Preclinical Models of Brain Metastases in Breast Cancer
Abstract
1. Background
2. Models of Breast Cancer Brain Metastases
3. Detection Methods of BCBM
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 231BR | MDA-MB-231BR |
| BBB | blood–brain barrier |
| BCBM | breast cancer brain metastasis |
| BCF | breast cancer specific frequencies |
| BLI | bioluminescence imaging |
| BC | breast cancer |
| CT | computed tomography |
| ER | estrogen receptor |
| FDG | fluorodeoxyglucose |
| FLI | fluorescence imaging |
| Gd | gadolinium |
| HER2 | human epidermal growth factor receptor 2 |
| IBCC | human immortalized breast cancer cell |
| MRI | magnetic resonance imaging |
| PDO | patient-derived organoids |
| PDOX | patient-derived organoid xenografts |
| PDX | patient-derived xenografts |
| PET | positron emission tomography |
| PR | progesterone receptor |
| SPIO | superparamagnetic iron oxide particles |
| TNBC | triple negative breast cancer |
| VEGF-A | vascular endothelial growth factor A |
| WBRT | whole brain radiotherapy |
| eGFP | enhanced green fluorescent protein |
References
- Malmgren, J.A.; Mayer, M.; Atwood, M.K.; Kaplan, H.G. Differential presentation and survival of de novo and recurrent metastatic breast cancer over time: 1990–2010. Breast Cancer Res. Treat. 2018, 167, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Tevaarwerk, A.J.; Gray, R.J.; Schneider, B.P.; Smith, M.L.; Wagner, L.I.; Fetting, J.H.; Davidson, N.; Goldstein, L.J.; Miller, K.D.; Sparano, J.A. Survival in patients with metastatic recurrent breast cancer after adjuvant chemotherapy: Little Evidence of Improvement over the Past 30 Years. Cancer 2013, 119, 1140–1148. [Google Scholar] [CrossRef]
- Barnholtz-Sloan, J.S.; Sloan, A.E.; Davis, F.G.; Vigneau, F.D.; Lai, P.; Sawaya, R.E. Incidence Proportions of Brain Metastases in Patients Diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J. Clin. Oncol. 2004, 22, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, E.M.; Anders, C.K. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann. Transl. Med. 2018, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, Y.; Liu, X.; Liao, X.; He, J.; Niu, L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer 2019, 19, 1091. [Google Scholar] [CrossRef]
- Rostami, R.; Mittal, S.; Rostami, P.; Tavassoli, F.; Jabbari, B. Brain metastasis in breast cancer: A comprehensive literature review. J. Neuro Oncol. 2016, 127, 407–414. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic Behavior of Breast Cancer Subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Niwińska, A.; Murawska, M.; Pogoda, K. Breast cancer brain metastases: Differences in survival depending on biological subtype, RPA RTOG prognostic class and systemic treatment after whole-brain radiotherapy (WBRT). Ann. Oncol. 2010, 21, 942–948. [Google Scholar] [CrossRef]
- Clayton, A.J.; Danson, S.; Jolly, S.; Ryder, W.D.J.; Burt, P.A.; Stewart, A.L.; Wilkinson, P.M.; Welch, R.S.; Magee, B.; Wilson, G.; et al. Incidence of Cerebral Metastases in Patients Treated with Trastuzumab for Metastatic Breast Cancer. Br. J. Cancer 2004, 91, 639–643. [Google Scholar] [CrossRef]
- Lo, H.-W.; Sirkisoon, S.R.; Carpenter, R.L.; Rimkus, T.; Miller, L.; Metheny-Barlow, L. EGFR and HER2 signaling in breast cancer brain metastasis. Front. Biosci. (Elite Ed.) 2016, 8, 245–263. [Google Scholar] [CrossRef]
- Ghajar, C.M.; Peinado, H.; Mori, H.; Matei, I.R.; Evason, K.J.; Brazier, H.; Almeida, D.; Koller, A.; Hajjar, K.A.; Stainier, D.Y.R.; et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 2013, 15, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.R.; Chiu, A.S.; Farrell, K.; Killelea, B.K.; Lannin, D.R. Why Has Breast Cancer Screening Failed to Decrease the Incidence of de Novo Stage IV Disease? Cancers 2019, 11, 500. [Google Scholar] [CrossRef]
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Prim. 2019, 5, 5. [Google Scholar] [CrossRef]
- Lorger, M.; Felding-Habermann, B. Capturing Changes in the Brain Microenvironment during Initial Steps of Breast Cancer Brain Metastasis. Am. J. Pathol. 2010, 176, 2958–2971. [Google Scholar] [CrossRef]
- Lockman, P.R.; Mittapalli, R.K.; Taskar, K.S.; Rudraraju, V.; Gril, B.; Bohn, K.A.; Adkins, C.E.; Roberts, A.; Thorsheim, H.R.; Gaasch, J.A.; et al. Heterogeneous Blood–Tumor Barrier Permeability Determines Drug Efficacy in Experimental Brain Metastases of Breast Cancer. Clin. Cancer Res. 2010, 16, 5664–5678. [Google Scholar] [CrossRef]
- Terrell-Hall, T.B.; Nounou, M.I.; El-Amrawy, F.; Griffith, J.; Lockman, P.R. Trastuzumab distribution in an in-vivo and in-vitro model of brain metastases of breast cancer. Oncotarget 2017, 8, 83734–83744. [Google Scholar] [CrossRef]
