Implication of Different Tumor Biomarkers in Drug Resistance and Invasiveness in Primary and Metastatic Colorectal Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Flow Cytometry Analysis
2.3. RNA Extraction and Reverse Transcriptase Assay
2.4. Gene Expression by qPCR
2.5. Treatment with Oxaliplatin and Growth Curve
2.6. Cell Viability Detection of Previously Oxaliplatin-Treated and Non-Treated Cells
2.7. Statistical Analysis
3. Results
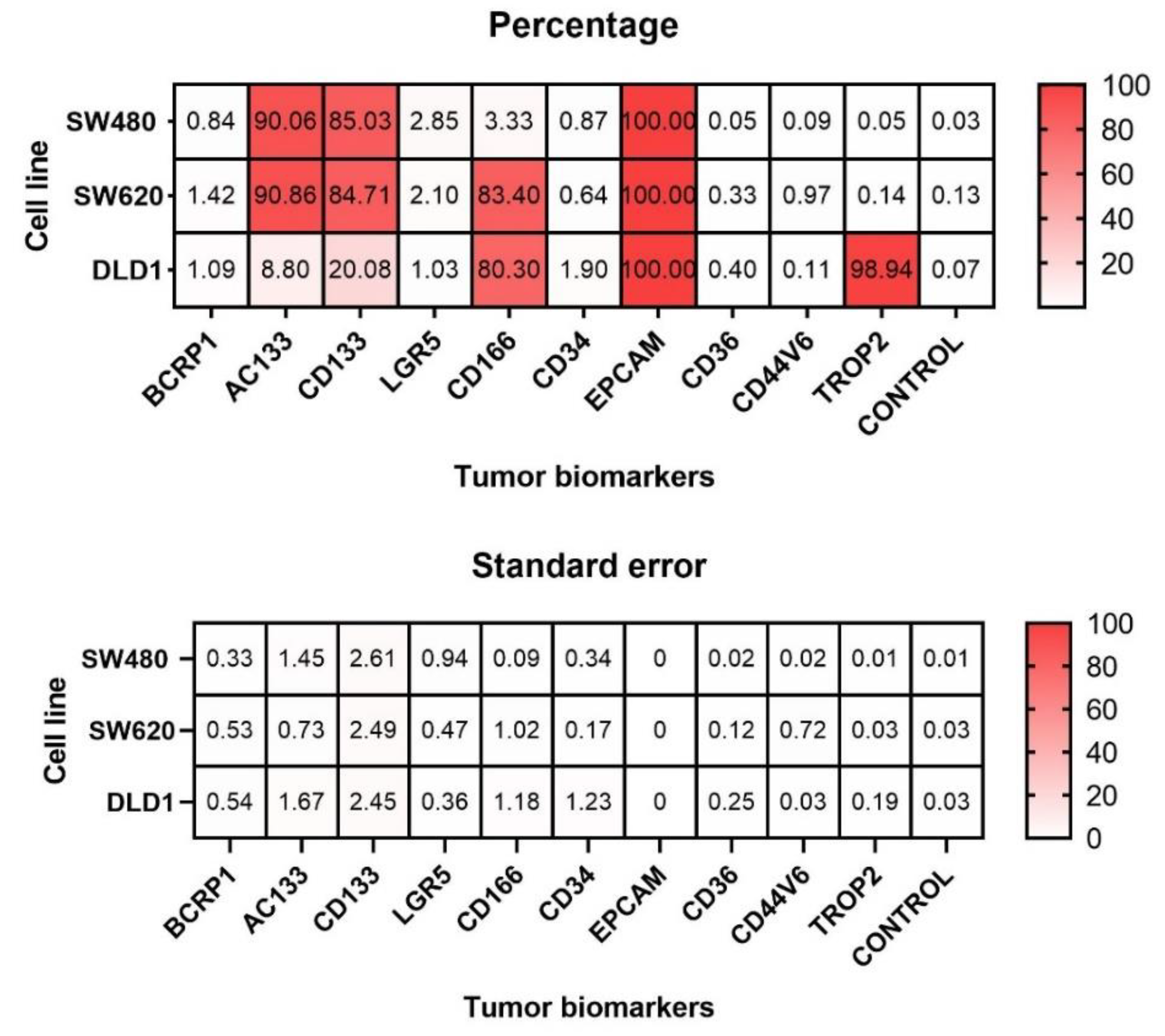
3.1. Expression of Tumoral Biomarkers in SW-480, SW-620 and DLD-1
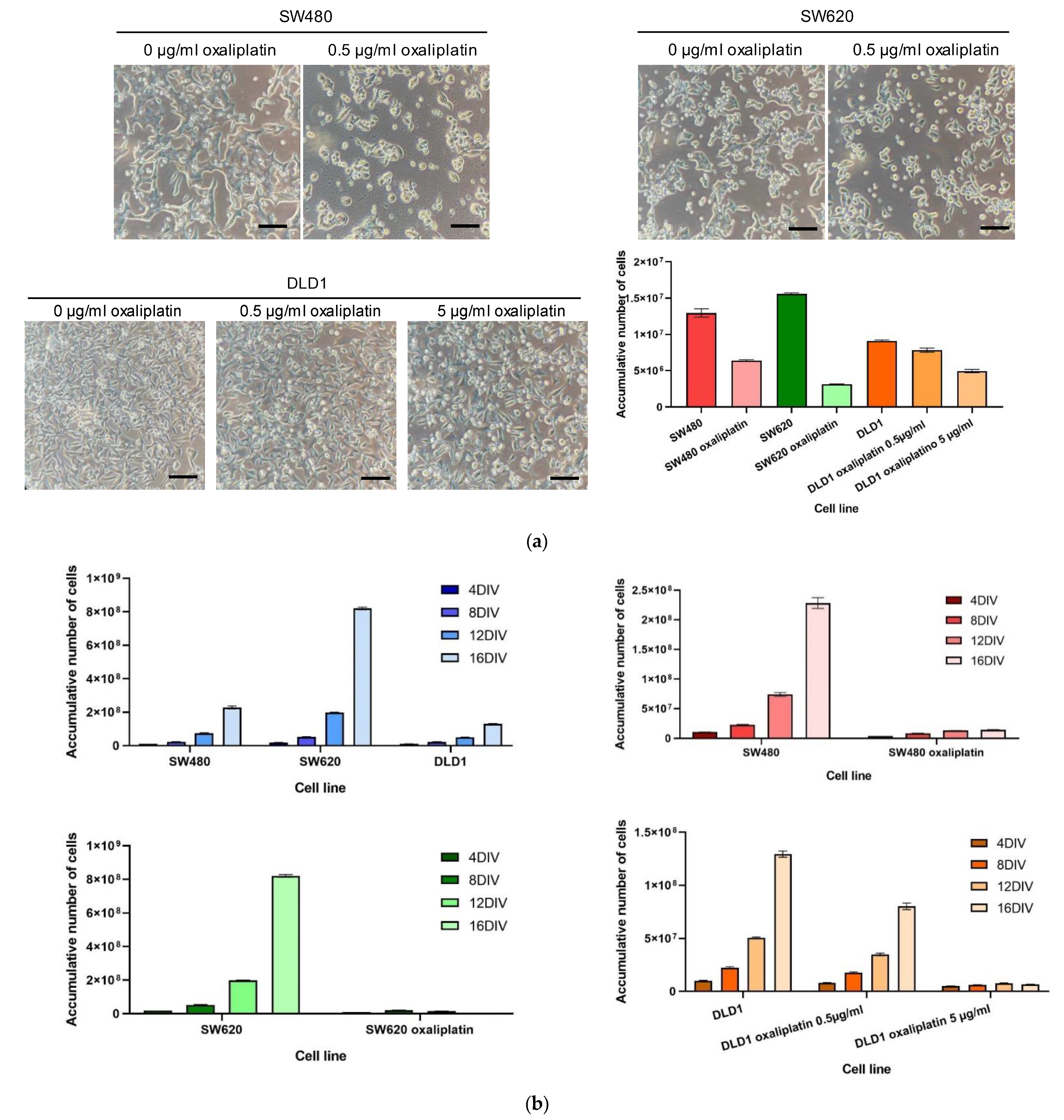
3.2. Colorectal Cell Line Growth in Exposure to Oxaliplatin Chemotherapy Drug
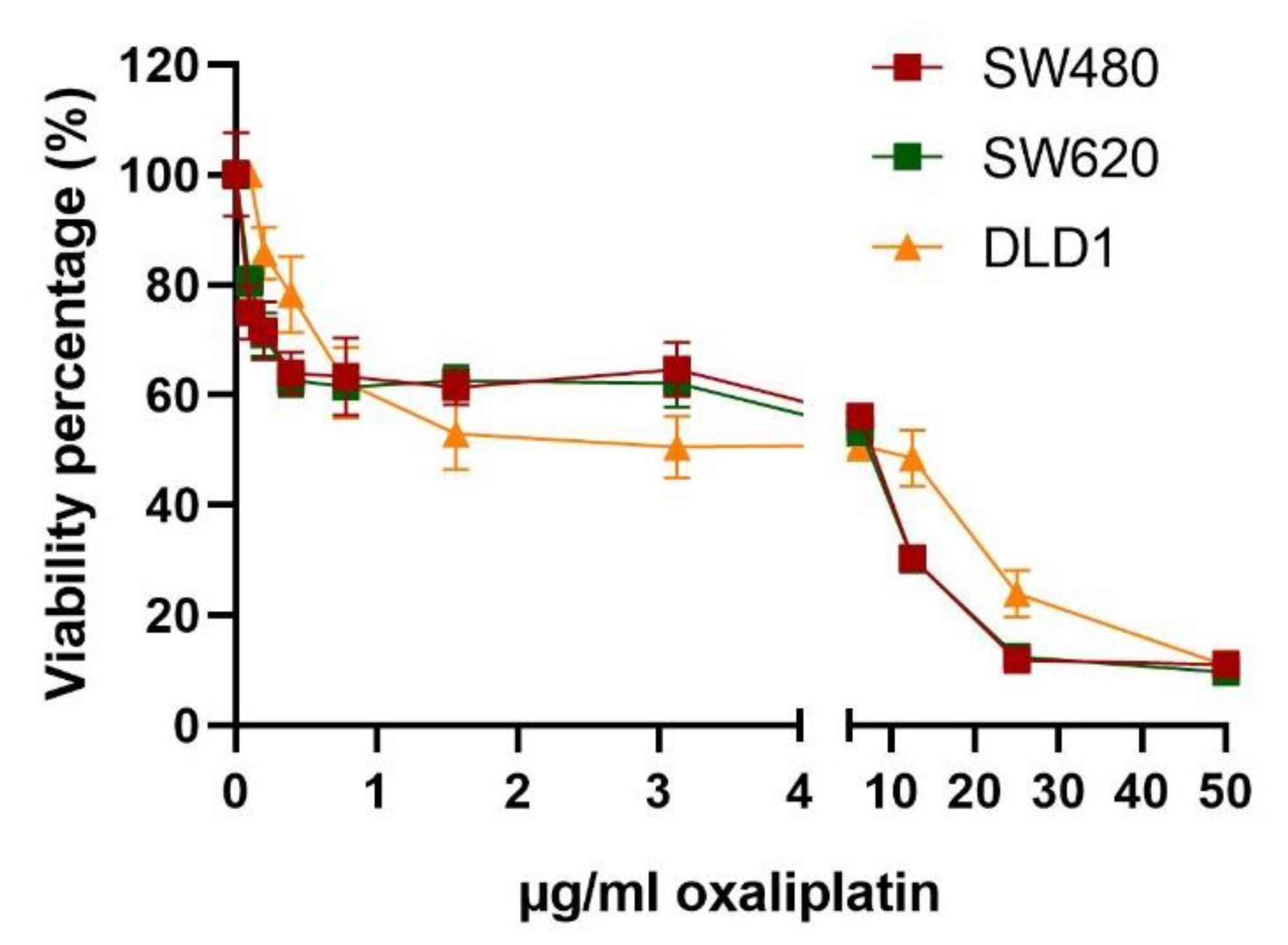
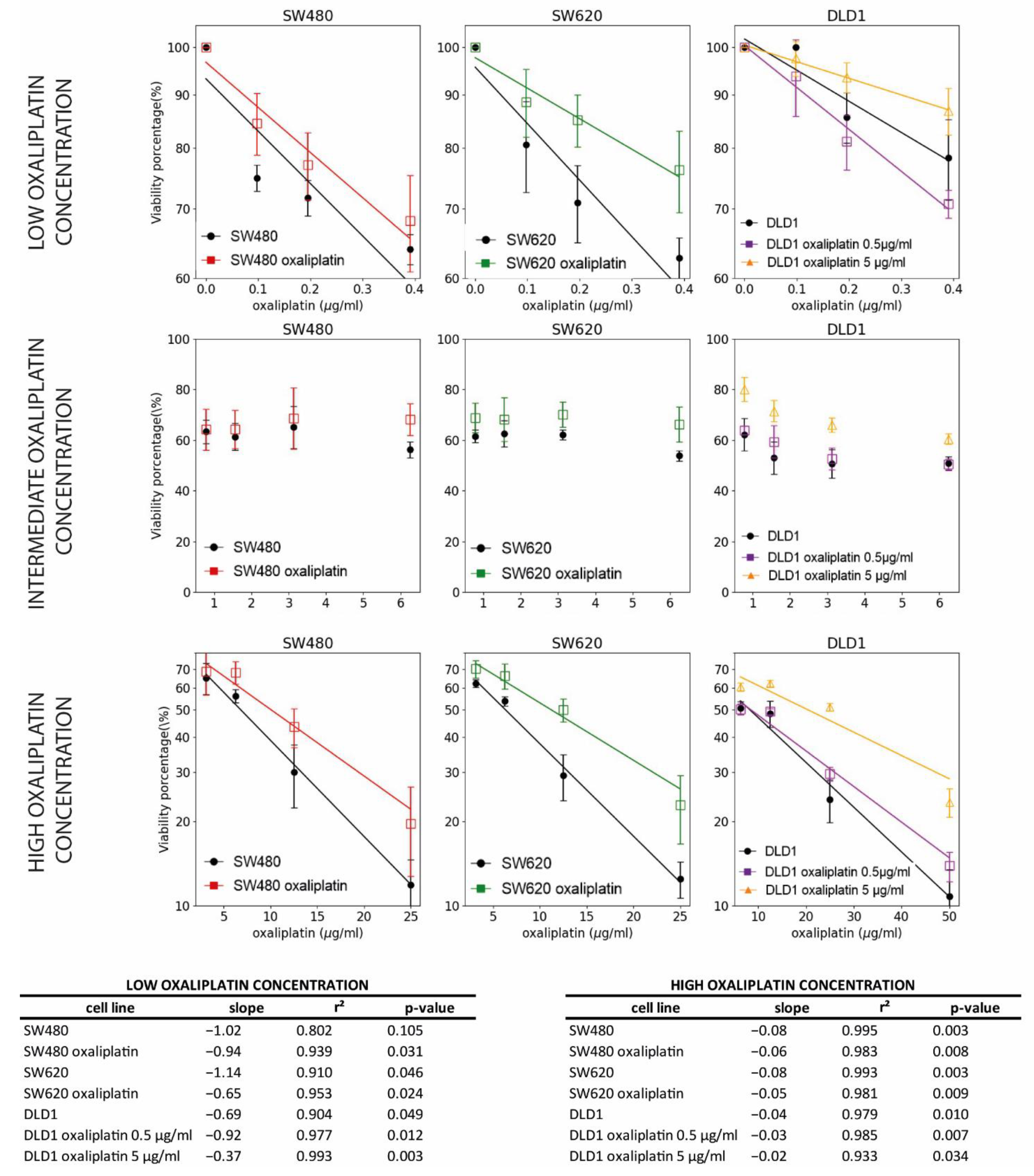
3.3. Resistance to Oxaliplatin in Non-Treated and Previously Oxaliplatin-Treated Cell Lines
3.4. TROP2 Role in Drug Resistance
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mattiuzzi, C.; Sanchis-Gomar, F.; Lippi, G. Concise update on colorectal cancer epidemiology. Ann. Transl. Med. 2019, 7, 609. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.R.; Chou, J.F.; Keshinro, A.; Chapman, W.C.; Bauer, P.S.; Mutch, M.G.; Parikh, P.J.; Cercek, A.; Saltz, L.B.; Gollub, M.J.; et al. Development and Assessment of a Clinical Calculator for Estimating the Likelihood of Recurrence and Survival among Patients with Locally Advanced Rectal Cancer Treated with Chemotherapy, Radiotherapy, and Surgery. JAMA Netw. Open 2021, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Wasmuth, H.H.; Gachabayov, M.; Bokey, L.; Fingerhut, A.; Orangio, G.R.; Remzi, F.H.; Bergamaschi, R. Statistical, Clinical, Methodological