Atutxa, A.; Japón, M.A.; Castaño, J.P.; Luque, R.M.; Cano, D.A.; et al. E-cadherin expression is associated with somatostatin analogue response in acromegaly. J. Cell. Mol. Med. 2019, 23, 3088–3096. [Google Scholar] [CrossRef] [Green Version]
- Zhou, K.; Jin, H.; Luo, Y. Expression and Significance of E-Cadherin and β-Catenins in Pituitary Adenoma. Int. J. Surg. Pathol. 2013, 21, 363–367. [Google Scholar] [CrossRef]
- Gil, J.; Jordà, M.; Soldevila, B.; Puig-Domingo, M. Epithelial-Mesenchymal Transition in the Resistance to Somatostatin Receptor Ligands in Acromegaly. Front. Endocrinol. 2021, 12, 646210. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Costa, A.; Korbonits, M. AIP and the somatostatin system in pituitary tumours. J. Endocrinol. 2017, 235, R101–R116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, S.; Carlsen, E.; Marques, P.; Stiles, C.E.; Gadaleta, E.; Berney, D.M.; Roncaroli, F.; Chelala, C.; Solomou, A.; Herincs, M.; et al. Tumor microenvironment defines the invasive phenotype of AIP-mutation-positive pituitary tumors. Oncogene 2019, 38, 5381–5395. [Google Scholar] [CrossRef]
- Luque, R.M.; Ibáñez-Costa, A.; Sanchez-Tejada, L.; Rivero-Cortés, E.; Robledo, M.; Madrazo-Atutxa, A.; Mora, M.; Alvarez, C.; Lucas-Morante, T.; Álvarez-Escolá, C.; et al. El Registro Molecular de Adenomas Hipofisarios (REMAH): Una apuesta de futuro de la Endocrinología española por la medicina individualizada y la investigación traslacional. Endocrinol. Nutr. 2016, 63, 274–284. [Google Scholar] [CrossRef]
- Gil, J.; Marques-Pamies, M.; Jordà, M.; Fajardo-Montañana, C.; García-Martínez, A.; Sampedro, M.; Serra, G.; Salinas, I.; Blanco, A.; Valassi, E.; et al. Molecular determinants of enhanced response to somatostatin receptor ligands after debulking in large GH-producing adenomas. Clin. Endocrinol. 2020, 94, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Zada, G.; Lin, N.; Laws, E.R. Patterns of extrasellar extension in growth hormone–secreting and nonfunctional pituitary macroadenomas. Neurosurg. Focus 2010, 29, E4. [Google Scholar] [CrossRef]
- Mallona, I.; Díez-Villanueva, A.; A Peinado, M. Methylation plotter: A web tool for dynamic visualization of DNA methylation data. Source Code Biol. Med. 2014, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [Green Version]
- Berx, G.; Van Roy, F. Involvement of Members of the Cadherin Superfamily in Cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a003129. [Google Scholar] [CrossRef]
- Lekva, T.; Berg, J.P.; Heck, A.; Fougner, S.L.; Olstad, O.K.; Ringstad, G.; Bollerslev, J.; Ueland, T. Attenuated RORC Expression in the Presence of EMT Progression in Somatotroph Adenomas following Treatment with Somatostatin Analogs Is Associated with Poor Clinical Recovery. PLoS ONE 2013, 8, e66927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, S.; Kato, T.; Kato, Y. EMT Involved in Migration of Stem/Progenitor Cells for Pituitary Development and Regeneration. J. Clin. Med. 2016, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Millán, M.I.P.; Brinkmeier, M.L.; Mortensen, A.H.; A Camper, S. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. eLife 2016, 5, e14470. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Zhu, J.; A Martin, T.A.; Jiang, A.; Sanders, A.J.; Jiang, W.G. Epithelial-mesenchymal Transition (EMT) Markers in Human Pituitary Adenomas Indicate a Clinical Course. Anticancer Res. 2015, 35, 2635–2643. [Google Scholar] [PubMed]
- Evang, J.A.; Berg, J.P.; Casar-Borota, O.; Lekva, T.; Kringen, M.K.; Ramm-Pettersen, J.; Bollerslev, J. Reduced levels of E-cadherin correlate with progression of corticotroph pituitary tumours. Clin. Endocrinol. 2011, 75, 811–818. [Google Scholar] [CrossRef]