- Sayyad, M.R.; Puchalapalli, M.; Vergara, N.G.; Wangensteen, S.M.; Moore, M.; Mu, L.; Edwards, C.; Anderson, A.; Kall, S.; Sullivan, M.; et al. Syndecan-1 facilitates breast cancer metastasis to the brain. Breast Cancer Res. Treat. 2019, 178, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-N.; Lohaus, R.; Hanisch, U.-K.; Binder, C.; Dehghani, F.; Pukrop, T. Coculture System with an Organotypic Brain Slice and 3D Spheroid of Carcinoma Cells. J. Vis. Exp. 2013, 9, e50881. [Google Scholar] [CrossRef] [PubMed]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Sultan, A.; Ling, B.; Zhang, H.; Ma, B.; Michel, D.; Alcorn, J.; Yang, J. Synergistic Effect between Sphingosine-1-Phosphate and Chemotherapy Drugs against Human Brain-metastasized Breast Cancer MDA-MB-361 cells. J. Cancer 2013, 4, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Cailleau, R.; Olivé, M.; Cruciger, Q.V.J. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Abuhussein, O.; Yang, J. Evaluating the antitumor activity of sphingosine-1-phosphate against human triple-negative breast cancer cells with basal-like morphology. Investig. New Drugs 2020, 38, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Nome, R.V.; Bratland, Å.; Harman, G.; Fodstad, Ø.; Andersson, Y.; Ree, A.H. Cell cycle checkpoint signaling involved in histone deacetylase inhibition and radiation-induced cell death. Mol. Cancer Ther. 2005, 4, 1231–1238. [Google Scholar] [CrossRef]
- Rye, P.D.; Norum, L.; Olsen, D.-R.; Garman-Vik, S.; Kaul, S.; Fodstad, Ø. Brain Metastasis Model in Athymic Nude Mice Using a Novel MUC1-Secreting Human Breast-Cancer Cell Line, MA11. Int. J. Cancer 1996, 68, 682–687. [Google Scholar] [CrossRef]
- Baschnagel, A.; Russo, A.; Burgan, W.E.; Carter, D.; Beam, K.; Palmieri, D.; Steeg, P.S.; Tofilon, P.; Camphausen, K. Vorinostat enhances the radiosensitivity of a breast cancer brain metastatic cell line grown in vitro and as intracranial xenografts. Mol. Cancer Ther. 2009, 8, 1589–1595. [Google Scholar] [CrossRef]
- McGowan, P.M.; Simedrea, C.; Ribot, E.J.; Foster, P.J.; Palmieri, D.; Steeg, P.S.; Allan, A.L.; Chambers, A.F. Notch1 Inhibition Alters the CD44hi/CD24lo Population and Reduces the Formation of Brain Metastases from Breast Cancer. Mol. Cancer Res. 2011, 9, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Hua, E.; Bisht, K.; Woditschka, S.; Skordos, K.W.; Liewehr, D.J.; Steinberg, S.M.; Brogi, E.; Akram, M.M.; Killian, J.K.; et al. Inhibition of Polo-like kinase 1 prevents the growth of metastatic breast cancer cells in the brain. Clin. Exp. Metastasis 2011, 28, 899–908. [Google Scholar] [CrossRef]
- Mittapalli, R.K.; Liu, X.; Adkins, C.E.; Nounou, M.I.; Bohn, K.A.; Terrell, T.B.; Qhattal, H.S.; Geldenhuys, W.J.; Palmieri, D.; Steeg, P.S.; et al. Paclitaxel–Hyaluronic NanoConjugates Prolong Overall Survival in a Preclinical Brain Metastases of Breast Cancer Model. Mol. Cancer Ther. 2013, 12, 2389–2399. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, W.-C.; Zhang, L.; Zhang, C.; Lowery, F.; Ding, Z.; Guo, H.; Wang, H.; Huang, S.; Sahin, A.A.; et al. Src Family Kinases as Novel Therapeutic Targets to Treat Breast Cancer Brain Metastases. Cancer Res. 2013, 73, 5764–5774. [Google Scholar] [CrossRef]
- Smart, D.D.K.; Glaessner, A.G.; Palmieri, D.; Wong-Goodrich, S.J.; Kramp, T.; Gril, B.; Shukla, S.; Lyle, T.; Hua, E.; Cameron, H.A.; et al. Analysis of radiation therapy in a model of triple-negative breast cancer brain metastasis. Clin. Exp. Metastasis 2015, 32, 717–727. [Google Scholar] [CrossRef]
- Crowe, W.; Wang, L.; Zhang, Z.; Varagic, J.; Bourland, J.D.; Chan, M.D.; Habib, A.A.; Zhao, D. MRI evaluation of the effects of whole brain radiotherapy on breast cancer brain metastasis. Int. J. Radiat. Biol. 2019, 95, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Wu, S.-Y.; Jimenez, H.; Xing, F.; Zhu, D.; Liu, Y.; Wu, K.; Tyagi, A.; Zhao, D.; Lo, H.-W.; et al. Ca2+ and CACNA1H mediate targeted suppression of breast cancer brain metastasis by AM RF EMF. EBioMedicine 2019, 44, 194–208. [Google Scholar] [CrossRef]
- Thomas, F.C.; Taskar, K.; Rudraraju, V.; Goda, S.; Thorsheim, H.R.; Gaasch, J.A.; Mittapalli, R.K.; Palmieri, D.; Steeg, P.S.; Lockman, P.R.; et al. Uptake of ANG1005, A Novel Paclitaxel Derivative, through the Blood-Brain Barrier into Brain and Experimental Brain Metastases of Breast Cancer. Pharm. Res. 2009, 26, 2486–2494. [Google Scholar] [CrossRef]
- Hamilton, A.M.; Aidoudi-Ahmed, S.; Sharma, S.; Kotamraju, V.R.; Foster, P.J.; Sugahara, K.N.; Ruoslahti, E.; Rutt, B.K. Nanoparticles coated with the tumor-penetrating peptide iRGD reduce experimental breast cancer metastasis in the brain. J. Mol. Med. 2015, 93, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.; Sprowls, S.; Szalai, G.; Arsiwala, T.; Saralkar, P.; Straight, B.; Hatcher, S.; Tyree, E.; Yost, M.; Kohler, W.J.; et al. Hypomethylating Agent Azacitidine Is Effective in Treating Brain Metastasis Triple-Negative Breast Cancer Through Regulation of DNA Methylation of Keratin 18 Gene. Transl. Oncol. 2020, 13, 100775. [Google Scholar] [CrossRef]
- Lee, H.-T.; Xue, J.; Chou, P.-C.; Zhou, A.; Yang, P.; Conrad, C.A.; Aldape, K.D.; Priebe, W.; Patterson, C.; Sawaya, R.; et al. Stat3 orchestrates interaction between endothelial and tumor cells and inhibition of Stat3 suppresses brain metastasis of breast cancer cells. Oncotarget 2015, 6, 10016–10029. [Google Scholar] [CrossRef] [PubMed]
- Delaney, L.J.; Ciraku, L.; Oeffinger, B.E.; Wessner, C.E.; Liu, J.-B.; Li, J.; Nam, K.; Forsberg, F.; Leeper, D.B.; O’Kane, P.; et al. Breast Cancer Brain Metastasis Response to Radiation After Microbubble Oxygen Delivery in a Murine Model. J. Ultrasound Med. 2019, 38, 3221–3228. [Google Scholar] [CrossRef]
- Zhao, H.; Cui, K.; Nie, F.; Wang, L.; Brandl, M.B.; Jin, G.; Li, F.; Mao, Y.; Xue, Z.; Rodriguez, A.A.; et al. The effect of mTOR inhibition alone or combined with MEK inhibitors on brain metastasis: An in vivo analysis in triple-negative breast cancer models. Breast Cancer Res. Treat. 2012, 131, 425–436. [Google Scholar] [CrossRef]