Evaluation of Local Recurrence Following Transanal Total Mesorectal Excision for Rectal Cancer: A Systematic Review. Dis. Colon Rectum 2021, 64, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Douillard, J.Y.; Cunningham, D.; Roth, A.D.; Navarro, M.; James, R.D.; Karasek, P.; Jandik, P.; Iveson, T.; Carmichael, J.; Alakl, M.; et al. Irinotecan combined with fluorouracil compared with fluorouracil alone. as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet 2000, 355, 1041–1047. [Google Scholar] [CrossRef]
- Erlichman, C. Efficacy of adjuvant fluorouracil and folinic acid in B2 colon cancer. J. Clin. Oncol. 1999, 17, 1356–1363. [Google Scholar] [CrossRef]
- Grothey, A.; Sobrero, A.F.; Shields, A.F.; Yoshino, T.; Paul, J.; Taieb, J.; Souglakos, J.; Shi, Q.; Kerr, R.; Labianca, R.; et al. Duration of adjuvant chemotherapy for stage III colon cancer. N. Engl. J. Med. 2019, 378, 1177–1188. [Google Scholar] [CrossRef]
- Townsend, D. Oxaliplatin. 2007. Available online: https://doi.org/10.1016/B978-008055232-3.62973-3 (accessed on 15 March 2022).
- Comella, P.; Casaretti, R.; Sandomenico, C.; Avallone, A.; Franco, L. Role of oxaliplatin in the treatment of colorectal cancer. Ther. Clin. Risk Manag. 2009, 5, 229–238. [Google Scholar] [CrossRef]
- Piccart, M.J.; Green, J.A.; Lacave, A.J.; Reed, N.; Vergote, I.; Benedetti-Panici, P.; Bonetti, A.; Kristeller-Tome, V.; Fernandez, C.M.; Curran, D.; et al. Oxaliplatin or Paclitaxel in Patients with Platinum-Pretreated Advanced Ovarian Cancer: A Randomized Phase II Study of the European Organization for Research and Treatment of Cancer Gynecology Group. J. Clin. Oncol. 2000, 18, 1193–1202. [Google Scholar] [CrossRef]
