Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Inclusion and Exclusion Criteria
2.2.1. Inclusion Criteria
2.2.2. Exclusion Criteria
2.3. Intervention
2.4. Tissue and Blood Samples Processing
2.5. Whole Genome Genotyping of Initial Cohorts (1st Phase of the Study)
2.5.1. Microarray Assay
2.5.2. Genotypes Calling
2.6. PCR-Genotyping of Large Cohorts (2nd Phase of the Study)
Statistical Analysis
3. Results
3.1. Basic Clinical Characteristics
3.1.1. Whole Genome Genotyping—Phase I
3.1.2. Extended Cohorts for PCR-Analysis—Phase II
3.2. Allelic Frequencies Comparison
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murji, A.; Bedaiwy, M.; Singh, S.S.; Bougie, O.; CAPTURE Registry Steering Committee. Influence of Ethnicity on Clinical Presentation and Quality of Life in Women with Uterine Fibroids: Results from a Prospective Observational Registry. J. Obstet. Gynaecol. Can. 2020, 42, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Adamyan, L.V. Uterine Fibroids: Diagnosis, Treatment and Rehabilitation. Clinic Recommendations; Moscow Russian Health Ministry: Moscow, Russia, 2015; (In Russian).
- Laughlin, S.K.; Schroeder, J.C.; Baird, D.D. New Directions in the Epidemiology of Uterine Fibroids. Semin. Reprod. Med. 2010, 28, 204–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, E.A.; Cookson, C.; Gandolfo, R.A.; Schulze-Rath, R. Epidemiology of uterine fibroids: A systematic review. BJOG Int. J. Obstet. Gynaecol. 2017, 124, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Wallach, E.E.; Buttram, V.C.; Reiter, R.C. Uterine leiomyomata: Etiology, symptomatology, and management. Fertil. Steril. 1981, 36, 433–445. [Google Scholar] [CrossRef]
- Pavone, D.; Clemenza, S.; Sorbi, F.; Fambrini, M.; Petraglia, F. Epidemiology and Risk Factors of Uterine Fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 46, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Yoo, E.-H.; Lee, P.I.; Huh, C.-Y.; Kim, D.-H.; Lee, B.S.; Lee, J.-K.; Kim, D. Predictors of leiomyoma recurrence after laparoscopic myomectomy. J. Minim. Invasive Gynecol. 2007, 14, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Laberge, P.Y.; Murji, A.; Vilos, G.A.; Allaire, C.; Leyland, N.; Singh, S.S. Guideline No. 389-Medical Management of Symptomatic Uterine Leiomyomas—An Addendum. J. Obstet. Gynaecol. Can. 2019, 41, 1521–1524. [Google Scholar] [CrossRef] [Green Version]
- Conconi, D.; Chiappa, V.; Perego, P.; Redaelli, S.; Bovo, G.; Lavitrano, M.; Milani, R.; Dalprà, L.; Lissoni, A.A. Potential role of BCL2 in the recurrence of uterine smooth muscle tumors of uncertain malignant potential. Oncol. Rep. 2016, 37, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Kotani, Y.; Tobiume, T.; Fujishima, R.; Shigeta, M.; Takaya, H.; Nakai, H.; Suzuki, A.; Tsuji, I.; Mandai, M.; Matsumura, N. Recurrence of uterine myoma after myomectomy: Open myomectomy versus laparoscopic myomectomy. J. Obstet. Gynaecol. Res. 2018, 44, 298–302. [Google Scholar] [CrossRef] [Green Version]