- Yoneda, T.; Williams, P.J.; Hiraga, T.; Niewolna, M.; Nishimura, R. A Bone-Seeking Clone Exhibits Different Biological Properties from the MDA-MB-231 Parental Human Breast Cancer Cells and a Brain-Seeking Clone in Vivo and in Vitro. J. Bone Miner. Res. 2001, 16, 1486–1495. [Google Scholar] [CrossRef]
- Kim, L.S.; Huang, S.; Lu, W.; Lev, D.C.; Price, J.E. Vascular endothelial growth factor expression promotes the growth of breast cancer brain metastases in nude mice. Clin. Exp. Metastasis 2004, 21, 107–118. [Google Scholar] [CrossRef]
- Palmieri, D.; Duchnowska, R.; Woditschka, S.; Hua, E.; Qian, Y.; Biernat, W.; Sosińska-Mielcarek, K.; Gril, B.; Stark, A.M.; Hewitt, S.M.; et al. Profound Prevention of Experimental Brain Metastases of Breast Cancer by Temozolomide in an MGMT-Dependent Manner. Clin. Cancer Res. 2014, 20, 2727–2739. [Google Scholar] [CrossRef] [PubMed]
- Ippen, F.M.; Grosch, J.K.; Subramanian, M.; Kuter, B.M.; Liederer, B.M.; Plise, E.G.; Mora, J.L.; Nayyar, N.; Schmidt, S.P.; Giobbie-Hurder, A.; et al. Targeting the PI3K/Akt/mTOR pathway with the pan-Akt inhibitor GDC-0068 in PIK3CA-mutant breast cancer brain metastases. Neuro Oncol. 2019, 21, 1401–1411. [Google Scholar] [CrossRef]
- Bos, P.D.; Zhang, X.H.F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Gril, B.; Palmieri, D.; Bronder, J.L.; Herring, J.M.; Vega-Valle, E.; Feigenbaum, L.; Liewehr, D.J.; Steinberg, S.M.; Merino, M.J.; Rubin, S.D.; et al. Effect of Lapatinib on the Outgrowth of Metastatic Breast Cancer Cells to the Brain. J. Natl. Cancer Inst. 2008, 100, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Gril, B.; Palmieri, D.; Qian, Y.; Smart, D.; Ileva, L.; Liewehr, D.J.; Steinberg, S.M.; Steeg, P.S. Pazopanib Reveals a Role for Tumor Cell B-Raf in the Prevention of HER2+ Breast Cancer Brain Metastasis. Clin. Cancer Res. 2011, 17, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, T.; Zhang, H.; Gou, X.; Han, C.; Wang, J.; Chen, A.T.; Ma, J.; Liu, J.; Chen, Z.; et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat. Cell Biol. 2020, 22, 1276–1285. [Google Scholar] [CrossRef]
- Murrell, D.H.; Zarghami, N.; Jensen, M.D.; Dickson, F.; Chambers, A.F.; Wong, E.; Foster, P.J. MRI surveillance of cancer cell fate in a brain metastasis model after early radiotherapy. Magn. Reson. Med. 2017, 78, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Bronder, J.L.; Herring, J.M.; Yoneda, T.; Weil, R.J.; Stark, A.M.; Kurek, R.; Vega-Valle, E.; Feigenbaum, L.; Halverson, D.; et al. Her-2 Overexpression Increases the Metastatic Outgrowth of Breast Cancer Cells in the Brain. Cancer Res. 2007, 67, 4190–4198. [Google Scholar] [CrossRef]
- Lyle, L.T.; Lockman, P.R.; Adkins, C.E.; Mohammad, A.S.; Sechrest, E.; Hua, E.; Palmieri, D.; Liewehr, D.J.; Steinberg, S.M.; Kloc, W.; et al. Alterations in Pericyte Subpopulations Are Associated with Elevated Blood–Tumor Barrier Permeability in Experimental Brain Metastasis of Breast Cancer. Clin. Cancer Res. 2016, 22, 5287–5299. [Google Scholar] [CrossRef]
- Hu, J.; Ljubimova, J.Y.; Inoue, S.; Konda, B.; Patil, R.; Ding, H.; Espinoza, A.; Wawrowsky, K.A.; Patil, C.; Ljubimov, A.V.; et al. Phosphodiesterase Type 5 Inhibitors Increase Herceptin Transport and Treatment Efficacy in Mouse Metastatic Brain Tumor Models. PLoS ONE 2010, 5, e10108. [Google Scholar] [CrossRef]
- Kodack, D.P.; Chung, E.; Yamashita, H.; Incio, J.; Duyverman, A.M.M.J.; Song, Y.; Farrar, C.T.; Huang, Y.; Ager, E.; Kamoun, W.; et al. Combined targeting of HER2 and VEGFR2 for effective treatment of HER2-amplified breast cancer brain metastases. Proc. Natl. Acad. Sci. USA 2012, 109, E3119–E3127. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, A.; Takagi, S.; Yusa, T.; Yaguchi, M.; Hayashi, A.; Tamura, T.; Kawakita, Y.; Ishikawa, T.; Ohta, Y. Antitumor Activity of TAK-285, an Investigational, Non-Pgp Substrate HER2/EGFR Kinase Inhibitor, in Cultured Tumor Cells, Mouse and Rat Xenograft Tumors, and in an HER2-Positive Brain Metastasis Model. J. Cancer 2013, 4, 557–565. [Google Scholar] [CrossRef]
- Gautam, S.K.; Kanchan, R.K.; Siddiqui, J.A.; Maurya, S.K.; Rauth, S.; Perumal, N.; Atri, P.; Venkata, R.C.; Mallya, K.; Mirza, S.; et al. Blocking c-MET/ERBB1 Axis Prevents Brain Metastasis in ERBB2+ Breast Cancer. Cancers 2020, 12, 2838. [Google Scholar] [CrossRef]
- Hall, D.G.; Stoica, G. Characterization of brain and bone-metastasizing clones selected from an ethylnitrosourea-induced rat mammary carcinoma. Clin. Exp. Metastasis 1994, 12, 283–295. [Google Scholar] [CrossRef]
- Erin, N.; Kale, Ş.; Tanrıöver, G.; Köksoy, S.; Duymuş, Ö.; Korcum, A.F. Differential characteristics of heart, liver, and brain metastatic subsets of murine breast carcinoma. Breast Cancer Res. Treat. 2013, 139, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Avraham, H.K.; Jiang, S.; Fu, Y.; Nakshatri, H.; Ovadia, H.; Avraham, S. Angiopoietin-2 mediates blood-brain barrier impairment and colonization of triple-negative breast cancer cells in brain. J. Pathol. 2014, 232, 369–381. [Google Scholar] [CrossRef]