- Jaxel, C.; Kohn, K.W.; Wani, M.C.; Wall, M.E.; Pommier, Y. Structure-Activity Study of the Actions of Camptothecin Derivatives on Mammalian Topoisomerase I: Evidence for a Specific Receptor Site and a Relation to Antitumor Activity. Cancer Res. 1989, 49, 1465–1469. [Google Scholar] [PubMed]
- Van Der Jeught, K.; Xu, H.C.; Li, Y.J.; Lu, X.B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Liu, H.; Liu, L.; Wang, Y.D.; Chen, W.D. Emerging Roles of Wnt Ligands in Human Colorectal Cancer. Front. Oncol. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, T.; Jin, Y.; Wei, J.; Yang, Y.; Zhang, H. ABCG2 is required for self-renewal and chemoresistance of CD133-positive human colorectal cancer cells. Tumor Biol. 2016, 37, 12889–12896. [Google Scholar] [CrossRef]
- Liu, C.; Liu, L.; Chen, X.; Cheng, J.; Zhang, H.; Shen, J.; Shan, J.; Xu, Y.; Yang, Z.; Lai, M.; et al. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 2016, 64, 117–129. [Google Scholar] [CrossRef]
- Horst, D.; Kriegl, L.; Engel, J.; Kirchner, T.; Jung, A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Investig. 2009, 27, 844–850. [Google Scholar] [CrossRef]
- Tampakis, A.; Weixler, B.; Rast, S.; Tampaki, E.-C.; Cremonesi, E.; Kancherla, V.; Tosti, N.; Kettelhack, C.; Ng, C.K.Y.; Delko, T.; et al. Nestin and CD34 expression in colorectal cancer predicts improved overall survival. Acta Oncol. 2021, 60, 727–734. [Google Scholar] [CrossRef]
