hTERT DNA Methylation Analysis Identifies a Biomarker for Retinoic Acid-Induced hTERT Repression in Breast Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and ATRA Treatment
2.2. RNA Extraction and Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
2.3. DNA Extraction and Sanger Sequencing
2.4. Bisulfite Modification
2.5. Statistical Analysis
3. Results
3.1. TERT mRNA Expression and Mutational Analysis in Breast Cancer Cell Lines
3.2. ATRA Induces Differential Modulations of hTERT Expression in Breast Cancer Cell Lines
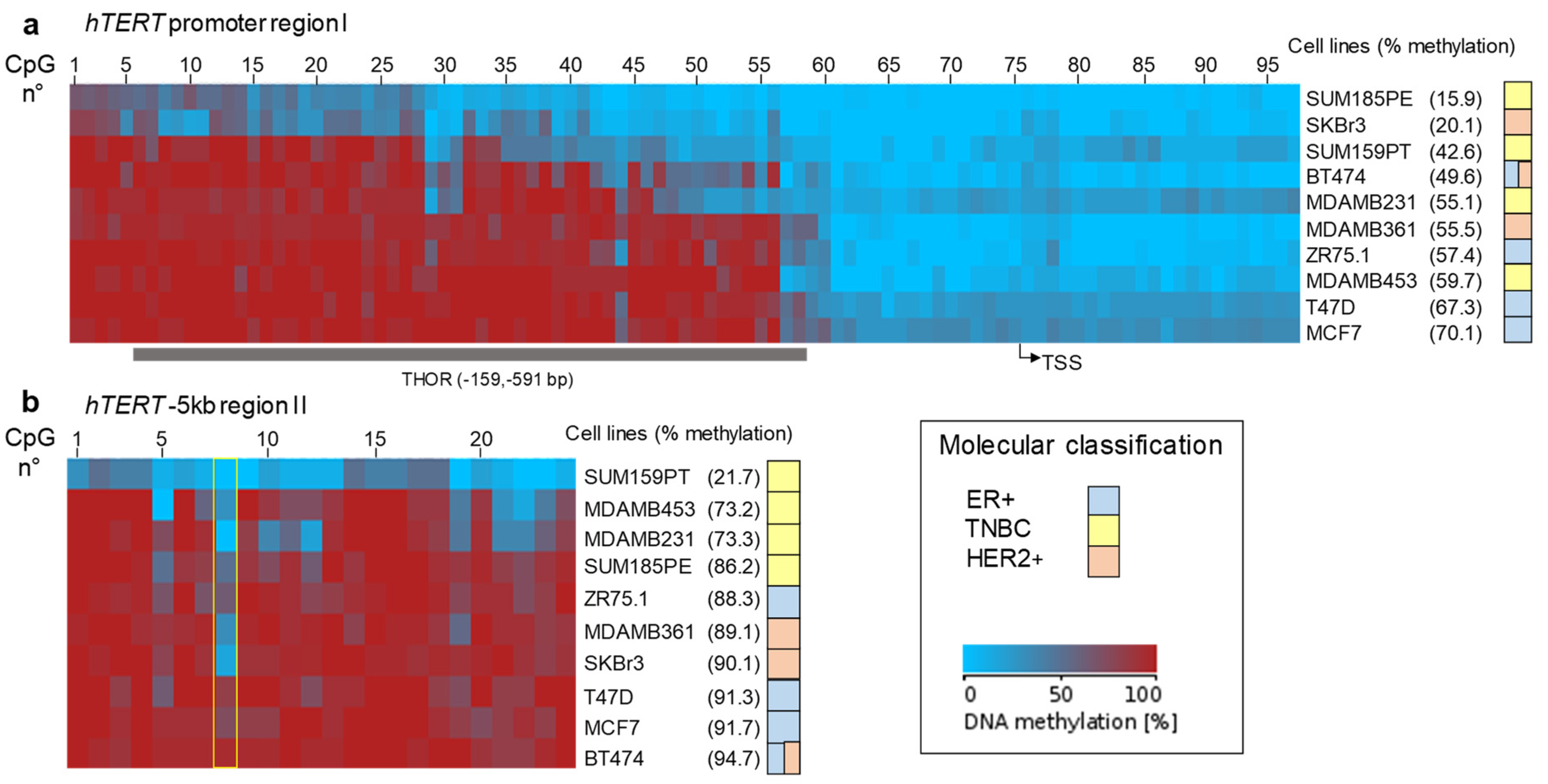
3.3. Pattern of DNA Methylation at hTERT Promoter Region I and Region II
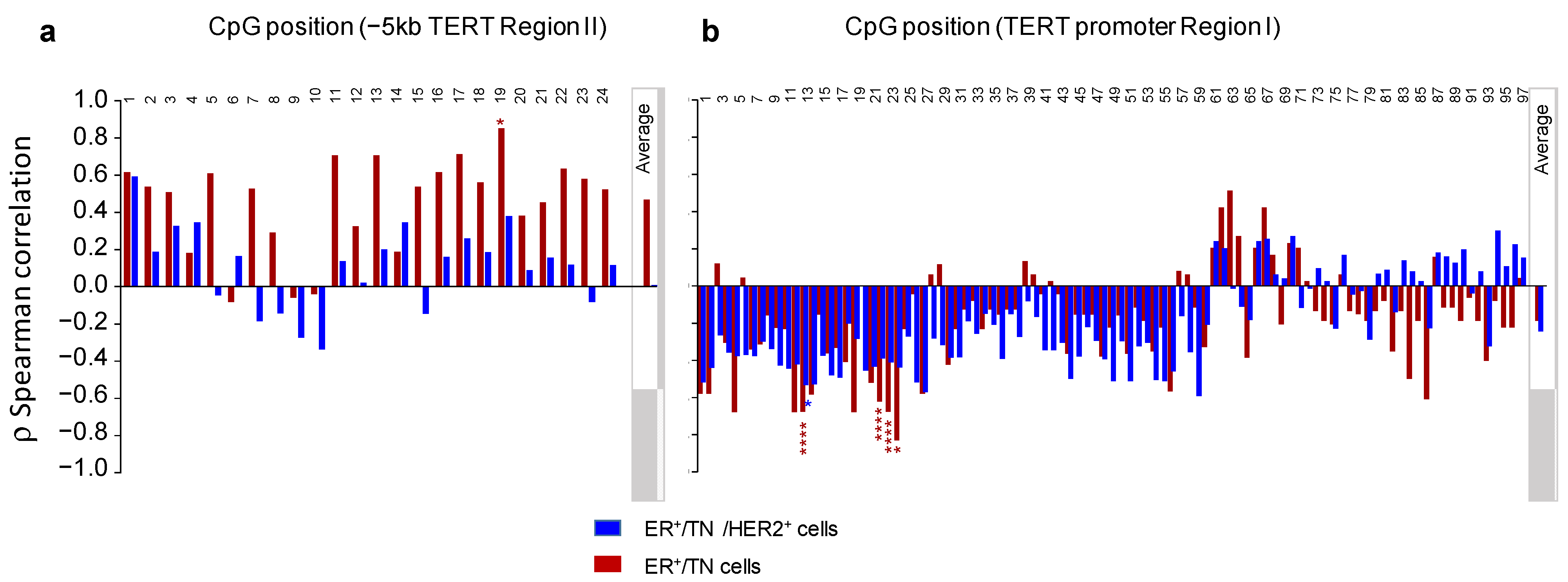
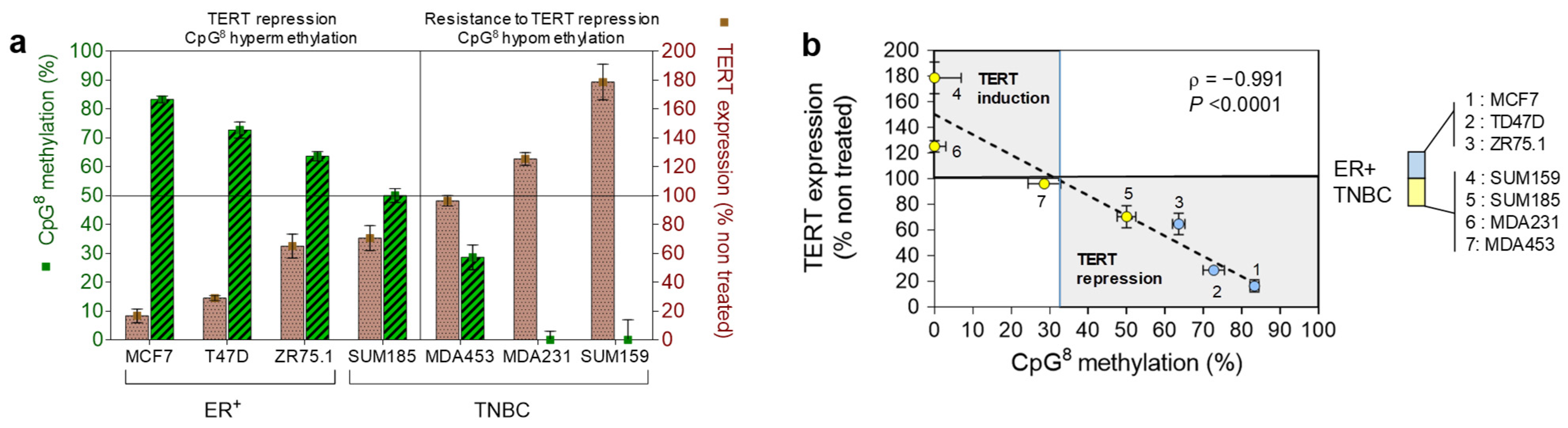
3.4. Relationship between DNA Methylation and Expression of hTERT at Baseline or after ATRA Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting Chromosomes against Genome Instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Pestana, A.; Vinagre, J.; Sobrinho-Simões, M.; Soares, P. TERT Biology and Function in Cancer: Beyond Immortalisation. J. Mol. Endocrinol. 2017, 58, R129–R146. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Funk, W.D.; Wang, S.S.; Weinrich, S.L.; Avilion, A.A.; Chiu, C.P.; Adams, R.R.; Chang, E.; Allsopp, R.C.; Yu, J. The RNA Component of Human Telomerase. Science 1995, 269, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Counter, C.M.; Eaton, E.N.; Ellisen, L.W.; Steiner, P.; Caddle, S.D.; Ziaugra, L.; Beijersbergen, R.L.; Davidoff, M.J.; Liu, Q.; et al. HEST2, the Putative Human Telomerase Catalytic Subunit Gene, Is up-Regulated in Tumor Cells and during Immortalization. Cell 1997, 90, 785–795. [Google Scholar] [CrossRef]
- Dudognon, C.; Pendino, F.; Hillion, J.; Saumet, A.; Lanotte, M.; Ségal-Bendirdjian, E. Death Receptor Signaling Regulatory Function for Telomerase: HTERT Abolishes TRAIL-Induced Apoptosis, Independently of Telomere Maintenance. Oncogene 2004, 23, 7469–7474. [Google Scholar] [CrossRef][Green Version]
- Ségal-Bendirdjian, E.; Geli, V. Non-Canonical Roles of Telomerase: Unraveling the Imbroglio. Front. Cell Dev. Biol. 2019, 7, 332. [Google Scholar] [CrossRef]