- Kim, S.-H.; Redvers, R.P.; Chi, L.H.; Ling, X.; Lucke, A.J.; Reid, R.C.; Fairlie, D.P.; Martin, A.C.B.M.; Anderson, R.L.; Denoyer, D.; et al. Identification of brain metastasis genes and therapeutic evaluation of histone deacetylase inhibitors in a clinically relevant model of breast cancer brain metastasis. Dis. Model. Mech. 2018, 11, DMM034850. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Xia, Y.; Feng, Z.; Lin, W.; Xue, Q.; Jiang, J.; Yu, X.; Peng, C.; Luo, M.; Yang, Y.; et al. Repositioning antipsychotic fluphenazine hydrochloride for treating triple negative breast cancer with brain metastases and lung metastases. Am. J. Cancer Res. 2019, 9, 459–478. [Google Scholar]
- Kijewska, M.; Viski, C.; Turrell, F.; Fitzpatrick, A.; Van Weverwijk, A.; Gao, Q.; Iravani, M.; Isacke, C.M. Using an in-vivo syngeneic spontaneous metastasis model identifies ID2 as a promoter of breast cancer colonisation in the brain. Breast Cancer Res. 2019, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, A.; Redvers, R.P.; Ling, X.; Ayton, S.; Fuentes, M.; Tavancheh, E.; Diala, I.; Lalani, A.; Loi, S.; David, S.; et al. Neoadjuvant neratinib promotes ferroptosis and inhibits brain metastasis in a novel syngeneic model of spontaneous HER2 + ve breast cancer metastasis. Breast Cancer Res. 2019, 21, 94. [Google Scholar] [CrossRef]
- Contreras-Zarate, M.J.; Ormond, D.R.; Gillen, A.E.; Hanna, C.; Day, N.L.; Serkova, N.J.; Jacobsen, B.M.; Edgerton, S.M.; Thor, A.D.; Borges, V.F.; et al. Development of Novel Patient-Derived Xenografts from Breast Cancer Brain Metastases. Front. Oncol. 2017, 7, 252. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Okano, M.; Maiti, A.; Rashid, O.M.; Saito, K.; Kono, K.; Matsuyama, R.; Endo, I.; Takabe, K. Novel Breast Cancer Brain Metastasis Patient-Derived Orthotopic Xenograft Model for Preclinical Studies. Cancers 2020, 12, 444. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Kabraji, S.; Xie, S.; Pan, P.; Liu, Z.; Ni, J.; Zhao, J.J. Improving orthotopic mouse models of patient-derived breast cancer brain metastases by a modified intracarotid injection method. Sci. Rep. 2019, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.H.; Alzubi, M.A.; Sohal, S.S.; Olex, A.L.; Dozmorov, M.G.; Harrell, J.C. Characterizing the efficacy of cancer therapeutics in patient-derived xenograft models of metastatic breast cancer. Breast Cancer Res. Treat. 2018, 170, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shen, D.; Shao, J.; Crowder, R.; Liu, W.; Prat, A.; He, X.; Liu, S.; Hoog, J.; Lu, C.; et al. Endocrine-Therapy-Resistant ESR1 Variants Revealed by Genomic Characterization of Breast-Cancer-Derived Xenografts. Cell Rep. 2013, 4, 1116–1130. [Google Scholar] [CrossRef]
- Ni, J.; Ramkissoon, S.H.; Xie, S.; Goel, S.; Stover, D.G.; Guo, H.; Luu, V.; Marco, E.; Ramkissoon, L.A.; Kang, Y.J.; et al. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat. Med. 2016, 22, 723–726. [Google Scholar] [CrossRef]
- Michelhaugh, S.K.; Muzik, O.; Guastella, A.R.; Klinger, N.V.; Polin, L.A.; Cai, H.; Xin, Y.; Mangner, T.J.; Zhang, S.; Juhász, C.; et al. Assessment of Tryptophan Uptake and Kinetics Using 1-(2-18F-Fluoroethyl)-l-Tryptophan and α-11C-Methyl-l-Tryptophan PET Imaging in Mice Implanted with Patient-Derived Brain Tumor Xenografts. J. Nucl. Med. 2017, 58, 208–213. [Google Scholar] [CrossRef]
- Xing, F.; Kobayashi, A.; Okuda, H.; Watabe, M.; Pai, S.K.; Pandey, P.R.; Hirota, S.; Wilber, A.; Mo, Y.Y.; Moore, B.E.; et al. Reactive astrocytes promote the metastatic growth of breast cancer stem-like cells by activating Notch signalling in brain. EMBO Mol. Med. 2013, 5, 384–396. [Google Scholar] [CrossRef]
- Louie, E.; Chen, X.F.; Coomes, A.; Ji, K.; Tsirka, S.; Chen, E. Neurotrophin-3 modulates breast cancer cells and the microenvironment to promote the growth of breast cancer brain metastasis. Oncogene 2013, 32, 4064–4077. [Google Scholar] [CrossRef]
- Zhang, R.D.; Price, J.E.; Fujimaki, T.; Bucana, C.D.; Fidler, I.J. Differential Permeability of the Blood-Brain Barrier in Experimental Brain Metastases Produced by Human Neoplasms Implanted into Nude Mice. Am. J. Pathol. 1992, 141, 1115–1124. [Google Scholar]
- Samoto, K.; Ikezaki, K.; Ono, M.; Shono, T.; Kohno, K.; Kuwano, M.; Fukui, M. Expression of vascular endothelial growth factor and its possible relation with neovascularization in human brain tumors. Cancer Res. 1995, 55, 1189–1193. [Google Scholar] [PubMed]
- Emens, L.A. Breast Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2018, 24, 511–520. [Google Scholar] [CrossRef]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; De Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 116. [Google Scholar] [CrossRef]
- Ramani, V.C.; Lemaire, C.A.; Triboulet, M.; Casey, K.M.; Heirich, K.; Renier, C.; Vilches-Moure, J.G.; Gupta, R.; Razmara, A.M.; Zhang, H.; et al. Investigating circulating tumor cells and distant metastases in patient-derived orthotopic xenograft models of triple-negative breast cancer. Breast Cancer Res. 2019, 21, 98. [Google Scholar] [CrossRef]
- Aviv, H.; Bartling, S.; Kieslling, F.; Margel, S. Radiopaque iodinated copolymeric nanoparticles for X-ray imaging applications. Biomaterials 2009, 30, 5610–5616. [Google Scholar] [CrossRef]
- Krause, W.; Leike, J.; Sachse, A.; Schuhmann-Giampieri, G. Characterization of Iopromide Liposomes. Investig. Radiol. 1993, 28, 1028–1032. [Google Scholar] [CrossRef]
