Reporting on the Value of Artificial Intelligence in Predicting the Optimal Embryo for Transfer: A Systematic Review including Data Synthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
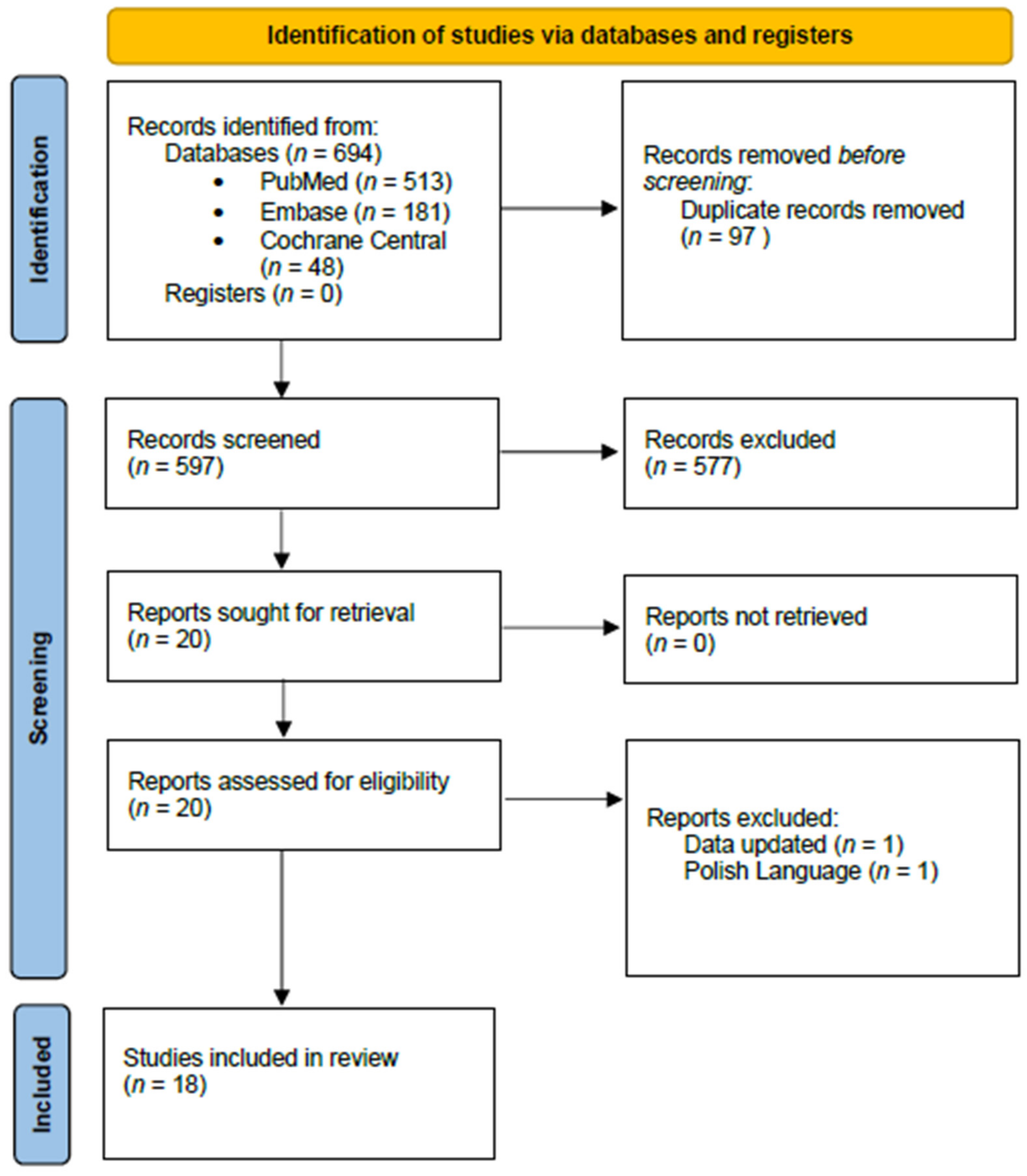
2.2. Study Selection
2.3. Excluded Studies
2.4. Data Extraction
2.5. Outcomes
2.6. Bias Assessment
2.7. Deviations from Protocol
Metrics and Measures
2.8. Statistical Analysis
3. Results
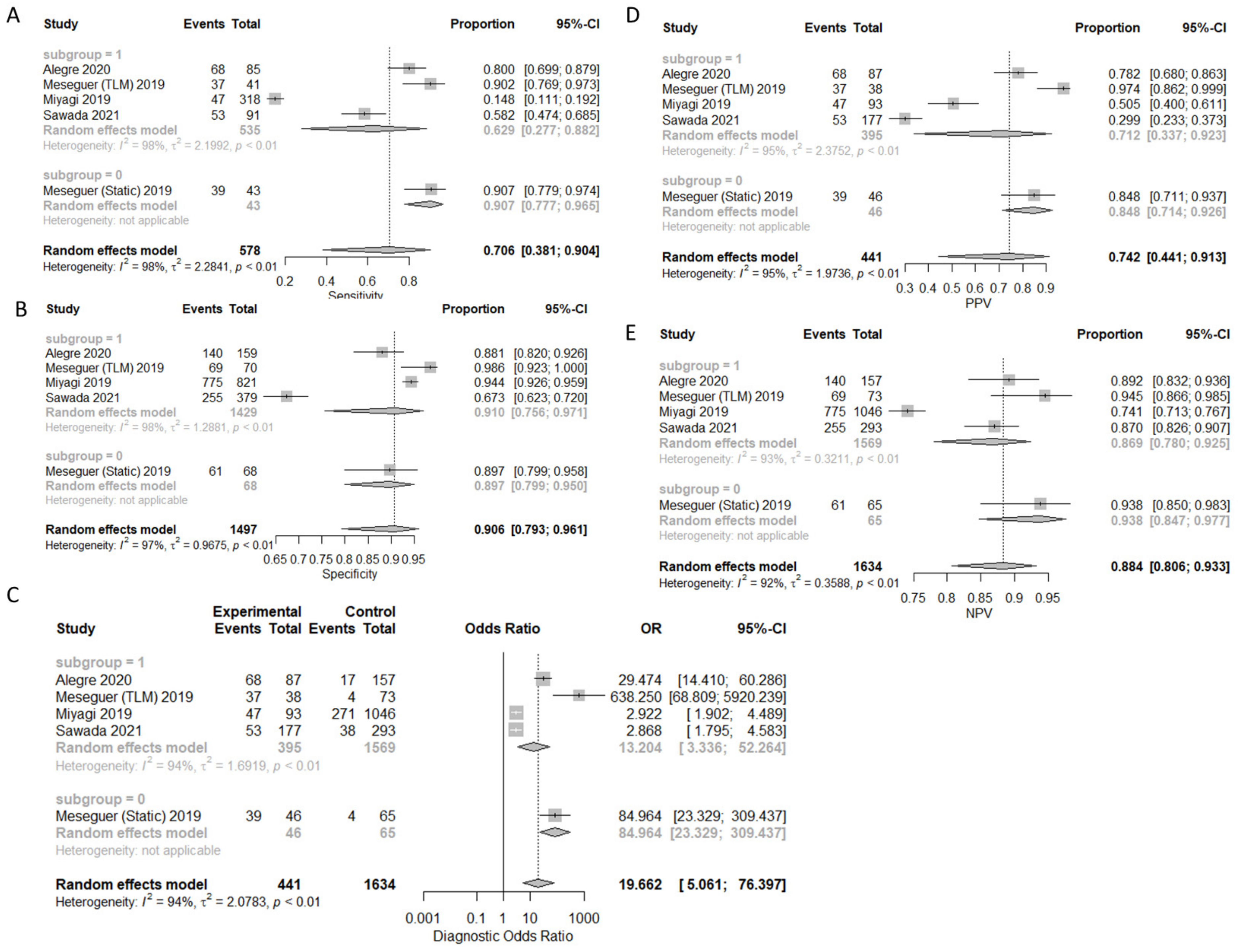
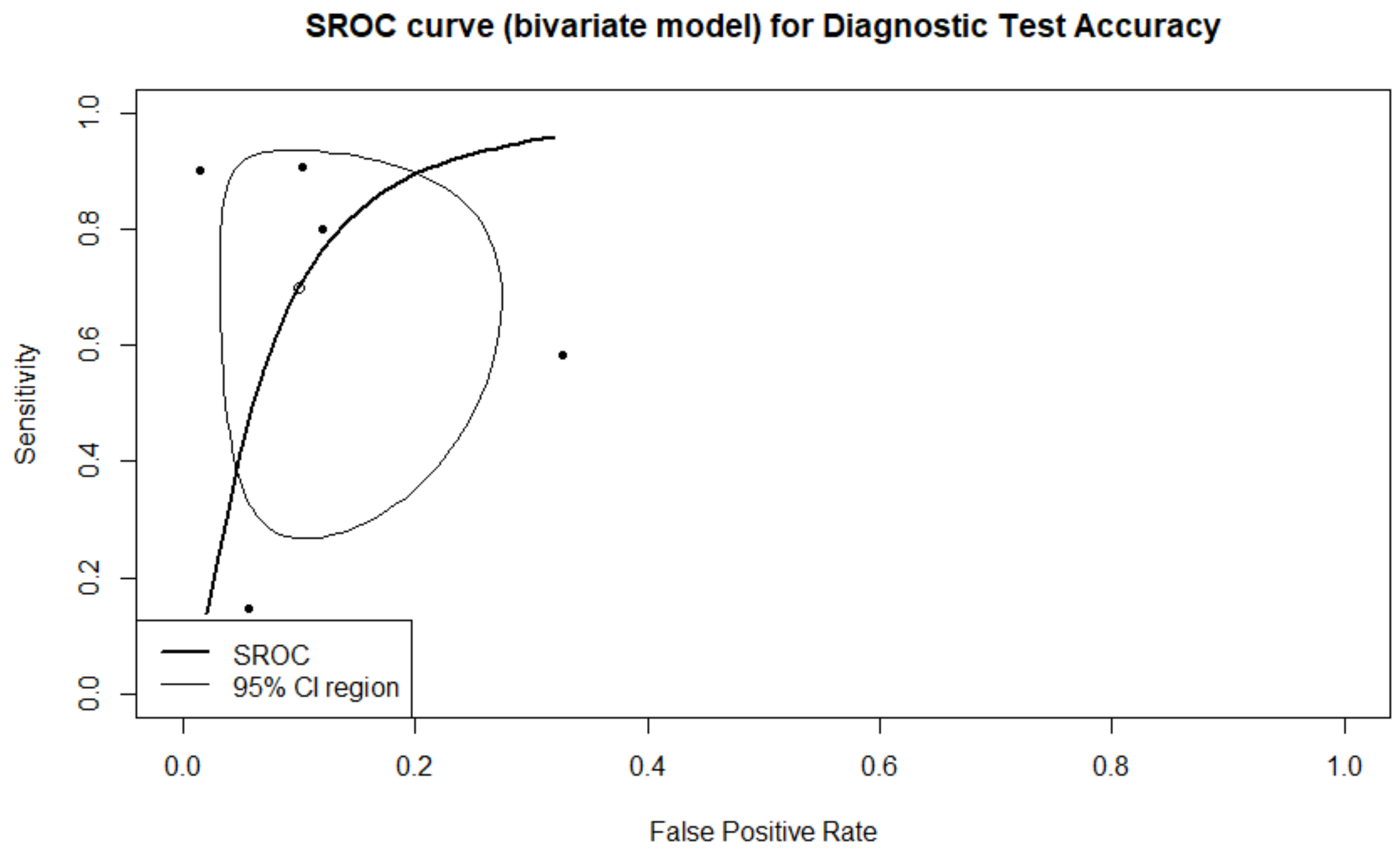
3.1. Prediction of Live-Birth
3.2. Sensitivity Analysis on Live Birth Prediction
3.3. Secondary Outcome Measures
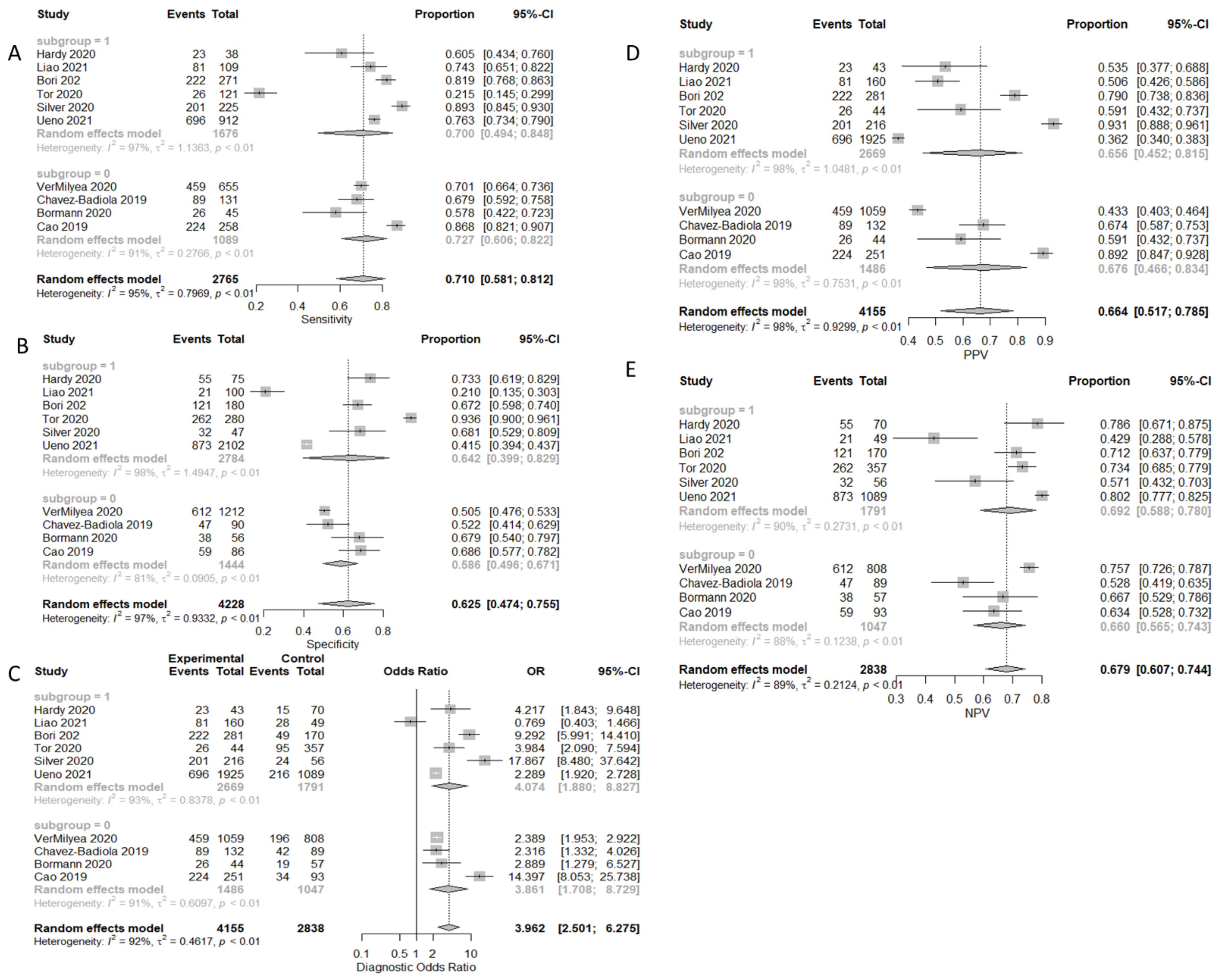
3.3.1. Prediction of Pregnancy
3.3.2. Prediction of Clinical Pregnancy with Fetal Heart-Beat
3.3.3. Prediction of Ploidy Status
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.L.; Callender, C.S.K.H.; Kulikoff, X.R.; Srinivasan, V.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbasi, N.; Abbastabar, H.; Abdela, J.; et al. Population and Fertility by Age and Sex for 195 Countries and Territories, 1950–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1995–2051. [Google Scholar] [CrossRef] [Green Version]
- Barash, O.; Ivani, K.; Huen, N.; Willman, S.; Weckstein, L. Morphology of the Blastocysts Is the Single Most Important Factor Affecting Clinical Pregnancy Rates in IVF PGS Cycles with Single Embryo Transfers. Fertil. Steril. 2017, 108, e99. [Google Scholar] [CrossRef]
- Machtinger, R.; Racowsky, C. Morphological Systems of Human Embryo Assessment and Clinical Evidence. Reprod. BioMed. Online 2013, 26, 210–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundin, K.; Ahlström, A. Quality Control and Standardization of Embryo Morphology Scoring and Viability Markers. Reprod. BioMed. Online 2015, 31, 459–471. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.-J.; Zheng, W.-L.; Liu, C.-H.; Huang, I.; Lai, H.-H.; Liu, M. Using Deep Learning with Large Dataset of Microscope Images to Develop an Automated Embryo Grading System. FandR 2019, 1, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Storr, A.; Venetis, C.A.; Cooke, S.; Kilani, S.; Ledger, W. Inter-Observer and Intra-Observer Agreement between Embryologists during Selection of a Single Day 5 Embryo for Transfer: A Multicenter Study. Hum. Reprod. 2017, 32, 307–314. [Google Scholar] [CrossRef]
- Kragh, M.F.; Rimestad, J.; Berntsen, J.; Karstoft, H. Automatic Grading of Human Blastocysts from Time-Lapse Imaging. Comput. Biol. Med. 2019, 115, 103494. [Google Scholar] [CrossRef]
- Cruz, M.; Garrido, N.; Herrero, J.; Pérez-Cano, I.; Muñoz, M.; Meseguer, M. Timing of Cell Division in Human Cleavage-Stage Embryos Is Linked with Blastocyst Formation and Quality. Reprod. BioMed. Online 2012, 25, 371–381. [Google Scholar] [CrossRef]