- Guterres, A.N.; Villanueva, J. Targeting Telomerase for Cancer Therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of Life-Span by Introduction of Telomerase into Normal Human Cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Krämer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase Activation by Genomic Rearrangements in High-Risk Neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zheng, C.; Lindvall, C.; Hou, M.; Ekedahl, J.; Lewensohn, R.; Yan, Z.; Yang, X.; Henriksson, M.; Blennow, E.; et al. Frequent Amplification of the Telomerase Reverse Transcriptase Gene in Human Tumors. Cancer Res. 2000, 60, 6230–6235. [Google Scholar] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef]
- Bell, R.J.A.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. Cancer. The Transcription Factor GABP Selectively Binds and Activates the Mutant TERT Promoter in Cancer. Science 2015, 348, 1036–1039. [Google Scholar] [CrossRef]
- Stern, J.L.; Theodorescu, D.; Vogelstein, B.; Papadopoulos, N.; Cech, T.R. Mutation of the TERT Promoter, Switch to Active Chromatin, and Monoallelic TERT Expression in Multiple Cancers. Genes Dev. 2015, 29, 2219–2224. [Google Scholar] [CrossRef]
- Guilleret, I.; Yan, P.; Grange, F.; Braunschweig, R.; Bosman, F.T.; Benhattar, J. Hypermethylation of the Human Telomerase Catalytic Subunit (HTERT) Gene Correlates with Telomerase Activity. Int. J. Cancer 2002, 101, 335–341. [Google Scholar] [CrossRef]
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S.; Ferrara, F.; Fazi, P.; Cicconi, L.; Di Bona, E.; et al. Retinoic Acid and Arsenic Trioxide for Acute Promyelocytic Leukemia. N. Engl. J. Med. 2013, 369, 111–121. [Google Scholar] [CrossRef]
- de Thé, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef]
- Pendino, F.; Flexor, M.; Delhommeau, F.; Buet, D.; Lanotte, M.; Segal-Bendirdjian, E. Retinoids Down-Regulate Telomerase and Telomere Length in a Pathway Distinct from Leukemia Cell Differentiation. Proc. Natl. Acad. Sci. USA 2001, 98, 6662–6667. [Google Scholar] [CrossRef]
- Pendino, F.; Sahraoui, T.; Lanotte, M.; Ségal-Bendirdjian, E. A Novel Mechanism of Retinoic Acid Resistance in Acute Promyelocytic Leukemia Cells through a Defective Pathway in Telomerase Regulation. Leukemia 2002, 16, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Pendino, F.; Hillion, J.; Dudognon, C.; Delaunay, J.; Mourah, S.; Podgorniak, M.-P.; Lafon, I.; Chomienne, C.; Lanotte, M.; Dombret, H.; et al. Telomerase Targeting by Retinoids in Cells from Patients with Myeloid Leukemias of Various Subtypes, Not Only APL. Leukemia 2006, 20, 599–603. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pendino, F.; Dudognon, C.; Delhommeau, F.; Sahraoui, T.; Flexor, M.; Bennaceur-Griscelli, A.; Lanotte, M.; Ségal-Bendirdjian, E. Retinoic Acid Receptor Alpha and Retinoid-X Receptor-Specific Agonists Synergistically Target Telomerase Expression and Induce Tumor Cell Death. Oncogene 2003, 22, 9142–9150. [Google Scholar] [CrossRef] [PubMed]