- Torchilin, V.P.; Frank-Kamenetsky, M.D.; Wolf, G.L. CT visualization of blood pool in rats by using long-circulating, iodine-containing micelles. Acad. Radiol. 1999, 6, 61–65. [Google Scholar] [CrossRef]
- Li, X.; Lee, S.C.; Zhang, S.; Akasaka, T. Biocompatibility and Toxicity of Nanobiomaterials. J. Nanomater. 2012, 2012, 591278. [Google Scholar] [CrossRef]
- O’Neill, K.; Lyons, S.K.; Gallagher, W.M.; Curran, K.M.; Byrne, A.T. Bioluminescent imaging: A critical tool in pre-clinical oncology research: Bioluminescent Imaging: A Critical Tool in Pre-Clinical Oncology Research. J. Pathol. 2010, 220, 317–327. [Google Scholar] [CrossRef]
- Imamura, T.; Saitou, T.; Kawakami, R. In vivo optical imaging of cancer cell function and tumor microenvironment. Cancer Sci. 2018, 109, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Troy, T.; Jekic-McMullen, D.; Sambucetti, L.; Rice, B. Quantitative Comparison of the Sensitivity of Detection of Fluorescent and Bioluminescent Reporters in Animal Models. Mol. Imaging 2004, 3, 9–23. [Google Scholar] [CrossRef]
- Choy, G.; O’Connor, S.; Diehn, F.E.; Costouros, N.; Alexander, H.R.; Choyke, P.; Libutti, S.K. Comparison of noninvasive fluorescent and bioluminescent small animal optical imaging. BioTechniques 2003, 35, 1022–1030. [Google Scholar] [CrossRef]
- Müller-Taubenberger, A.; Ishikawa-Ankerhold, H.C. Fluorescent Reporters and Methods to Analyze Fluorescent Signals. In Dictyostelium Discoideum Protocols; Eichinger, L., Rivero, F., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 983, pp. 93–112. ISBN 978-1-62703-301-5. [Google Scholar]
- Hoffman, R.M. Application of GFP imaging in cancer. Lab. Investig. 2015, 95, 432–452. [Google Scholar] [CrossRef] [PubMed]
- Kocher, B.; Piwnica-Worms, D. Illuminating Cancer Systems with Genetically Engineered Mouse Models and Coupled Luciferase Reporters In Vivo. Cancer Discov. 2013, 3, 616–629. [Google Scholar] [CrossRef] [PubMed]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjugate Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Iwano, S.; Sugiyama, M.; Hama, H.; Watakabe, A.; Hasegawa, N.; Kuchimaru, T.; Tanaka, K.Z.; Takahashi, M.; Ishida, Y.; Hata, J.; et al. Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Ashton, J.R.; West, J.L.; Badea, C.T. In vivo small animal micro-CT using nanoparticle contrast agents. Front. Pharmacol. 2015, 6, 256. [Google Scholar] [CrossRef]
- de Crespigny, A.; Bou-Reslan, H.; Nishimura, M.C.; Phillips, H.; Carano, R.A.D.; D’Arceuil, H.E. 3D micro-CT imaging of the postmortem brain. J. Neurosci. Methods 2008, 171, 207–213. [Google Scholar] [CrossRef]
- Engelhorn, T.; Eyüpoglu, I.Y.; Schwarz, M.A.; Karolczak, M.; Bruenner, H.; Struffert, T.; Kalender, W.; Doerfler, A. In vivo micro-CT imaging of rat brain glioma: A comparison with 3T MRI and histology. Neurosci. Lett. 2009, 458, 28–31. [Google Scholar] [CrossRef]
- Cormode, D.P.; Roessl, E.; Thran, A.; Skajaa, T.; Gordon, R.E.; Schlomka, J.-P.; Fuster, V.; Fisher, E.A.; Mulder, W.J.M.; Proksa, R.; et al. Atherosclerotic Plaque Composition: Analysis with Multicolor CT and Targeted Gold Nanoparticles. Radiology 2010, 256, 774–782. [Google Scholar] [CrossRef]
- Hallouard, F.; Briançon, S.; Anton, N.; Li, X.; Vandamme, T.; Fessi, H. Iodinated nano-emulsions as contrast agents for preclinical X-ray imaging: Impact of the free surfactants on the pharmacokinetics. Eur. J. Pharm. Biopharm. 2013, 83, 54–62. [Google Scholar] [CrossRef]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Control. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]
- Li, X.; Anton, N.; Zuber, G.; Vandamme, T. Contrast agents for preclinical targeted X-ray imaging. Adv. Drug Deliv. Rev. 2014, 76, 116–133. [Google Scholar] [CrossRef]
- Hainfeld, J.F.; O’Connor, M.J.; Dilmanian, F.A.; Slatkin, D.N.; Adams, D.J.; Smilowitz, H.M. Micro-CT enables microlocalisation and quantification of Her2-targeted gold nanoparticles within tumour regions. Br. J. Radiol. 2011, 84, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zheng, L.; Peng, C.; Shen, M.; Shi, X.; Zhang, G. Folic acid-modified dendrimer-entrapped gold nanoparticles as nanoprobes for targeted CT imaging of human lung adencarcinoma. Biomaterials 2013, 34, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Groheux, D.; Espié, M.; Giacchetti, S.; Hindié, E. Performance of FDG PET/CT in the Clinical Management of Breast Cancer. Radiology 2013, 266, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Popovtzer, R.; Reuveni, T.; Motiei, M.; Romman, Z.; Popovtzer, A. Targeted gold nanoparticles enable molecular CT imaging of cancer: An in vivo study. Int. J. Nanomed. 2011, 6, 2859–2864. [Google Scholar] [CrossRef]
- Chen, W.; Silverman, D.H.S.; Delaloye, S.; Czernin, J.; Kamdar, N.; Pope, W.; Satyamurthy, N.; Schiepers, C.; Cloughesy, T. 18F-FDOPA PET Imaging of Brain Tumors: Comparison Study with 18F-FDG PET and Evaluation of Diagnostic Accuracy. J. Nucl. Med. 2006, 47, 904–911. [Google Scholar]
- Wolf, G.; Abolmaali, N. Preclinical Molecular Imaging Using PET and MRI. In Molecular Imaging in Oncology; Schober, O., Riemann, B., Eds.; Recent Results in Cancer Research; Springer: Berlin/Heidelberg, Germany, 2013; pp. 257–310. ISBN 978-3-642-10853-2. [Google Scholar]