- Välimäki, N.; Kuisma, H.; Pasanen, A.; Heikinheimo, O.; Sjöberg, J.; Bützow, R.; Sarvilinna, N.; Heinonen, H.-R.; Tolvanen, J.; Bramante, S.; et al. Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. eLife 2018, 18, 7. [Google Scholar] [CrossRef]
- Vikhlyaeva, E.; Khodzhaeva, Z.; Fantschenko, N. Familial predisposition to uterine leiomyomas. Int. J. Gynaecol. Obstet. 1995, 51, 127–131. [Google Scholar] [CrossRef]
- Van Voorhis, B.J.; Romitti, P.A.; Jones, M.P. Family history as a risk factor for development of uterine leiomyomas. Results of a pilot study. J. Reprod. Med. 2002, 47, 663–669. [Google Scholar] [PubMed]
- Bondagji, N.S.; Morad, F.A.; Al-Nefaei, A.A.A.; Khan, I.A.; Elango, R.; Abdullah, L.S.; Al-Mansouri, N.M.; Sabir, J.; Banaganapalli, B.; Edris, S.; et al. Replication of GWAS loci revealed the moderate effect of TNRC6B locus on susceptibility of Saudi women to develop uterine leiomyomas. J. Obstet. Gynaecol. Res. 2016, 43, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Ordulu, Z. Fibroids. Clin. Obstet. Gynecol. 2016, 59, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Sogoyan, N.; Kuznetsova, M.V.; Asaturova, A.V.; Adamyan, L.V.; Trofimov, D.Y. Somatic mutations in MED12 gene exon 2 in women with a single uterine fibroid or multiple ones. Obst. Ginekol. 2018, 12, 63–70. [Google Scholar] [CrossRef]
- Holdsworth-Carson, S.; Zaitseva, M.; Vollenhoven, B.J.; Rogers, P. Clonality of smooth muscle and fibroblast cell populations isolated from human fibroid and myometrial tissues. Mol. Hum. Reprod. 2014, 20, 250–259. [Google Scholar] [CrossRef] [Green Version]
- Mäkinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef]
- Mehine, M.; Kaasinen, E.; Mäkinen, N.; Katainen, R.; Kämpjärvi, K.; Pitkänen, E.; Heinonen, H.-R.; Bützow, R.; Kilpivaara, O.; Kuosmanen, A.; et al. Characterization of uterine leiomyomas by whole-genome sequencing. N. Engl. J. Med. 2013, 369, 43–53. [Google Scholar] [CrossRef]
- Osinovskaya, N.S.; Malysheva, O.V.; Shved, N.Y.; Ivashchenko, T.E.; Sultanov, I.Y.; Efimova, O.; Yarmolinskaya, M.I.; Bezhenar, V.F.; Baranov, V.S. Frequency and Spectrum of MED12 Exon 2 Mutations in Multiple Versus Solitary Uterine Leiomyomas from Russian Patients. Int. J. Gynecol. Pathol. 2016, 35, 509–515. [Google Scholar] [CrossRef]
- Ajabnoor, G.M.A.; Mohammed, N.A.; Banaganapalli, B.; Abdullah, L.S.; Bondagji, O.N.; Mansouri, N.; Sahly, N.N.; Vaidyanathan, V.; Bondagji, N.; Elango, R.; et al. Expanded Somatic Mutation Spectrum of MED12 Gene in Uterine Leiomyomas of Saudi Arabian Women. Front. Genet. 2018, 9, 552. [Google Scholar] [CrossRef] [Green Version]