- Wang, C.; Xie, J.; Guo, J.; Manning, H.C.; Gore, J.C.; Guo, N. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol. Rep. 2012, 28, 1301–1308. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, J.; Shi, H.; Gao, B.; Zhang, L. LncRNA TINCR/microRNA-107/CD36 regulates cell proliferation and apoptosis in colorectal cancer via PPAR signaling pathway based on bioinformatics analysis. Biol. Chem. 2019, 400, 663–675. [Google Scholar] [CrossRef]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Fang, Y.J.; Lu, Z.H.; Wang, G.Q.; Pan, Z.Z.; Zhou, Z.W.; Yun, J.P.; Zhang, M.F.; Wan, D.S. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int. J. Color. Dis. 2009, 24, 875. [Google Scholar] [CrossRef]
- Barker, N.; Van Es, J.H.; Kuipers, J.; Kujala, P.; Van Den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- De Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5 + stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wu, J.; Meng, X.; Zuo, Y.; Xia, Q.; Chen, J.; Feng, Y.; Liu, R.; Li, L.; Huang, W. AC133 expression associated with poor prognosis in stage II colorectal cancer. Med. Oncol. 2013, 30, 356. [Google Scholar] [CrossRef] [PubMed]
- Durmus, S.; van der Valk, M.; Teunissen, S.F.; Song, J.Y.; Wagenaar, E.; Beijnen, J.H.; Schinkel, A.H. ABC transporters Mdr1a/1b, Bcrp1, Mrp2 and Mrp3 determine the sensitivity to PhIP/DSS-induced colon carcinogenesis and inflammation. Arch. Toxicol. 