- Bormann, C.L.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Kandula, H.; Souter, I.; Dimitriadis, I.; Gupta, R.; Pooniwala, R.; Shafiee, H. Consistency and Objectivity of Automated Embryo Assessments Using Deep Neural Networks. Fertil. Steril. 2020, 113, 781–787.e1. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, S.; Bhide, P.; Jordan, V.; Pacey, A.; Marjoribanks, J.; Farquhar, C. Time-lapse Systems for Embryo Incubation and Assessment in Assisted Reproduction. Cochrane Database Syst. Rev. 2019, 29, CD011320. [Google Scholar] [CrossRef]
- Martínez-Granados, L.; Serrano, M.; González-Utor, A.; Ortiz, N.; Badajoz, V.; López-Regalado, M.L.; Boada, M.; Castilla, J.A. Reliability and Agreement on Embryo Assessment: 5 Years of an External Quality Control Programme. Reprod. BioMed. Online 2018, 36, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.I.; Ferreira, A.S.; Cecílio, M.H.M.; Chéles, D.S.; de Souza, R.C.M.; Nogueira, M.F.G.; Rocha, J.C. Artificial Intelligence in the IVF Laboratory: Overview through the Application of Different Types of Algorithms for the Classification of Reproductive Data. J. Assist. Reprod. Genet. 2020, 37, 2359–2376. [Google Scholar] [CrossRef]
- Matusevičius, A.; Dirvanauskas, D.; Maskeliūnas, R.; Raudonis, V. Embryo Cell Detection Using Regions with Convolutional Neural Networks. In Proceedings of the IVUS International Conference on Information Technology (IVUS 2017), Kaunas, Lithuania, 28 April 2017. [Google Scholar]
- Suk, H.-I. Chapter 1—An Introduction to Neural Networks and Deep Learning. In Deep Learning for Medical Image Analysis; Zhou, S.K., Greenspan, H., Shen, D., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 3–24. ISBN 978-0-12-810408-8. [Google Scholar]
- Curchoe, C.L.; Bormann, C.L. Artificial Intelligence and Machine Learning for Human Reproduction and Embryology Presented at ASRM and ESHRE 2018. J. Assist. Reprod. Genet. 2019, 36, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.; Zitnick, C.; Bala, K.; Girshick, R. Inside-Outside Net: Detecting Objects in Context with Skip Pooling and Recurrent Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 2016; pp. 2874–2883. [Google Scholar]
- Uyar, A.; Bener, A.; Ciray, H.N.; Bahceci, M. A Frequency Based Encoding Technique for Transformation of Categorical Variables in Mixed IVF Dataset. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 6214–6217. [Google Scholar]
- Kim, K.G. Book Review: Deep Learning. Healthc. Inf. Res. 2016, 22, 351–354. [Google Scholar] [CrossRef] [Green Version]
- Simopoulou, M.; Sfakianoudis, K.; Maziotis, E.; Antoniou, N.; Rapani, A.; Anifandis, G.; Bakas, P.; Bolaris, S.; Pantou, A.; Pantos, K.; et al. Are Computational Applications the “Crystal Ball” in the IVF Laboratory? The Evolution from Mathematics to Artificial Intelligence. J. Assist. Reprod. Genet. 2018, 35, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.; Cooke, S.; Illingworth, P.J.; Gardner, D.K. Deep Learning as a Predictive Tool for Fetal Heart Pregnancy Following Time-Lapse Incubation and Blastocyst Transfer. Hum. Reprod. 2019, 34, 1011–1018. [Google Scholar] [CrossRef] [Green Version]