- Azouz, A.; Wu, Y.-L.; Hillion, J.; Tarkanyi, I.; Karniguian, A.; Aradi, J.; Lanotte, M.; Chen, G.-Q.; Chehna, M.; Ségal-Bendirdjian, E. Epigenetic Plasticity of HTERT Gene Promoter Determines Retinoid Capacity to Repress Telomerase in Maturation-Resistant Acute Promyelocytic Leukemia Cells. Leukemia 2010, 24, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, P.; Choufani, S.; Mack, S.; Gallagher, D.; Zhang, C.; Lipman, T.; Zhukova, N.; Walker, E.J.; Martin, D.; Merino, D.; et al. Methylation of the TERT Promoter and Risk Stratification of Childhood Brain Tumours: An Integrative Genomic and Molecular Study. Lancet Oncol. 2013, 14, 534–542. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Leão, R.; Lipman, T.; Campbell, B.; Lee, D.; Price, A.; Zhang, C.; Heidari, A.; Stephens, D.; Boerno, S.; et al. A Cancer Specific Hypermethylation Signature of the TERT Promoter Predicts Biochemical Relapse in Prostate Cancer: A Retrospective Cohort Study. Oncotarget 2016, 7, 57726–57736. [Google Scholar] [CrossRef]
- Faleiro, I.; Apolónio, J.D.; Price, A.J.; De Mello, R.A.; Roberto, V.P.; Tabori, U.; Castelo-Branco, P. The TERT Hypermethylated Oncologic Region Predicts Recurrence and Survival in Pancreatic Cancer. Future Oncol. Lond. Engl. 2017, 13, 2045–2051. [Google Scholar] [CrossRef]
- Leão, R.; Lee, D.; Figueiredo, A.; Hermanns, T.; Wild, P.; Komosa, M.; Lau, I.; Mistry, M.; Nunes, N.M.; Price, A.J.; et al. Combined Genetic and Epigenetic Alterations of the TERT Promoter Affect Clinical and Biological Behavior of Bladder Cancer. Int. J. Cancer 2019, 144, 1676–1684. [Google Scholar] [CrossRef]
- Garsuault, D.; Bouyer, C.; Nguyen, E.; Kandhari, R.; Prochazkova-Carlotti, M.; Chevret, E.; Forgez, P.; Ségal-Bendirdjian, E. Complex Context Relationships between DNA Methylation and Accessibility, Histone Marks, and HTERT Gene Expression in Acute Promyelocytic Leukemia Cells: Perspectives for All-Trans Retinoic Acid in Cancer Therapy. Mol. Oncol. 2020, 14, 1310–1326. [Google Scholar] [CrossRef]