2019, 93, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Pandiyan, S.M.; Shanmugaraj, P.; Manoharan, J.P.; Vidyalakshmi, S. A network pharmacological approach to reveal the multidrug resistance reversal and associated mechanisms of acetogenins against colorectal cancer. J. Biomol. Struct. Dyn. 2021, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Michl, M.; Taverna, F.; Kumbrink, J.; Schiergens, T.S.; Heinemann, V.; Engel, J.; Kirchner, T.; Neumann, J. Biomarker alterations associated with distinct patterns of metastatic spread in colorectal cancer. Virchows Arch. 2021, 478, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, Z.; Mansoori, B.; Mohammadi, A.; Kazemi, T.; Mokhtarzadeh, A.; Shanehbandi, D.; Hemmat, N.; Derakhshani, A.; Brunetti, O.; Safaei, S.; et al. The combination effect of Prominin1 (CD133) suppression and Oxaliplatin treatment in colorectal cancer therapy. Biomed. Pharmacother. 2021, 137, 111364. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.L.; Peng, J.Y.; Zhang, P.; Liu, W.J.; Huang, L.; Qin, H.L. Immunohistochemical analysis revealed CD34 and Ki67 protein expression as significant prognostic factors in colorectal cancer. Med. Oncol. 2010, 27, 304–309. [Google Scholar] [CrossRef]
- Toma, S.C.; Uscatu, C.D.; Ungureanu, B.S.; Mirea, C.S.; Dumitrescu, T.; Georgescu, E.F.; Georgescu, I. Correlations between CD34 Immunolabelled Blood Vessels and CD34 mRNA Expression in Colorectal Cancer. Curr. Health Sci. J. 2018, 44, 60–63. [Google Scholar]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Wahab, S.M.R.; Islam, F.; Gopalan, V.; Lam, A.K.