- VerMilyea, M.; Hall, J.M.M.; Diakiw, S.M.; Johnston, A.; Nguyen, T.; Perugini, D.; Miller, A.; Picou, A.; Murphy, A.P.; Perugini, M. Development of an Artificial Intelligence-Based Assessment Model for Prediction of Embryo Viability Using Static Images Captured by Optical Light Microscopy during IVF. Hum. Reprod. 2020, 35, 770–784. [Google Scholar] [CrossRef] [Green Version]
- Geller, J.; Collazo, I.; Pai, R.; Hendon, N.; Lokeshwar, S.D.; Arora, H.; Gerkowicz, S.A.; Palmerola, K.L.; Ramasamy, R. Development of an Artificial Intelligence-Based Assessment Model for Prediction of Pregnancy Success Using Static Images Captured by Optical Light Microscopy during IVF. Fertil. Steril. 2020, 114, e171. [Google Scholar] [CrossRef]
- Stone, D.L.; Rosopa, P.J. The Advantages and Limitations of Using Meta-Analysis in Human Resource Management Research. Hum. Resour. Manag. Rev. 2017, 27, 1–7. [Google Scholar] [CrossRef]
- Milewski, R.; Jamiołkowski, J.; Milewska Anna, J.; Domitrz, J.; Szamatowicz, J.; Wołczyński, S. Prognosis of the IVF ICSI/ET procedure efficiency with the use of artificial neural networks among patients of the Department of Reproduction and Gynecological Endocrinology. Ginekol. Pol. 2009, 80, 900–906. [Google Scholar]
- Ueno, S.; Berntsen, J.; Ito, M.; Uchiyama, K.; Okimura, T.; Yabuuchi, A.; Kato, K. Pregnancy Prediction Performance of an Annotation-Free Embryo Scoring System on the Basis of Deep Learning after Single Vitrified-Warmed Blastocyst Transfer: A Single-Center Large Cohort Retrospective Study. Fertil. Steril. 2021, 116, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.M.; Athanasiou, T. Summary Receiver Operating Characteristic Curve Analysis Techniques in the Evaluation of Diagnostic Tests. Ann. Thorac. Surg. 2005, 79, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.D. Properties of the Summary Receiver Operating Characteristic (SROC) Curve for Diagnostic Test Data. Stat. Med. 2002, 21, 1237–1256. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.D. The Partial Area under the Summary ROC Curve. Stat. Med. 2005, 24, 2025–2040. [Google Scholar] [CrossRef]
- Debray, T.P.A.; Damen, J.A.A.G.; Snell, K.I.E.; Ensor, J.; Hooft, L.; Reitsma, J.B.; Riley, R.D.; Moons, K.G.M. A Guide to Systematic Review and Meta-Analysis of Prediction Model Performance. BMJ 2017, 356, i6460. [Google Scholar] [CrossRef] [Green Version]
- Debray, T.P.; Damen, J.A.; Riley, R.D.; Snell, K.; Reitsma, J.B.; Hooft, L.; Collins, G.S.; Moons, K.G. A Framework for Meta-Analysis of Prediction Model Studies with Binary and Time-to-Event Outcomes. Stat. Methods Med. Res. 2019, 28, 2768–2786. [Google Scholar] [CrossRef]