- Grigore, A.D.; Jolly, M.K.; Jia, D.; Farach-Carson, M.C.; Levine, H. Tumor Budding: The Name is EMT. Partial EMT. J. Clin. Med. 2016, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Oh, T.G.; Wang, S.-C.M.; Acharya, B.R.; Goode, J.M.; Graham, J.D.; Clarke, C.L.; Yap, A.; Muscat, G.E. The Nuclear Receptor, RORγ, Regulates Pathways Necessary for Breast Cancer Metastasis. eBioMedicine 2016, 6, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Petrossians, P.; Borges-Martins, L.; Espinoza, C.; Daly, A.; Betea, D.; Valdes-Socin, H.; Stevenaert, A.; Chanson, P.; Beckers, A. Gross total resection or debulking of pituitary adenomas improves hormonal control of acromegaly by somatostatin analogs. Eur. J. Endocrinol. 2005, 152, 61–66. [Google Scholar] [CrossRef]
- Colao, A.; Attanasio, R.; Pivonello, R.; Cappabianca, P.; Cavallo, L.M.; Lasio, G.; Lodrini, A.; Lombardi, G.; Cozzi, R. Partial Surgical Removal of Growth Hormone-Secreting Pituitary Tumors Enhances the Response to Somatostatin Analogs in Acromegaly. J. Clin. Endocrinol. Metab. 2006, 91, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Unutmaz, D.; Zou, Y.-R.; Sunshine, M.J.; Pierani, A.; Brenner-Morton, S.; Mebius, R.E.; Littman, D.R. Requirement for RORγ in Thymocyte Survival and Lymphoid Organ Development. Science 2000, 288, 2369–2373. [Google Scholar] [CrossRef] [Green Version]
- Eberl, G.; Marmon, S.; Sunshine, M.J.; Rennert, P.D.; Choi, Y.; Littman, D.R. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004, 5, 64–73. [Google Scholar] [CrossRef]
- Akashi, M.; Takumi, T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005, 12, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.A.; Piedrafita, F.J.; Pfahl, M.; Maki, R. TOR: A new orphan receptor expressed in the thymus that can modulate retinoid and thyroid hormone signals. Mol. Endocrinol. 1995, 9, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Hu, Y.; Ren, J.; Yu, N.; Li, Z.; Xu, Z. Circulating Th22 cells, as well as Th17 cells, are elevated in patients with renal cell carcinoma. Int. J. Med Sci. 2021, 18, 99–108. [Google Scholar] [CrossRef]
- Lytle, N.K.; Ferguson, L.P.; Rajbhandari, N.; Gilroy, K.; Fox, R.G.; Deshpande, A.; Schürch, C.M.; Hamilton, M.; Robertson, N.; Lin, W.; et al. A Multiscale Map of the Stem Cell State in Pancreatic Adenocarcinoma. Cell 2019, 177, 572–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, D.; Wang, J.; Gao, B.; Li, J.; Wu, F.; Zou, J.X.; Xu, J.; Jiang, Y.; Zou, H.; Huang, Z.; et al. RORγ is a targetable master regulator of cholesterol biosynthesis in a cancer subtype. Nat. Commun. 2019, 10, 462. [Google Scholar] [CrossRef]
- Kim, S.M.; Choi, J.E.; Hur, W.; Kim, J.-H.; Hong, S.W.; Lee, E.B.; Lee, J.H.; Li, T.Z.; Sung, P.S.; Yoon, S.K. RAR-Related Orphan Receptor Gamma (ROR-γ) Mediates Epithelial-Mesenchymal Transition Of Hepatocytes During Hepatic Fibrosis. J. Cell. Biochem. 2017, 118, 2026–2036. [Google Scholar] [CrossRef] [Green Version]
- Slominski, A.T.; Kim, T.-K.; Takeda, Y.; Janjetovic, Z.; Brożyna, A.A.; Skobowiat, C.; Wang, J.; Postlethwaite, A.; Li, W.; Tuckey, R.C.; et al. RORα and ROR γ are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. FASEB J. 2014, 28, 2775–2789. [Google Scholar] [CrossRef] [Green Version]