- Zhou, H.; Chen, M.; Zhao, D. Longitudinal MRI Evaluation of Intracranial Development and Vascular Characteristics of Breast Cancer Brain Metastases in a Mouse Model. PLoS ONE 2013, 8, e62238. [Google Scholar] [CrossRef]
- Heyn, C.; Ronald, J.A.; Ramadan, S.S.; Snir, J.A.; Barry, A.M.; MacKenzie, L.T.; Mikulis, D.J.; Palmieri, D.; Bronder, J.L.; Steeg, P.S.; et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2006, 56, 1001–1010. [Google Scholar] [CrossRef]
- Perera, M.; Ribot, E.J.; Percy, D.B.; McFadden, C.; Simedrea, C.; Palmieri, D.; Chambers, A.F.; Foster, P.J. In Vivo Magnetic Resonance Imaging for Investigating the Development and Distribution of Experimental Brain Metastases due to Breast Cancer. Transl. Oncol. 2012, 5, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.N.; Chen, Y.; McFadden, C.D.; Simedrea, F.C.; Foster, P.J. In-vivo longitudinal MRI Study: An Assessment of Melanoma Brain Metastases in a Clinically Relevant Mouse Model. Melanoma Res. 2015, 25, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Murrell, D.H.; Hamilton, A.M.; Mallett, C.L.; van Gorkum, R.; Chambers, A.F.; Foster, P.J. Understanding Heterogeneity and Permeability of Brain Metastases in Murine Models of HER2-Positive Breast Cancer through Magnetic Resonance Imaging: Implications for Detection and Therapy. Transl. Oncol. 2015, 8, 176–184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Economopoulos, V.; Chen, Y.; McFadden, C.; Foster, P.J. MRI Detection of Nonproliferative Tumor Cells in Lymph Node Metastases Using Iron Oxide Particles in a Mouse Model of Breast Cancer. Transl. Oncol. 2013, 6, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Mellor, H.R.; Ferguson, D.J.P.; Callaghan, R. A model of quiescent tumour microregions for evaluating multicellular resistance to chemotherapeutic drugs. Br. J. Cancer 2005, 93, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Prunier, C.; Baker, D.; Dijke, P.T.; Ritsma, L. TGF-β Family Signaling Pathways in Cellular Dormancy. Trends Cancer 2019, 5, 66–78. [Google Scholar] [CrossRef]
- De Meulenaere, V.; Neyt, S.; Vandeghinste, B.; Mollet, P.; De Wever, O.; Decrock, E.; Leybaert, L.; Goethals, I.; Vanhove, C.; Descamps, B.; et al. Species-dependent extracranial manifestations of a brain seeking breast cancer cell line. PLoS ONE 2018, 13, e0208340. [Google Scholar] [CrossRef]
- Parkins, K.M.; Hamilton, A.M.; Makela, A.V.; Chen, Y.; Foster, P.J.; Ronald, J.A. A multimodality imaging model to track viable breast cancer cells from single arrest to metastasis in the mouse brain. Sci. Rep. 2016, 6, 35889. [Google Scholar] [CrossRef]
- Zhou, Z.; Vaidyanathan, G.; McDougald, D.; Kang, C.M.; Balyasnikova, I.; Devoogdt, N.; Ta, A.N.; McNaughton, B.R.; Zalutsky, M.R. Fluorine-18 Labeling of the HER2-Targeting Single-Domain Antibody 2Rs15d Using a Residualizing Label and Preclinical Evaluation. Mol. Imaging Biol. 2017, 19, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef]
- Gillet, J.-P.; Calcagno, A.M.; Varma, S.; Marino, M.; Green, L.J.; Vora, M.I.; Patel, C.; Orina, J.N.; Eliseeva, T.A.; Singal, V.; et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 18708–18713. [Google Scholar] [CrossRef]
- Johnson, J.I.; Decker, S.; Zaharevitz, D.; Rubinstein, L.V.; Venditti, J.M.; Schepartz, S.; Kalyandrug, S.; Christian, M.; Arbuck, S.; Hollingshead, M.; et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer 2001, 84, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.J.; Kang, Y.; Serganova, I.; Gupta, G.P.; Giri, D.D.; Doubrovin, M.; Ponomarev, V.; Gerald, W.L.; Blasberg, R.; Massagué, J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Investig. 2005, 115, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Daniel, V.C.; Marchionni, L.; Hierman, J.S.; Rhodes, J.T.; Devereux, W.L.; Rudin, C.M.; Yung, R.; Parmigiani, G.; Dorsch, M.; Peacock, C.D.; et al. A Primary Xenograft Model of Small-Cell Lung Cancer Reveals Irreversible Changes in Gene Expression Imposed by Culture In vitro. Cancer Res. 2009, 69, 3364–3373. [Google Scholar] [CrossRef]
- DeRose, Y.S.; Wang, G.; Lin, Y.-C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.W.; Factor, R.; Matsen, C.; Milash, B.A.; Nelson, E.; et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Claerhout, S.; Prat, A.; Dobrolecki, L.E.; Petrovic, I.; Lai, Q.; Landis, M.D.; Wiechmann, L.; Schiff, R.; Giuliano, M.; et al. A Renewable Tissue Resource of Phenotypically Stable, Biologically and Ethnically Diverse, Patient-Derived Human Breast Cancer Xenograft Models. Cancer Res. 2013, 73, 4885–4897. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef]
- Guillen, K.P.; Fujita, M.; Butterfield, A.J.; Scherer, S.D.; Bailey, M.H.; Chu, Z.; DeRose, Y.S.; Zhao, L.; Cortes-Sanchez, E.; Yang, C.-H.; et al. A Breast Cancer Patient-Derived Xenograft and Organoid Platform for Drug Discovery and Precision Oncology. bioRxiv 2021, 2, 433268. [Google Scholar] [CrossRef]
- Cosgrove, N.; Varešlija, D.; Keelan, S.; Elangovan, A.; Atkinson, J.M.; Cocchiglia, S.; Bane, F.T.; Singh, V.; Furney, S.; Hu, C.; et al. Mapping molecular subtype specific alterations in breast cancer brain metastases identifies clinically relevant vulnerabilities. Nat. Commun. 2022, 13, 514. [Google Scholar] [CrossRef] [PubMed]
- Shumakovich, M.A.; Mencio, C.P.; Siglin, J.S.; Moriarty, R.A.; Geller, H.M.; Stroka, K.M. Astrocytes from the brain microenvironment alter migration and morphology of metastatic breast cancer cells. FASEB J. 2017, 31, 5049–5067. [Google Scholar] [CrossRef] [PubMed]