- Markowski, D.N.; Nimzyk, R.; Belge, G.; Löning, T.; Helmke, B.M.; Bullerdiek, J. Molecular topography of the MED12-deleted region in smooth muscle tumors: A possible link between non-B DNA structures and hypermutability. Mol. Cytogenet. 2013, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäkinen, N.; Heinonen, H.-R.; Sjöberg, J.; Taipale, J.; Vahteristo, P.; Aaltonen, L.A. Mutation analysis of components of the Mediator kinase module in MED12 mutation-negative uterine leiomyomas. Br. J. Cancer 2014, 110, 2246–2249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lac, V.; Verhoef, L.; Aguirre-Hernandez, R.; Nazeran, T.M.; Tessier-Cloutier, B.; Praetorius, T.; Orr, N.L.; Noga, H.; Lum, A.; Khattra, J.; et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Hum. Reprod. 2019, 34, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Munro, D.; Ghersi, D.; Singh, M. Two critical positions in zinc finger domains are heavily mutated in three human cancer types. PLoS Comput. Biol. 2018, 14, e1006290. [Google Scholar] [CrossRef] [PubMed]
- Malentacchi, F.; Turrini, I.; Sorbi, F.; Projetto, E.; Castiglione, F.; Fambrini, M.; Petraglia, F.; Pillozzi, S.; Noci, I. Pilot investigation of the mutation profile of PIK3CA/PTEN genes (PI3K pathway) in grade 3 endometrial cancer. Oncol. Rep. 2018, 41, 1560–1574. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Mäkinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef] [Green Version]
- Baranov, V.S.; Osinovskaya, N.S.; Yarmolinskaya, M.I. Pathogenomics of Uterine Fibroids Development. Int. J. Mol. Sci. 2019, 6, 6151. [Google Scholar] [CrossRef] [Green Version]
- Mortezaee, F.T.; Tabatabaiefar, M.A.; Chaleshtori, M.H.; Miraj, S. Lack of Association between ESR1 and CYP1A1 Gene Polymorphisms and Susceptibility to Uterine Leiomyoma in Female Patients of Iranian Descent. Cell J. 2014, 16, 225–230. [Google Scholar]
- Cha, P.-C.; Takahashi, A.; Hosono, N.; Low, S.-K.; Kamatani, N.; Kubo, M.; Nakamura, Y. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat. Genet. 2011, 43, 447–450. [Google Scholar] [CrossRef]
- Gallagher, C.S.; Mäkinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019, 10, 4857. [Google Scholar] [CrossRef] [Green Version]
- Ciavattini, A.; Di Giuseppe, J.; Stortoni, P.; Montik, N.; Giannubilo, S.R.; Litta, P.; Islam, S.; Tranquilli, A.L.; Reis, F.M.; Ciarmela, P. Uterine fibroids: Pathogenesis and interactions with endometrium and endomyometrial junction. Obstet. Gynecol. Int. 2013, 2013, 173184. [Google Scholar] [CrossRef] [PubMed]
- Sparic, R.; Mirkovic, L.; Malvasi, A.; Tinelli, A. Epidemiology of Uterine Myomas: A Review. Int. J. Fertil. Steril. 2016, 9, 424–435. [Google Scholar] [PubMed]