- Solt, L.A.; Burris, T.P. Action of RORs and their ligands in (patho)physiology. Trends Endocrinol. Metab. 2012, 23, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahalingam, D.; Wang, J.S.; Hamilton, E.P.; Sarantopoulos, J.; Nemunaitis, J.; Weems, G.; Carter, L.L.; Hu, X.; Schreeder, M.; Wilkins, H.J. Phase 1 Open-Label, Multicenter Study of First-in-Class RORγ Agonist LYC-55716 (Cintirorgon): Safety, Tolerability, and Preliminary Evidence of Antitumor Activity. Clin. Cancer Res. 2019, 25, 3508–3516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Sano, T.; Yoshimoto, K.; Yamada, S. Downregulation of E-cadherin and its undercoat proteins in pituitary growth hormone cell adenomas with prominent fibrous bodies. Endocr. Pathol. 2002, 13, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.R.; Sano, T.; Yoshimoto, K.; Asa, S.L.; Yamada, S.; Mizusawa, N.; Kudo, E. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod. Pathol. 2007, 20, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [Green Version]
- Claus, R.; Wilop, S.; Hielscher, T.; Sonnet, M.; Dahl, E.; Galm, O.; Jost, E.; Plass, C. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics 2012, 7, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Pedraza-Arévalo, S.; Gahete, M.D.; Alors-Pérez, E.; Luque, R.M.; Castaño, J.P. Multilayered heterogeneity as an intrinsic hallmark of neuroendocrine tumors. Rev. Endocr. Metab. Disord. 2018, 19, 179–192. [Google Scholar] [CrossRef]
- Sampedro Nuñez, M.; Marazuela, M. Spanish Molecular Registry of Pituitary Adenomas (REMAH Nacional): Orphanet. Available online: https://www.orpha.net/consor/cgi-bin/ResearchTrials_RegistriesMaterials.php?lng=EN&data_id=83067&Nombredelregistro/material=Registro-Molecular-de-Adenomas-Hipofisarios--REMAH-Nacional-&title=RegistroMoleculardeAdenomasHipofisariosREMAHNacional&sea (accessed on 30 December 2021).







| Patients’ Characteristics | ||
|---|---|---|
| Cohort (n) | 57 | |
| Male/Female | 25/32 | |
| Age, mean ± SD | 45.74 ± 12.35 | |
| Body mass index (BMI) ± SD | 27.7 ± 5.21 | |
| Medical Treatment | ||
| SRL response | Pre * (46) | Non-pre ** (11) |
| Nonresponders | 17 | 0 |
| Partial responders | 9 | 1 |
| Complete responders | 12 | 4 |
| NA | 8 | 6 |
| Tumor Characteristics | ||
| Macroadenoma | 47 (82%) | |
| Extrasellar growth | 39 (68%) | |
| Cavernous sinus invasion | 27 (48%) | |
| Maximum tumor diameter (mm), mean ± SD | 19.49 ± 10.03 | |
| GNAS mutation | 23 (41%) | |
| Hypointense T2 signal | 19 (33%) | |
| Comorbidities | ||
| Hypopituitarism | 19 (33%) | |
| Visual alterations | 12 (21%) | |
| Diabetes | 17 (30%) | |
| Dislipemia | 18 (31%) | |
| HBP | 25 (44%) | |
| Cardiovasular incident | 18 (32%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, J.; Marques-Pamies, M.; Valassi, E.; García-Martínez, A.; Serra, G.; Hostalot, C.; Fajardo-Montañana, C.; Carrato, C.; Bernabeu, I.; Marazuela, M.; et al. Implications of Heterogeneity of Epithelial-Mesenchymal States in Acromegaly Therapeutic Pharmacologic Response. Biomedicines 2022, 10, 460. https://doi.org/10.3390/biomedicines10020460
Gil J, Marques-Pamies M, Valassi E, García-Martínez A, Serra G, Hostalot C, Fajardo-Montañana C, Carrato C, Bernabeu I, Marazuela M, et al. Implications of Heterogeneity of Epithelial-Mesenchymal States in Acromegaly Therapeutic Pharmacologic Response. Biomedicines. 2022; 10(2):460. https://doi.org/10.3390/biomedicines10020460
Chicago/Turabian StyleGil, Joan, Montserrat Marques-Pamies, Elena Valassi, Araceli García-Martínez, Guillermo Serra, Cristina Hostalot, Carmen Fajardo-Montañana, Cristina Carrato, Ignacio Bernabeu, Mónica Marazuela, and et al. 2022. "Implications of Heterogeneity of Epithelial-Mesenchymal States in Acromegaly Therapeutic Pharmacologic Response" Biomedicines 10, no. 2: 460. https://doi.org/10.3390/biomedicines10020460