- Knier, N.N.; Hamilton, A.M.; Foster, P.J. Comparing the fate of brain metastatic breast cancer cells in different immune compromised mice with cellular magnetic resonance imaging. Clin. Exp. Metastasis 2020, 37, 465–475. [Google Scholar] [CrossRef] [PubMed]

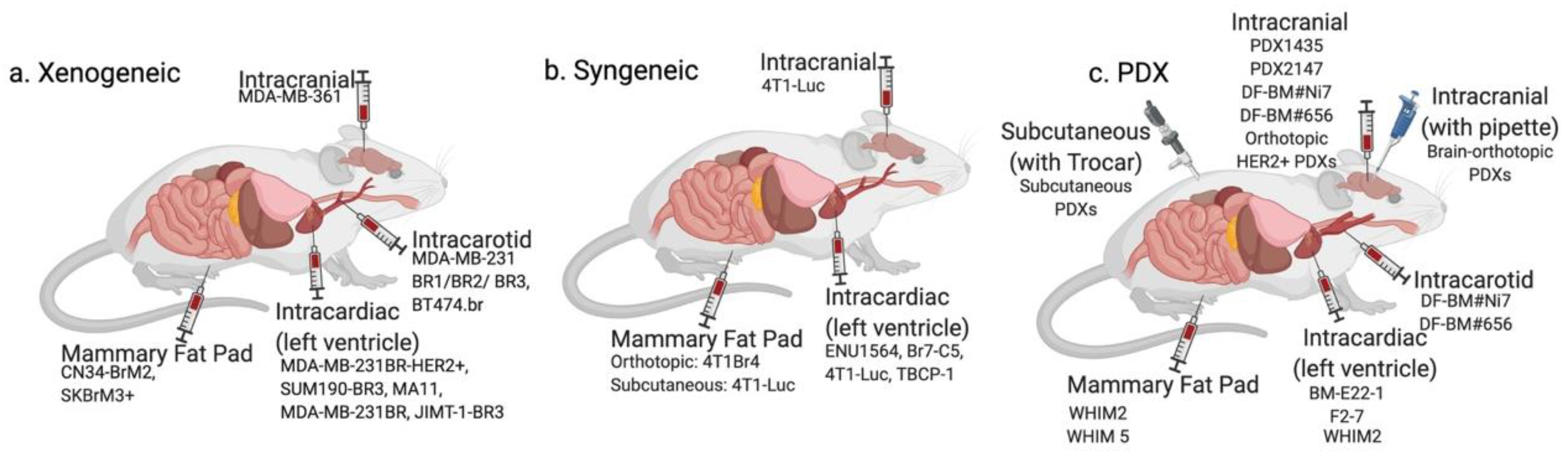
| Cell Type | Origin | Subtype | Animal Model | Injection Method | Detection Method | Drugs Studied | Original Reference | |
|---|---|---|---|---|---|---|---|---|
| Xenogeneic | MDA-MB-361 | Brain metastasis | ER+/PR+/HER2+ | Nude mice | Intracarotid | Histology | Docetaxel, doxorubicin and cyclophosphamide [20] | [21] |
| MDA-MB-468 | Pleural effusion | TNBC | Nude mice | Intracarotid | Histology | Docetaxel [22] | [21] | |
| MA11 | Bone marrow aspirate | TNBC | BALB/C nu/nu nude mice | Intracardiac | Autopsy, Histology, and MRI | Ionizing radiation and trichostatin A (HDAC inhibitor) [23] | [24] | |
| MDA-MB-231BR | Pleural effusion | TNBC | Nude mice | Intracardiac | Histology | Vorinostat [25] DAPT [26] GSK461364A [27] HA-paclitaxel nanoconjugate [28] Saracatinib with lapatinib [29] Whole brain radiotherapy [30,31] BCF [32] ANG1005/GRN1005 [33] iRGD nanoparticles [34] Azacitidine [35] WP1066 [36] Radiation with ultrasound-ruptured oxygen microbubbles [37] mTOR inhibitors (rapamycin, Temsirolimus-CCI-779) [38] | [39] | |
| MDA-MB-231BR1, -BR2, -BR3 | Pleural effusion | TNBC | Athymic NCr-nu/nu mice | Intracarotid | Histology | PTK787/Z 222584 [40] Temozolomide [41] | [40] | |
| MDA-MB- 231-BrM2 | Pleural effusion | TNBC | Athymic nude mice | Intracardiac | BLI, MRI, Histology | GDC-0068 [42] | [43] | |
| MDA-MB-231BR-HER2+ | Pleural effusion, then brain metastases in mice | ER-/PR-/HER2+ | BALB/c nude mice | Intracardiac | Immunofluorescence | Lapatinib [44] Pazopanib [45] LRRC31 nanoparticles with radiation [46] Whole brain radiotherapy [47] | [48] | |
| CN34-BrM2 | Pleural effusion | TNBC | Beige nude mice | Intracardiac | BLI, MRI, Histology | mTOR inhibitors (rapamycin, Temsirolimus-CCI-779) [38] | [43] | |
| JIMT-1-BR3 | Pleural effusion | HER2+ | NRC nu/nu mice | Intracardiac | Histology | Temozolomide [41] | [41] | |
| SUM190-BR3 | Primary tumor | HER2+ | Athymic NIH nu/nu mice | Intracardiac | Immunofluorescence | N/A | [49] | |
| BT474.br/Br.2/Br.3 | Primary tumor | ER+/PR+/HER2+ | Swiss nude mice | Intracarotid | Confocal microscopy, Immunofluorescence | Vardenafil and trastuzumab [50] Lapatinib and trastuzumab [51] TAK-285 [52] Saracatinib with lapatinib [29] | [29] | |
| SKBrM3+ | Plural effusion | ER-/PR-/HER2+ | Athymic nude mice | Mammary fat pad | BLI, Histology | Cabozantinib and Neratinib [53] | [53] | |
| Syngeneic | Br7-C5 | N-ethyl-N nitrosourea-induced mammary adenocarci- noma | Unspecified | Berlin–Druckrey IV rat | Intracardiac | Histology | N/A | [54] |
| 4T1BM | Murine mammary carcinoma | TNBC | Syngeneic BALB/c mice | Mammary fat pad | Histology | N/A | [55] | |
| 4T1Br4 | Murine mammary carcinoma | TNBC | Syngeneic BALB/c mice | Mammary fat pad | Histology | Trebananib [56] | [57] | |
| 4T1-Luc | Murine mammary carcinoma | TNBC | Syngeneic BALB/c mice | Intracranial, intracardiac, spontaneous | BLI | Fluphenazine hydrochloride [58] | [59] | |
| TBCP-1 | Spontaneous BALB/C mammary tumor | ER-/PR-/HER2+ | Syngeneic BALB/C mice | Intracardiac | Histology | Neratinib [60] | [60] | |
| Patient-Derived | F2-7 | Patient brain metastases | TNBC | NSG mice | Intracardiac | BLI | N/A | [61] |
| Brain-orthotopic PDXs | Patient brain metastases | TNBC and ER+ varied | NSG mice | Intracranial (pipette method) | Histology | N/A | [62] | |
| BM-E22-1 | Patient brain metastases | TNBC | NSG mice | Intracardiac | MRI | N/A | [61] | |
| DF-BM#Ni7, DF-BM#656 | Patient brain metastases | ER+ HER2+ (DF-BM#Ni7), TNBC (DF-BM#656) | NOD/SCID mice | Intracarotid (ligation method) | BLI | N/A | [63] | |
| WHIM 2/WHIM5 | Primary tumor/patient brain metastases | TNBC | NOD/SCID mice | Mammary fat pad | Histology | Carboplatin, cyclophosphamide, bortezomib, dacarbazine [64] | [65] | |
| PDX1435/PDX2147 | Patient brain metastases (PDX1435), primary tumor (PDX 2147) | TNBC | NOD/SCID mice | Intracranial | MRI | BCF [32] | [32] | |
| Orthotopic HER2+ PDXs | Patient brain metastases | HER2+, ER/PR status varied | NOD/SCID mice | Intracranial | BLI, MRI | Combination of PI3K inhibitor (BKM120) and mTORC1 inhibitor (RAD001) [66] | [66] | |
| Subcutaneous PDXs | Patient brain metastases | Unspecified | SCID BALB/c mice | Subcutaneous (trocar method) | PET/CT | N/A | [67] |
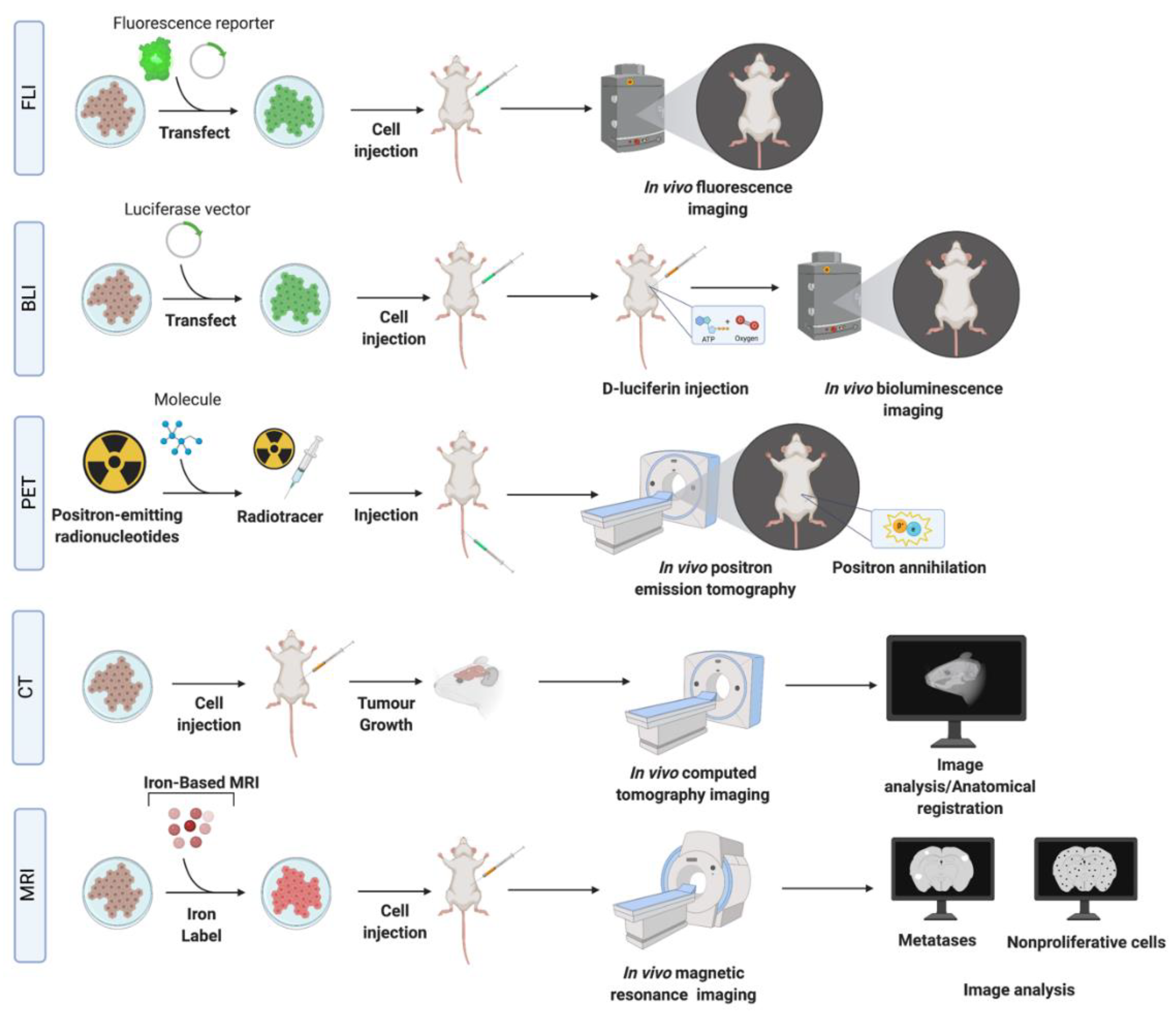
| Imaging Modality | Principles | Reporters /Detection Used | SR/S/HS/Sp | Information | Advantages | Disadvantages and Limitations for Imaging |
|---|---|---|---|---|---|---|
| BLI | Optical detection of light emitted from BLI reporters. | Genetically expressed proteins such as luciferase | SR—~1 mm S—Medium (1000 s of cells) HS—one cell Sp—High | Probe uptake, cell presence, and cell viability. | Minimally invasive, inexpensive, allows for signal quantification, whole mouse imaging and has high throughput. BLI signal is only produced by viable cancer cells permitting distinction between viable and dead cells. | Requires stable transfection of the reporter into cancer cells and injection of substrate into a mouse a. Limited depth penetration and therefore, not clinically translatable. Challenging to determine depth of a tumor within the body based on the signal. False negative effects can occur in areas where the substrate cannot easily accumulate, such as the brain, or in tumors with compromised vasculature. Probe uptake in the brain and limited imaging depth in biological tissues. |