- Deville, L.; Hillion, J.; Pendino, F.; Samy, M.; Nguyen, E.; Ségal-Bendirdjian, E. HTERT Promotes Imatinib Resistance in Chronic Myeloid Leukemia Cells: Therapeutic Implications. Mol. Cancer Ther. 2011, 10, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Stebbing, J.; Giamas, G.; Murphy, J. Endocrine Resistance in Hormone Receptor Positive Breast Cancer-From Mechanism to Therapy. Front. Endocrinol. 2019, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Moasser, M.M. The Oncogene HER2: Its Signaling and Transforming Functions and Its Role in Human Cancer Pathogenesis. Oncogene 2007, 26, 6469–6487. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, E.J.; Kitsberg, D.I.; Leder, P. A Mouse Model for Breast Cancer Induced by Amplification and Overexpression of the Neu Promoter and Transgene. Mol. Med. Camb. Mass 2000, 6, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Joe, A. Oncogene Addiction. Cancer Res. 2008, 68, 3077–3080. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.A.; Oza, G.; Sharma, A.; Arriaga, L.G.; Hernández Hernández, J.M.; Rotello, V.M.; Ramirez, J.T. Triple-Negative Breast Cancer: A Review of Conventional and Advanced Therapeutic Strategies. Int. J. Environ. Res. Public. Health 2020, 17, 2078. [Google Scholar] [CrossRef]
- Hiyama, E.; Gollahon, L.; Kataoka, T.; Kuroi, K.; Yokoyama, T.; Gazdar, A.F.; Hiyama, K.; Piatyszek, M.A.; Shay, J.W. Telomerase Activity in Human Breast Tumors. J. Natl. Cancer Inst. 1996, 88, 116–122. [Google Scholar] [CrossRef]
- Poremba, C.; Heine, B.; Diallo, R.; Heinecke, A.; Wai, D.; Schaefer, K.-L.; Braun, Y.; Schuck, A.; Lanvers, C.; Bànkfalvi, A.; et al. Telomerase as a Prognostic Marker in Breast Cancer: High-Throughput Tissue Microarray Analysis of HTERT and HTR. J. Pathol. 2002, 198, 181–189. [Google Scholar] [CrossRef]
- Kulić, A.; Plavetić, N.D.; Gamulin, S.; Jakić-Razumović, J.; Vrbanec, D.; Sirotković-Skerlev, M. Telomerase Activity in Breast Cancer Patients: Association with Poor Prognosis and More Aggressive Phenotype. Med. Oncol. Northwood Lond. Engl. 2016, 33, 23. [Google Scholar] [CrossRef]
- Elkak, A.; Mokbel, R.; Wilson, C.; Jiang, W.G.; Newbold, R.F.; Mokbel, K. HTERT MRNA Expression Is Associated with a Poor Clinical Outcome in Human Breast Cancer. Anticancer Res. 2006, 26, 4901–4904. [Google Scholar]
- Garattini, E.; Bolis, M.; Garattini, S.K.; Fratelli, M.; Centritto, F.; Paroni, G.; Gianni’, M.; Zanetti, A.; Pagani, A.; Fisher, J.N.; et al. Retinoids and Breast Cancer: From Basic Studies to the Clinic and Back Again. Cancer Treat. Rev. 2014, 40, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Garattini, E.; Paroni, G.; Terao, M. Retinoids and Breast Cancer: New Clues to Increase Their Activity and Selectivity. Breast Cancer Res. BCR 2012, 14, 111. [Google Scholar] [CrossRef] [PubMed]