- Alba, A.C.; Agoritsas, T.; Walsh, M.; Hanna, S.; Iorio, A.; Devereaux, P.J.; McGinn, T.; Guyatt, G. Discrimination and Calibration of Clinical Prediction Models: Users’ Guides to the Medical Literature. JAMA 2017, 318, 1377–1384. [Google Scholar] [CrossRef]
- Alegre, L.; Bori, L.; de los Ángeles Valera, M.; Nogueira, M.F.G.; Ferreira, A.S.; Rocha, J.C.; Meseguer, M. First Application of Artificial Neuronal Networks for Human Live Birth Prediction on Geri Time-Lapse Monitoring System Blastocyst Images. Fertil. Steril. 2020, 114, e140. [Google Scholar] [CrossRef]
- Meseguer, M.; Hickman, C.; Arnal, L.B.; Alegre, L.; Toschi, M.; Gallego, R.D.; Rocha, J.C. Is There Any Room to Improve Embryo Selection? Artificial Intelligence Technology Applied for Ive Birth Prediction on Blastocysts. Fertil. Steril. 2019, 112, e77. [Google Scholar] [CrossRef]
- Miyagi, Y.; Habara, T.; Hirata, R.; Hayashi, N. Feasibility of Deep Learning for Predicting Live Birth from a Blastocyst Image in Patients Classified by Age. Reprod. Med. Biol. 2019, 18, 190–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawada, Y.; Sato, T.; Nagaya, M.; Saito, C.; Yoshihara, H.; Banno, C.; Matsumoto, Y.; Matsuda, Y.; Yoshikai, K.; Sawada, T.; et al. Evaluation of Artificial Intelligence Using Time-Lapse Images of IVF Embryos to Predict Live Birth. Reprod. BioMed. Online 2021, 43, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Hardy, C.; Theodoratos, S.; Panitsa, G.; Walker, J.; Harbottle, S.; Sfontouris, I.A. External Validation of an ai System for Blastocyst Implantation Prediction. Fertil. Steril. 2020, 114, e140. [Google Scholar] [CrossRef]
- Chavez-Badiola, A.; Flores-Saiffe Farias, A.; Mendizabal-Ruiz, G.; Garcia-Sanchez, R.; Drakeley, A.J.; Garcia-Sandoval, J.P. Predicting Pregnancy Test Results after Embryo Transfer by Image Feature Extraction and Analysis Using Machine Learning. Sci. Rep. 2020, 10, 4394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Q.; Zhang, Q.; Feng, X.; Huang, H.; Xu, H.; Tian, B.; Liu, J.; Yu, Q.; Guo, N.; Liu, Q.; et al. Development of Deep Learning Algorithms for Predicting Blastocyst Formation and Quality by Time-Lapse Monitoring. Commun. Biol. 2021, 4, 415. [Google Scholar] [CrossRef] [PubMed]
- Bori, L.; Paya, E.; Alegre, L.; Viloria, T.A.; Remohi, J.A.; Naranjo, V.; Meseguer, M. Novel and Conventional Embryo Parameters as Input Data for Artificial Neural Networks: An Artificial Intelligence Model Applied for Prediction of the Implantation Potential. Fertil. Steril. 2020, 114, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Kan-Tor, Y.; Zabari, N.; Erlich, I.; Szeskin, A.; Amitai, T.; Richter, D.; Or, Y.; Shoham, Z.; Hurwitz, A.; Har-Vardi, I.; et al. Automated Evaluation of Human Embryo Blastulation and Implantation Potential Using Deep-Learning. Adv. Intell. Syst. 2020, 2, 2000080. [Google Scholar] [CrossRef]