-Y. The Identifications and Clinical Implications of Cancer Stem Cells in Colorectal Cancer. Clin. Colorectal. Cancer 2017, 16, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Kataki, A.; Giannakoulis, V.G.; Derventzi, A.; Papiris, K.; Koniaris, E.; Konstadoulakis, M. Membranous CD44v6 is upregulated as an early event in colorectal cancer: Downregulation is associated with circulating tumor cells and poor prognosis. Oncol. Lett. 2021, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E. The Concept of “Cancer Stem Cells” in the Context of Classic Carcinogenesis Hypotheses and Experimental Findings. Life 2021, 11, 1308. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Iglesias, L.; Barcia-Castro, L.; Rodríguez-Quiroga, M.; De La Cadena, M.P.; Rodŕguez-Berrocal, J.; Cordero, O.J. Surface expression marker profile in colon cancer cell lines and sphere-derived cells suggests complexity in CD26+ cancer stem cells subsets. Biol. Open 2019, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Swart, G.W.M. Activated Leukocyte Cell Adhesion Molecule/CD166, a Marker of Tumor Progression in Primary Malignant Melanoma of the Skin. Am. J. Pathol. 2000, 156, 769–774. [Google Scholar]
- Bartolomé, R.A.; Pintado-Berninches, L.; Jaén, M.; de Los Ríos, V.; Imbaud, J.I.; Casal, J.I. SOSTDC1 promotes invasion and liver metastasis in colorectal cancer via interaction with ALCAM/CD166. Oncogene 2020, 39, 6085–6098. [Google Scholar] [CrossRef] [PubMed]
- Gilsanz, A.; Sánchez-Martín, L.; Gutiérrez-López, M.D.; Ovalle, S.; Machado-Pineda, Y.; Reyes, R.; Swart, G.W.; Figdor, C.G.; Lafuente, E.M.; Cabañas, C. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol. Life Sci. 2013, 70, 475–493. [Google Scholar] [CrossRef]