- Bolis, M.; Garattini, E.; Paroni, G.; Zanetti, A.; Kurosaki, M.; Castrignanò, T.; Garattini, S.K.; Biancardi, F.; Barzago, M.M.; Gianni’, M.; et al. Network-Guided Modeling Allows Tumor-Type Independent Prediction of Sensitivity to All-Trans-Retinoic Acid. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Centritto, F.; Paroni, G.; Bolis, M.; Garattini, S.K.; Kurosaki, M.; Barzago, M.M.; Zanetti, A.; Fisher, J.N.; Scott, M.F.; Pattini, L.; et al. Cellular and Molecular Determinants of All-Trans Retinoic Acid Sensitivity in Breast Cancer: Luminal Phenotype and RARα Expression. EMBO Mol. Med. 2015, 7, 950–972. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, G.A.; Kats, L.; Pandolfi, P.P. Synergy against PML-RARa: Targeting Transcription, Proteolysis, Differentiation, and Self-Renewal in Acute Promyelocytic Leukemia. J. Exp. Med. 2013, 210, 2793–2802. [Google Scholar] [CrossRef]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.-L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef]
- Segal-Bendirdjian, E.; Jacquemin-Sablon, A. Cisplatin Resistance in a Murine Leukemia Cell Line Is Associated with a Defective Apoptotic Process. Exp. Cell Res. 1995, 218, 201–212. [Google Scholar] [CrossRef]
- Tusnády, G.E.; Simon, I.; Váradi, A.; Arányi, T. BiSearch: Primer-Design and Search Tool for PCR on Bisulfite-Treated Genomes. Nucleic Acids Res. 2005, 33, e9. [Google Scholar] [CrossRef]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing Primers for Methylation PCRs. Bioinform. Oxf. Engl. 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification Tool for Methylation Analysis. Nucleic Acids Res. 2008, 36, W170–W175. [Google Scholar] [CrossRef]
- Eldholm, V.; Haugen, A.; Zienolddiny, S. CTCF Mediates the TERT Enhancer-Promoter Interactions in Lung Cancer Cells: Identification of a Novel Enhancer Region Involved in the Regulation of TERT Gene. Int. J. Cancer 2014, 134, 2305–2313. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, J.; Rønneberg, J.A.; Tost, J.; Kristensen, V. The Epigenetics of Breast Cancer. Mol. Oncol. 2010, 4, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Moran, S.; Gomez, A.; Sayols, S.; Arribas-Jorba, C.; Sandoval, J.; Hilmarsdottir, H.; Olafsdottir, E.; Tryggvadottir, L.; Jonasson, J.G.; et al. A DNA Methylation-Based Definition of Biologically Distinct Breast Cancer Subtypes. Mol. Oncol. 