- Bormann, C.L.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Gupta, R.; Pooniwala, R.; Kandula, H.; Hariton, E.; Souter, I.; Dimitriadis, I.; Ramirez, L.B.; et al. Performance of a Deep Learning Based Neural Network in the Selection of Human Blastocysts for Implantation. eLife 2020, 9, e55301. [Google Scholar] [CrossRef]
- Silver, D.H.; Feder, M.; Gold-Zamir, Y.; Polsky, A.L.; Rosentraub, S.; Shachor, E.; Weinberger, A.; Mazur, P.; Zukin, V.D.; Bronstein, A.M. Data-Driven Prediction of Embryo Implantation Probability Using IVF Time-Lapse Imaging. arXiv 2020, arXiv:2006.01035. [Google Scholar]
- Cao, Q.; Liao, S.S.; Meng, X.; Ye, H.; Yan, Z.; Wang, P. Identification of Viable Embryos Using Deep Learning for Medical Image. In Proceedings of the 2018 5th International Conference on Bioinformatics Research and Applications; Association for Computing Machinery, New York, NY, USA, 27 December 2018; pp. 69–72. [Google Scholar]
- Bori, L.; Valera, M.Á.; Gilboa, D.; Maor, R.; Kottel, I.; Remohí, J.; Seidman, D.; Meseguer, M. O-084 Computer Vision Can Distinguish between Euploid and Aneuploid Embryos. A Novel Artificial Intelligence (AI) Approach to Measure Cell Division Activity Associated with Chromosomal Status. Hum. Reprod. 2021, 36, deab125.014. [Google Scholar] [CrossRef]
- Aparicio Ruiz, B.; Bori, L.; Paya, E.; Valera, M.A.; Quiñonero, A.; Dominguez, F.; Meseguer, M. P-203 Applying Artificial Intelligence for Ploidy Prediction: The Concentration of IL-6 in Spent Culture Medium, Blastocyst Morphological Grade and Embryo Morphokinetics as Variables under Consideration. Hum. Reprod. 2021, 36, deab127.066. [Google Scholar] [CrossRef]
- Lee, C.I.; Su, Y.R.; Chen, C.H.; Chang, T.A.; Kuo, E.E.S.; Hsieh, W.T.; Huang, C.C.; Lee, M.S.; Liu, M. O-086 End-to-End Deep Learning for Recognition of Ploidy Status Using Time-Lapse Videos. Hum. Reprod. 2021, 36, deab125.016. [Google Scholar] [CrossRef]
- Chavez-Badiola, A.; Flores-Saiffe-Farias, A.; Mendizabal-Ruiz, G.; Drakeley, A.J.; Cohen, J. Embryo Ranking Intelligent Classification Algorithm (ERICA): Artificial Intelligence Clinical Assistant Predicting Embryo Ploidy and Implantation. Reprod. BioMed. Online 2020, 41, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Bori, L.; Dominguez, F.; Fernandez, E.I.; Gallego, R.D.; Alegre, L.; Hickman, C.; Quiñonero, A.; Nogueira, M.F.G.; Rocha, J.C.; Meseguer, M. An Artificial Intelligence Model Based on the Proteomic Profile of Euploid Embryos and Blastocyst Morphology: A Preliminary Study. Reprod. BioMed. Online 2021, 42, 340–350. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Rosenwaks, Z. Artificial Intelligence in Reproductive Medicine: A Fleeting Concept or the Wave of the Future? Fertil. Steril. 2020, 114, 905–907. [Google Scholar] [CrossRef]