| FLI | Optical detection of light emitted from fluorescent reporters. | GFP, eGFP, EYFP, mCherry, TagRFP, Dendra2, tdTomato. | SR—~1 mm S—Medium Sp—High | Probe uptake, cell presence and cell viability. | Minimally invasive, inexpensive, allows for whole mouse imaging and has high throughput. Does not require injection of substrate. The signal is quantifiable. | Requires stable transfection/transduction of the reporter into cancer cells and excitation by an external light source. Background autofluoresence decreases sensitivity. Challenging to determine depth of a tumor within the body based on the signal. Probe uptake in the brain and limited imaging depth in biological tissues. |
| CT (with and without contrast) | Combinations of multiple X-ray measurements taken from different angles to produce tomographic images. With a contrast agent, CT images can reveal the location and density of vessels (early), and contrast agent accumulation in the tissue (late). | Iodine-containing polymers [77], liposomes [78] or micelles [79] and gold nanoparticles [80]. | SR—~ 100 um S—Low Sp—Medium | Tomographic images, vessel density, and agent accumulation. | Low cost, fast acquisition and high spatial resolution of 3D volumes. | Radiation exposure, low contrast can make certain pathologies difficult to discern; contrast-enhanced micro-CT is more commonly applied. Low contrast does not allow for visualization of tumor detail, often needs contrast enhancement. |
| PET | Detection of γ rays from positron emitting radioisotopes b. | FDG, 18F-FMISO. | SR—~1 mm S—High picomolar (100–1000 s of cells) Sp—High | Tracer uptake; biological and biochemical. Direct cell quantification, and signal specific to cells. | Can monitor tissue metabolism (glycolysis, DNA synthesis, amino acid transport and oxygenation state) in brain metastases, with excellent depth penetration. | Requires tracers, normal brain tissue has a high rate of glucose metabolism and therefore high FDG accumulation which decreases specificity. Signal decays over time (t1/2), and cells are exposed to radioactivity. Low radiotracer uptake in brain. |
| MRI (proton) | Detection of water proton relaxation after RF absorption. | See below. | SR—500–2000 microns S—Low millimolar Sp—Medium | Anatomical information, morphology, and tissue composition. | No ionizing radiation exposure, provides excellent soft tissue contrast. | Potential tissue heating during long scans, risk of peripheral nerve stimulation, sensitive to motion. Poor sensitivity in detecting micrometastases. |
| MRI (contrast) | MRI with use of contrast agents, administered to improve signal differences between normal and cancerous tissue. | Most common contrasts—gadolinium-based, manganese-based. | SR—500–2000 microns S—medium Sp—Medium | Improved visibility of tumors, inflammation, and blood supply. | No radiation exposure. Clinically, dynamic contrast enhanced (DCE) MRI can be used to image the tumor vasculature by acquiring sequential images during the passage of gadolinium through tissues and provides quantitative measures of perfusion, permeability and blood volume. | Requires administration of contrast. Heterogeneity of metastasis permeability in early and late stages of development. |
| MRI (iron nanoparticles) | Detection of intracellular iron particles via distortion of the magnetic field. | SPIO nanoparticles labeling via co-incubation with cancer cells. | SR—200–1000 microns S—High picomolar HS—one cell Sp—Medium | Cell location and presence, including nonproliferative cells. | High sensitivity, non-proliferative, cancer cells do not dilute the SPIO and can be identified by MRI as persistent signal voids by virtue of their retaining iron. | SPIO are diluted in the progeny of proliferative cells and therefore labeled cells become undetectable by MRI after repeated cell divisions. Poor cell quantification. Other structures in brain appear with low signal (i.e., blood, air, bone). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knier, N.N.; Pellizzari, S.; Zhou, J.; Foster, P.J.; Parsyan, A. Preclinical Models of Brain Metastases in Breast Cancer. Biomedicines 2022, 10, 667. https://doi.org/10.3390/biomedicines10030667
Knier NN, Pellizzari S, Zhou J, Foster PJ, Parsyan A. Preclinical Models of Brain Metastases in Breast Cancer. Biomedicines. 2022; 10(3):667. https://doi.org/10.3390/biomedicines10030667
Chicago/Turabian StyleKnier, Natasha N., Sierra Pellizzari, Jiangbing Zhou, Paula J. Foster, and Armen Parsyan. 2022. "Preclinical Models of Brain Metastases in Breast Cancer" Biomedicines 10, no. 3: 667. https://doi.org/10.3390/biomedicines10030667
APA StyleKnier, N. N., Pellizzari, S., Zhou, J., Foster, P. J., & Parsyan, A. (2022). Preclinical Models of Brain Metastases in Breast Cancer. Biomedicines, 10(3), 667. https://doi.org/10.3390/biomedicines10030667