2015, 9, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Leão, R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolónio, J.D.; et al. DNA Hypermethylation within TERT Promoter Upregulates TERT Expression in Cancer. J. Clin. Investig. 2019, 129, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Komosa, M.; Nunes, N.M.; Tabori, U. DNA Methylation of the TERT Promoter and Its Impact on Human Cancer. Curr. Opin. Genet. Dev. 2020, 60, 17–24. [Google Scholar] [CrossRef]



| Cell Line | MCF7 | T47D | ZR75.1 | BT474 | SKBR3 | MDAMB361 | MDAMB453 1 | MDAMB231 | SUM159PT | SUM185PE |
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor type | Adeno carcinoma | Ductal carcinoma | Ductal carcinoma | Ductal carcinoma | Adeno carcinoma | Adeno carcinoma | Metaplastic carcinoma | Adeno carcinoma | Anaplastic carcinoma | Ductal carcinoma |
| Epithelial | Epithelial | Epithelial | Epithelial | Epithelial | Epithelial | Epithelial | Mesenchymal | Mesenchymal | Epithelial | |
| Molecular 2 classification | ERα+ | ERα+/ HER2+ | HER2+ | HER2+ | TNBC | |||||
| hTERT promoter status | Wild-type | Wild-type SNP T349C | Wild-type SNP T349C | Wild-type SNP T349C | Wild-type | Wild-type | Wild-type | Mutated C228T | Mutated C228T SNP T349C | Wild-type SNP T349C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, E.; Richerolle, A.; Sánchez-Bellver, J.; Varennes, J.; Ségal-Bendirdjian, E. hTERT DNA Methylation Analysis Identifies a Biomarker for Retinoic Acid-Induced hTERT Repression in Breast Cancer Cell Lines. Biomedicines 2022, 10, 695. https://doi.org/10.3390/biomedicines10030695
Nguyen E, Richerolle A, Sánchez-Bellver J, Varennes J, Ségal-Bendirdjian E. hTERT DNA Methylation Analysis Identifies a Biomarker for Retinoic Acid-Induced hTERT Repression in Breast Cancer Cell Lines. Biomedicines. 2022; 10(3):695. https://doi.org/10.3390/biomedicines10030695
Chicago/Turabian StyleNguyen, Eric, Andréa Richerolle, Júlia Sánchez-Bellver, Jacqueline Varennes, and Evelyne Ségal-Bendirdjian. 2022. "hTERT DNA Methylation Analysis Identifies a Biomarker for Retinoic Acid-Induced hTERT Repression in Breast Cancer Cell Lines" Biomedicines 10, no. 3: 695. https://doi.org/10.3390/biomedicines10030695
APA StyleNguyen, E., Richerolle, A., Sánchez-Bellver, J., Varennes, J., & Ségal-Bendirdjian, E. (2022). hTERT DNA Methylation Analysis Identifies a Biomarker for Retinoic Acid-Induced hTERT Repression in Breast Cancer Cell Lines. Biomedicines, 10(3), 695. https://doi.org/10.3390/biomedicines10030695