- Emin, E.I.; Emin, E.; Papalois, A.; Willmott, F.; Clarke, S.; Sideris, M. Artificial Intelligence in Obstetrics and Gynaecology: Is This the Way Forward? In Vivo 2019, 33, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Zaninovic, N.; Rosenwaks, Z. Artificial Intelligence in Human in Vitro Fertilization and Embryology. Fertil. Steril. 2020, 114, 914–920. [Google Scholar] [CrossRef]
- Khosravi, P.; Kazemi, E.; Zhan, Q.; Malmsten, J.E.; Toschi, M.; Zisimopoulos, P.; Sigaras, A.; Lavery, S.; Cooper, L.A.D.; Hickman, C.; et al. Deep Learning Enables Robust Assessment and Selection of Human Blastocysts after in Vitro Fertilization. NPJ Digit. Med. 2019, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Ahlstrom, A.; Park, H.; Bergh, C.; Selleskog, U.; Lundin, K. Conventional Morphology Performs Better than Morphokinetics for Prediction of Live Birth after Day 2 Transfer. Reprod. BioMed. Online 2016, 33, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Petersen, B.M.; Boel, M.; Montag, M.; Gardner, D.K. Development of a Generally Applicable Morphokinetic Algorithm Capable of Predicting the Implantation Potential of Embryos Transferred on Day 3. Hum. Reprod. 2016, 31, 2231–2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simopoulou, M.; Sfakianoudis, K.; Maziotis, E.; Tsioulou, P.; Grigoriadis, S.; Rapani, A.; Giannelou, P.; Asimakopoulou, M.; Kokkali, G.; Pantou, A.; et al. PGT-A: Who and When? A Systematic Review and Network Meta-Analysis of RCTs. J. Assist. Reprod. Genet. 2021, 38, 1939–1957. [Google Scholar] [CrossRef] [PubMed]
- London, A.J. Artificial Intelligence and Black-Box Medical Decisions: Accuracy versus Explainability. Hastings Cent. Rep. 2019, 49, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Beil, M.; Proft, I.; van Heerden, D.; Sviri, S.; van Heerden, P.V. Ethical Considerations about Artificial Intelligence for Prognostication in Intensive Care. Intensive Care Med. Exp. 2019, 7, 70. [Google Scholar] [CrossRef] [Green Version]
- Swain, J.; VerMilyea, M.T.; Meseguer, M.; Ezcurra, D. Fertility AI Forum Group AI in the Treatment of Fertility: Key Considerations. J. Assist. Reprod. Genet. 2020, 37, 2817–2824. [Google Scholar] [CrossRef]
- van de Wiel, L.; Wilkinson, J.; Athanasiou, P.; Harper, J. The Prevalence, Promotion and Pricing of Three IVF Add-Ons on Fertility Clinic Websites. Reprod. BioMed. Online 2020, 41, 801–806. [Google Scholar] [CrossRef]


| Study | Outcome | Type of Input (TL/Static Images) | Sample Size | Type of AI Algorithm Employed | Model Optimization |
|---|---|---|---|---|---|
| Alegre 2020 [33] | Live-birth | TL | 244 | ANN | NP |
| Meseguer 2019 [34] | Live-birth | TL STATIC IMAGE | TL: 111 SI: 111 | ANN | NP |