- Vidal-Mazo, C.; Forero-Diaz, C.; Lopez-Gonzalez, E.; Yera-Gilabert, M.; Machancoses, F.H. Clinical recurrence of submucosal myoma after a mechanical hysteroscopic myomectomy: Review after 5 years follow up. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 243, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Gingold, J.A.; Gueye, N.-A.; Falcone, T. Minimally Invasive Approaches to Myoma Management. J. Minim. Invasive Gynecol. 2018, 25, 237–250. [Google Scholar] [CrossRef]
- Tanos, V.; Berry, K.E.; Frist, M.; Campo, R.; Dewilde, R.L. Prevention and Management of Complications in Laparoscopic Myomectomy. BioMed Res. Int. 2018, 2018, 8250952. [Google Scholar] [CrossRef] [Green Version]
- Tinelli, A.; Farghaly, S.A. Morcellation of occulted sarcomas during laparoscopic myomectomy and hysterectomy for patients with large fibroid uterus. Minerva Ginecol. 2018, 70, 84–88. [Google Scholar] [CrossRef]
- Fauconnier, A.; Chapron, C.; Babaki-Fard, K.; Dubuisson, J.-B. Recurrence of leiomyomata after myomectomy. Hum. Reprod. Update 2000, 6, 595–602. [Google Scholar] [CrossRef]
- Baretta, Z.; Mocellin, S.; Goldin, E.; Olopade, O.I.; Huo, D. Effect of BRCA germline mutations on breast cancer prognosis: A systematic review and meta-analysis. Medicine 2016, 95, e4975. [Google Scholar] [CrossRef]
- Gallotta, V.; Bruno, M.; Conte, C.; Giudice, M.T.; Davià, F.; Moro, F.; Zannoni, G.F.; Fagotti, A.; De Bonis, M.; Capoluongo, E.; et al. Salvage lymphadenectomy in recurrent ovarian cancer patients: Analysis of clinical outcome and BRCA1/2 gene mutational status. Eur. J. Surg. Oncol. 2020, 46, 1327–1333. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, W.; Liu, Y.; Liu, Y.; Wang, J.; Jiang, H. Association between the FMR1 CGG repeat lengths and the severity of idiopathic primary ovarian insufficiency: A meta analysis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3116–3122. [Google Scholar] [CrossRef] [Green Version]
- Özdemir, T.R.; Kırbıyık, Ö.; Dündar, B.N.; Abacı, A.; Kaya, Ö.Ö.; Çatlı, G.; Ozyilmaz, B.; Acar, S.; Koç, A.; Güvenç, M.S.; et al. Targeted next generation sequencing in patients with maturity-onset diabetes of the young (MODY). J. Pediatr. Endocrinol. Metab. 2018, 31, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
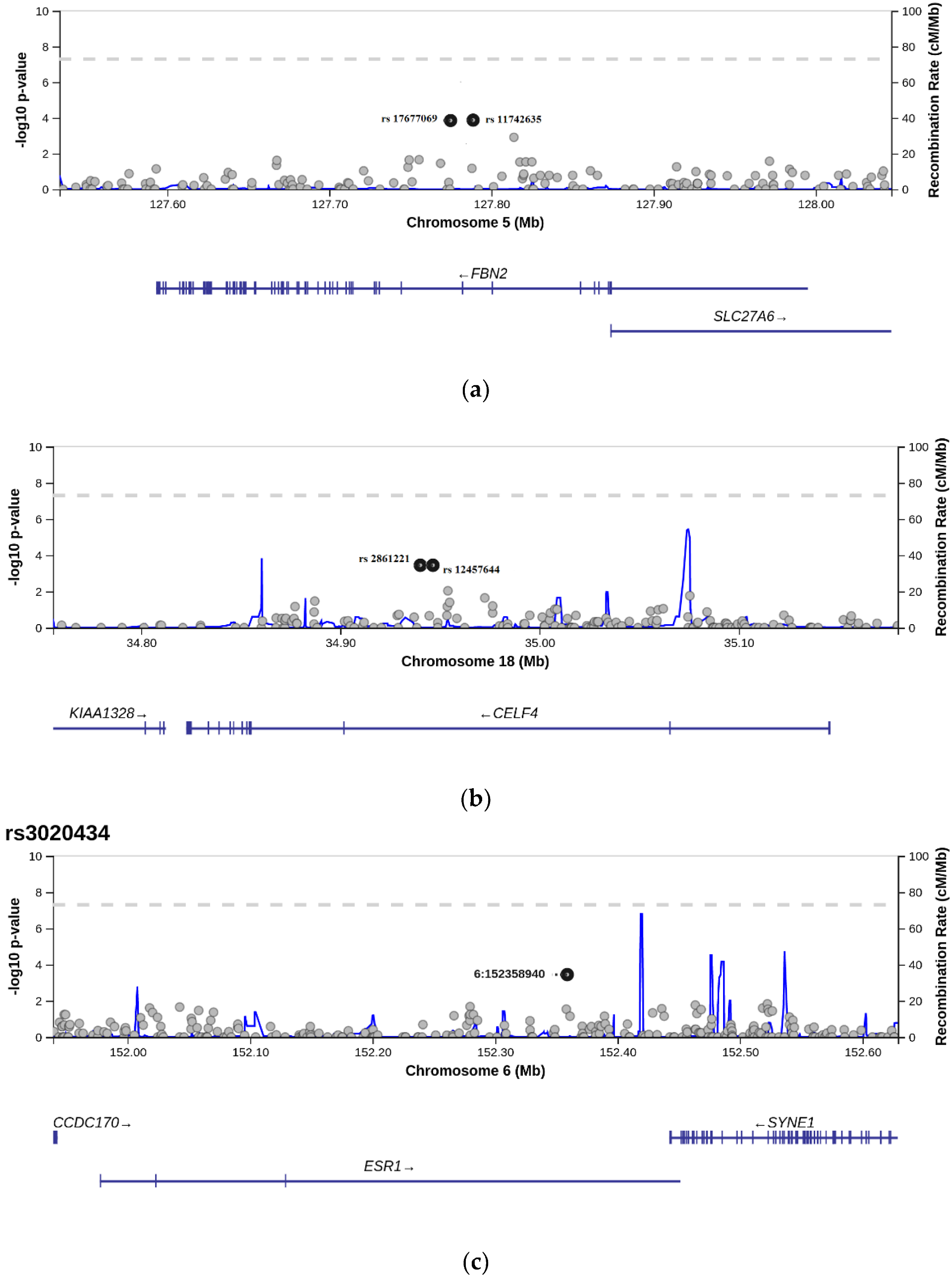

| NCBI rs ID | Gene | Chromosome Location/Position | Alleles | Frequency (1000Gen) | Location |
|---|---|---|---|---|---|
| rs12637801 | KCNMB2 | GRCh38.p12 chr 3p13 3:178661712 | C > A | A = 0.144 | Intron |
| rs2861221 | CELF4 | GRCh38.p12 chr 18 18:37360216 | C > G | G = 0.188 | Intron |
| rs3020434 | ESR1 | GRCh38.p12 chr 6 6:152037805 | C > T | T = 0.136 | Intron |
| rs11742635 | FBN2 | GRCh38.p12 chr 5 5:128453101 | G > T | T = 0.135 | Intron |
| rs12457644 | CELF4 | GRCh38.p12 chr 18 18:37365013 | G > A | A = 0.170 | Intron |
| rs17677069 | FBN2 | GRCh38.p12 chr 5 5:128438445 | A > G | G = 0.134 | Intron |
| Gene | SNP ID | Genotype/Allele | Total | UL Patients without FP | UL Patients with FP | Control | p-Value Controls vs. All LM/ Controls vs. FP | OR (95%CI) All LM/FP |