| Miyagi 2019 [35] | Live-birth | TL | 1139 | CNN | 5-fold cross validation |
| Sawada 2021 [36] | Live-birth | TL | 376 | CNN with Attention Branch Network | Back-propagation for 5 epochs |
| Hardy 2020 [37] | Clinical Pregnancy with FHB | TL | 113 | CNN | NP |
| VerMilyea 2020 [22] | Clinical Pregnancy with FHB | STATIC IMAGES | 1667 | ResNet | Back-propagation and SGD for 5 epochs |
| Chavez-Badiola 2020 [38] | Clinical Pregnancy | STATIC IMAGES | 221 | SVM | 10-fold cross validation |
| Liao 2021 [39] | Clinical Pregnancy with FHB | TL | 209 | DNN | NP |
| Bori 2020 [40] | Clinical Pregnancy with FHB | TL | 451 | ANN | 5-fold cross validation |
| Kan-Tor 2020 [41] | Clinical Pregnancy | TL | 401 | DNN | 20–60 epochs validation |
| Bormann 2020 [42] | Clinical Pregnancy with FHB | STATIC IMAGES | 102 | CNN | Genetic algorithm per 100 samples for a dataset of 3469 embryos |
| Silver 2020 [43] | Clinical Pregnancy with FHB | TL | 272 | CNN | NP |
| Cao 2018 [44] | Clinical Pregnancy | STATIC IMAGES | 344 | CNN | NP |
| Ueno 2021 [26] | Clinical Pregnancy with FHB | TL | 3014 | DNN | Back-propagation for 20 epochs and 5-fold cross validation |
| Bori 2021 [45] | Ploidy | TL | 331 | ANN | Back-propagation |
| Aparicio Ruiz 2021 [46] | Ploidy | TL | 319 | ANN | NP |
| Lee 2021 [47] | Ploidy | TL | 138 | CNN (3D ConvNets) | NP |
| Chavez-Badiola 2020 [48] | Ploidy | STATIC IMAGES | 84 | DNN | 10-fold cross validation |
| Study | Participants | Predictors | Outcomes | Analysis | Overall |
|---|---|---|---|---|---|
| Alegre 2020 [33] | - | + | + | + | - |
| Meseguer 2019 [34] | - | + | + | + | - |
| Miyagi 2019 [35] | + | + | + | - | - |
| Sawada 2021 [36] | + | + | + | + | + |
| Hardy 2020 [37] | - | + | + | + | - |
| VerMilyea 2020 [22] | + | - | + | + | - |
| Chavez-Badiola 2020 [38] | - | - | + | + | - |
| Liao 2021 [39] | - | + | + | + | - |
| Bori 2020 [40] | + | + | + | + | + |
| Kan-Tor 2020 [41] | + | + | + | + | + |
| Bormann 2020 [42] | - | - | + | + | - |
| Silver 2020 [43] | - | + | + | + | - |
| Cao 2018 [44] | + | - | + | + | - |
| Ueno 2021 [26] | + | + | + | - | - |
| Bori 2021 [49] | + | + | + | - | - |
| Aparicio Ruiz 2021 [46] | + | + | + | + | + |
| Lee 2021 [47] | - | + | + | + | - |
| Chavez-Badiola 2020 [48] | - | + | + | + | - |
| Outcomes | Sensitivity | Specificity | PPV | NPV | DOR |
|---|---|---|---|---|---|
| Live-Birth | 70.6% (38.1–90.4%) | 90.6% (79.3–96.1%) | 74.2% (44.1–91.3%) | 88.4% (80.6–93.3%) | 19.662 (5.061–76.397) |
| Live-Birth SI | 90.7% (77.7–96.5%) | 89.7% (79.9–95.0%) | 84.8% (71.4–92.6%) | 93.8% (84.7–97.7%) | 84.964 (23.329–309.437) |
| Live-Birth TL | 62.9% (27.7–88.2%) | 91.0% (75.6–97.1%) | 71.2% (33.7–92.3%) | 86.9% (78.0–92.5%) | 13.204 (3.336–52.264) |
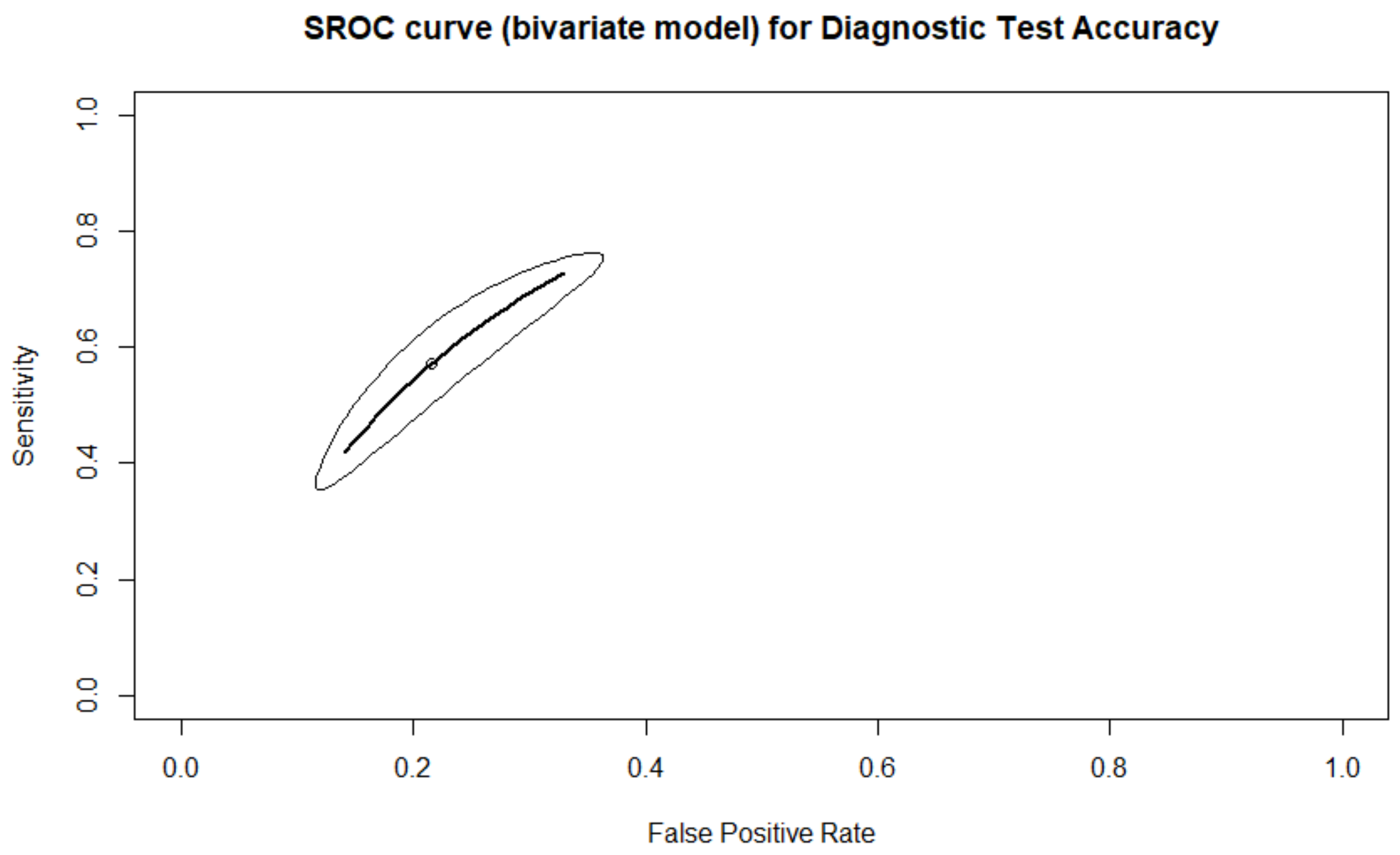
| Clinical Pregnancy | 71.0% (58.1–81.2%) | 62.5% (47.4–75.5%) | 66.4% (51.7–78.5%) | 67.9% (60.7–74.4%) | 3.962 (2.501–6.275) |
| Clinical Pregnancy SI | 72.7% (60.6–82.2%) | 58.6% (49.6–67.1%) | 67.6% (46.6–83.4%) | 66.0% (56.5–74.3%) | 3.861 (1.708–8.729) |
| Clinical Pregnancy TL | 70.0% (49.4–84.8%) | 64.2% (39.9–82.9%) | 65.6% (45.2–81.5%) | 69.2% (58.8–78.0%) | 4.074 (1.880–8.827) |
| Clinical Pregnancy with FHB | 75.2% (66.8–82.0%) | 55.3% (41.2–68.7%) | 62.5% (43.9–78.0%) | 69.5% (60.4–77.2%) | 3.549 (2.113–5.961) |
| Clinical Pregnancy with FHB SI | 69.3% (65.8–72.6%) | 56.7% (43.9–68.7%) | 44.0% (41.1–46.9%) | 75.1% (72.2–77.9%) | 2.415 (1.986–2.937) |
| Clinical Pregnancy with FHB TL | 78.7% (70.3–85.2%) | 53.9% (35.1–71.6%) | 66.8% (42.7–84.5%) | 68.1% (55.1–78.7%) | 4.101 (1.636–10.276) |
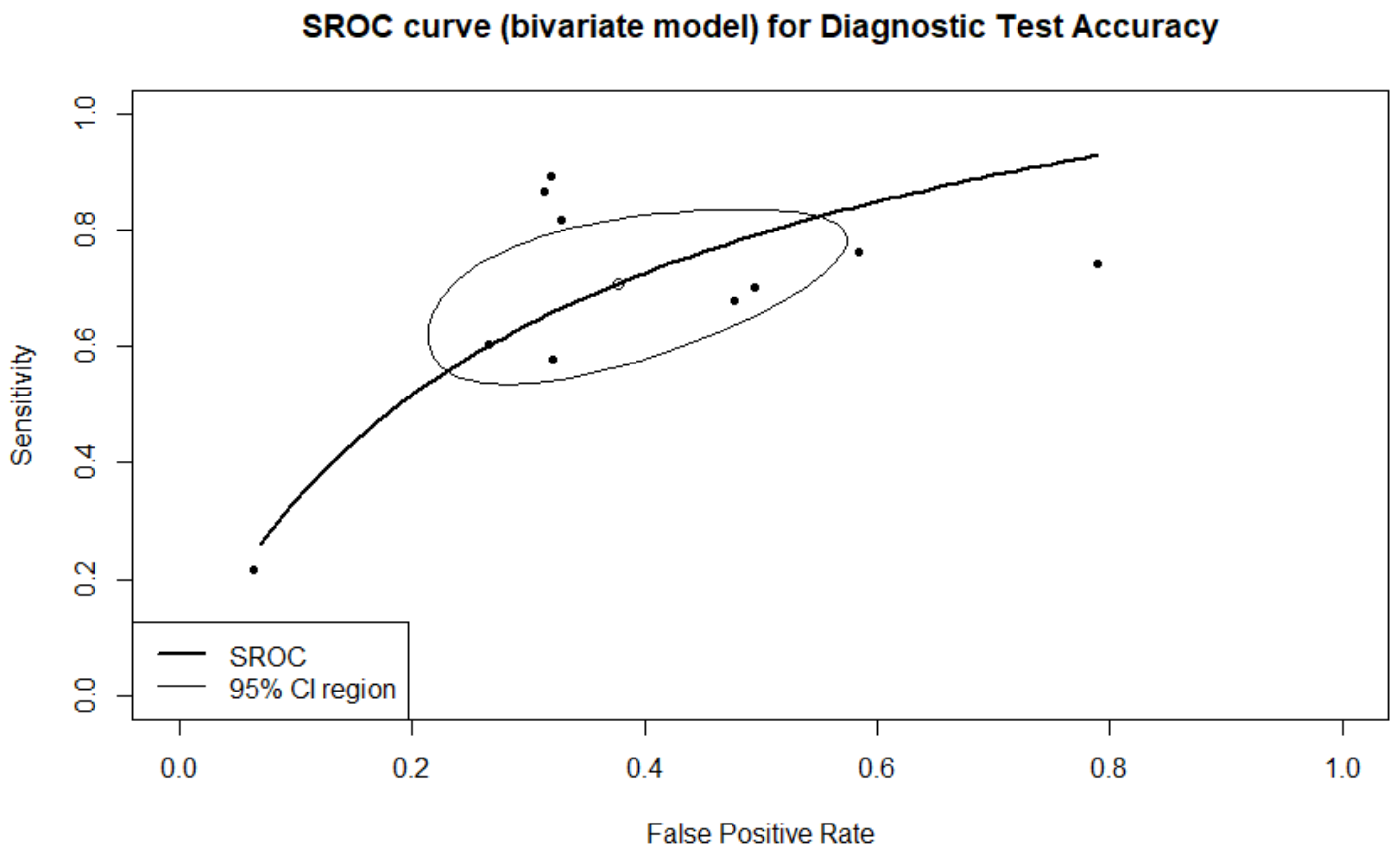
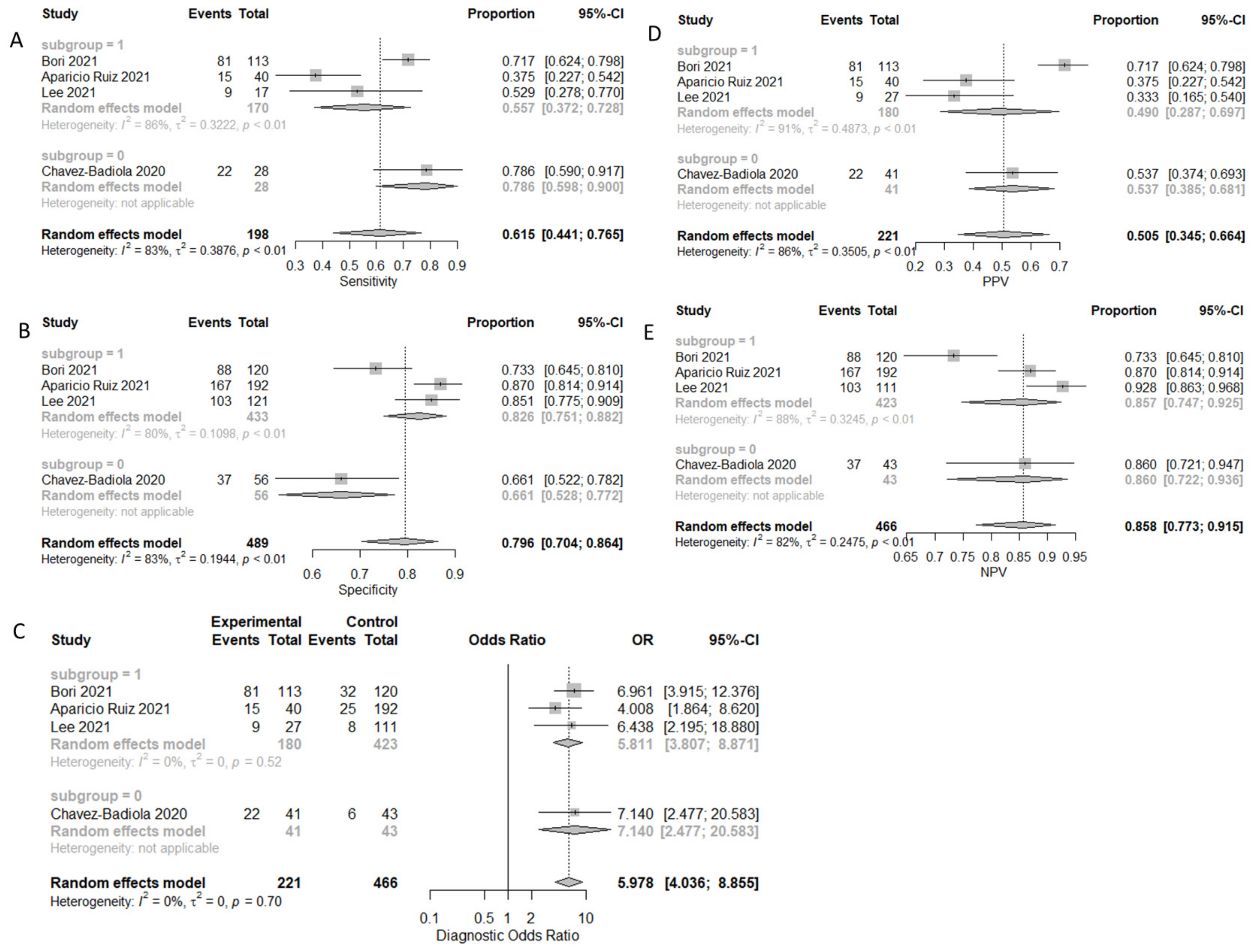
| Ploidy | 61.5% (44.1–76.5%) | 79.6% (70.4–86.4%) | 50.5% (34.5–68.1%) | 85.8% (77.3–91.5%) | 5.978 (4.036–8.855) |
| Ploidy TL | 55.7% (37.2–72.8%) | 82.6% (75.1–88.2%) | 49.0% (28.7–69.7%) | 85.7% (74.7–92.5%) | 5.811 (3.807–8.871) |
| Ploidy SI | 78.6% (59.0–90.0%) | 66.1% (52.8–77.2%) | 53.7% (38.5–68.1%) | 86.0% (72.2–93.6%) | 7.140 (2.477–20.583) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sfakianoudis, K.; Maziotis, E.; Grigoriadis, S.; Pantou, A.; Kokkini, G.; Trypidi, A.; Giannelou, P.; Zikopoulos, A.; Angeli, I.; Vaxevanoglou, T.; et al. Reporting on the Value of Artificial Intelligence in Predicting the Optimal Embryo for Transfer: A Systematic Review including Data Synthesis. Biomedicines 2022, 10, 697. https://doi.org/10.3390/biomedicines10030697
Sfakianoudis K, Maziotis E, Grigoriadis S, Pantou A, Kokkini G, Trypidi A, Giannelou P, Zikopoulos A, Angeli I, Vaxevanoglou T, et al. Reporting on the Value of Artificial Intelligence in Predicting the Optimal Embryo for Transfer: A Systematic Review including Data Synthesis. Biomedicines. 2022; 10(3):697. https://doi.org/10.3390/biomedicines10030697
Chicago/Turabian StyleSfakianoudis, Konstantinos, Evangelos Maziotis, Sokratis Grigoriadis, Agni Pantou, Georgia Kokkini, Anna Trypidi, Polina Giannelou, Athanasios Zikopoulos, Irene Angeli, Terpsithea Vaxevanoglou, and et al. 2022. "Reporting on the Value of Artificial Intelligence in Predicting the Optimal Embryo for Transfer: A Systematic Review including Data Synthesis" Biomedicines 10, no. 3: 697. https://doi.org/10.3390/biomedicines10030697
APA StyleSfakianoudis, K., Maziotis, E., Grigoriadis, S., Pantou, A., Kokkini, G., Trypidi, A., Giannelou, P., Zikopoulos, A., Angeli, I., Vaxevanoglou, T., Pantos, K., & Simopoulou, M. (2022). Reporting on the Value of Artificial Intelligence in Predicting the Optimal Embryo for Transfer: A Systematic Review including Data Synthesis. Biomedicines, 10(3), 697. https://doi.org/10.3390/biomedicines10030697