|---|---|---|---|---|---|---|---|---|
| CELF4 | rs2861221 | CC/CG/GG | 0.70/0.25/0.05 | 0.65/0.32/0.03 | 0.79/0.19/0.02 | 0.60/0.30/0.10 | 0.087/0.017 | 1.33/2.33 |
| rs12457644 | GG/AG/AA | 0.69/0.22/0.09 | 0.64/0.27/0.08 | 0.79/0.17/0.04 | 0.53/0.33/0.13 | 0.045/0.013 | 2.21/3.64 | |
| FBN2 | rs11742635 | GG/GT/TT | 0.73/0.24/0.03 | 0.74/0.24/0.02 | 0.77/0.22/0.01 | 0.57/0.37/0.07 | 0.137/0.025 | 1.99/3.33 |
| rs17677069 | AA/AG/GG | 0.69/0.25/0.05 | 0.60/0.40/0 | 0.79/0.17/0.04 | 0.53/0.30/0.17 | 0.007/0.001 | 2.6/3.95 | |
| KCNMB2 | rs12637801 | CC/CA/AA | 0.75/0.23/0.02 | 0.76/0.22/0.02 | 0.79/0.18/0.03 | 0.53/0.47/0 | 0.006/0.010 | 4.4/3.95 |
| ESR1 | rs3020434 | CC/CT/TT | 0.62/0.33/0.05 | 0.59/0.27/0.08 | 0.71/0.27/0.03 | 0.43/0.50/0.07 | 0.020/0.005 | 2.84/4.09 |
| Gene | Genotype | Single Myoma, n = 81 | Multiple Myoma, n = 134 | Distribution of Alleles, χ2 Test, p Value |
|---|---|---|---|---|
| rs3020434–ESR1 | CC | 46 (56.7%) | 87 (64.9%) | 0.492 |
| CT | 30 (37%) | 40 (29.8%) | ||
| TT | 5 (6%) | 7 (5.2%) | ||
| rs11742635–FBN2 | GG | 59 (73%) | 98 (73.1%) | 0.609 |
| GT | 14 (24%) | 33 (24.6%) | ||
| TT | 2 (3%) | 3 (2.2%) | ||
| rs124577644–CELF4 | AA | 5 (6%) | 3 (2%) | 0.232 |
| AG | 22 (26.8%) | 27 (20%) | ||
| GG | 54 (67%) | 94 (70.1%) | ||
| rs12637801–KCWMB2 | AA | 3 (4%) | 1 (0.7%) | 0.031 |
| AC | 10 (12%) | 33 (25%) | ||
| CC | 67 (83.5%) | 96 (71.6%) | ||
| rs2861221–CELF4 | CC | 53 (65.6%) | 96 (71.6%) | 0.138 |
| CG | 26 (32.8%) | 31 (23%) | ||
| GG | 1 (2%) | 7 (5%) | ||
| rs17677069–FBN2 | AA AG GG | 57 (70%) 13 (16%) 11 (14 5) | 100 (74.6%) 25 (18.7%) 9 (6.7%) | 0.238 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, M.V.; Sogoyan, N.S.; Donnikov, A.J.; Trofimov, D.Y.; Adamyan, L.V.; Mishina, N.D.; Shubina, J.; Zelensky, D.V.; Sukhikh, G.T. Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors. Biomedicines 2022, 10, 508. https://doi.org/10.3390/biomedicines10020508
Kuznetsova MV, Sogoyan NS, Donnikov AJ, Trofimov DY, Adamyan LV, Mishina ND, Shubina J, Zelensky DV, Sukhikh GT. Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors. Biomedicines. 2022; 10(2):508. https://doi.org/10.3390/biomedicines10020508
Chicago/Turabian StyleKuznetsova, Maria V., Nelly S. Sogoyan, Andrew J. Donnikov, Dmitry Y. Trofimov, Leila V. Adamyan, Natalia D. Mishina, Jekaterina Shubina, Dmitry V. Zelensky, and Gennady T. Sukhikh. 2022. "Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors" Biomedicines 10, no. 2: 508. https://doi.org/10.3390/biomedicines10020508
APA StyleKuznetsova, M. V., Sogoyan, N. S., Donnikov, A. J., Trofimov, D. Y., Adamyan, L. V., Mishina, N. D., Shubina, J., Zelensky, D. V., & Sukhikh, G. T. (2022). Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors. Biomedicines, 10(2), 508. https://doi.org/10.3390/biomedicines10020508