- Oh, B.Y.; Kim, S.-Y.; Lee, Y.S.; Hong, H.K.; Kim, T.W.; Kim, S.H.; Lee, W.Y.; Cho, Y.B. Twist1-induced epithelial-mesenchymal transition according to microsatellite instability status in colon cancer cells. Oncotarget 2016, 7, 57066–57076. [Google Scholar] [CrossRef]
- Cubas, R.; Li, M.; Chen, C.; Yao, Q. Trop2: A possible therapeutic target for late stage epithelial carcinomas. Biochim. Biophys. Acta-Rev. Cancer 2009, 1796, 309–314. [Google Scholar] [CrossRef]
- Kuai, X.; Jia, L.; Yang, T.; Huang, X.; Zhao, W.; Zhang, M.; Chen, Y.; Zhu, J.; Feng, Z.; Tang, Q. TROP2 promotes multidrug resistance by regulating Notch1 signaling pathway in gastric cancer cells. Med. Sci. Monit. 2020, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guerra, E.; Trerotola, M.; Tripaldi, R.; Aloisi, A.L.; Simeone, P.; Sacchetti, A.; Relli, V.; Amore, A.D.; La Sorda, R.; Lattanzio, R.; et al. Trop-2 induces tumor growth through AKT and determines sensitivity to AKT inhibitors. Clin. Cancer Res. 2016, 22, 4197–4205. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xing, G.; Zhang, C.; Lu, K.; Wang, Y.; He, X. Knockdown of Trop2 inhibits proliferation and migration and induces apoptosis of endometrial cancer cells via AKT/β-catenin pathway. Cell Biochem. Funct. 2020, 38, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jia, L.; Wang, T.; Zhang, Y.; Zhao, W.; Wang, X.; Chen, H. Trop2 binding IGF2R induces gefitinib resistance in NSCLC by remodeling the tumor microenvironment. J. Cancer 2021, 12, 5310–5319. [Google Scholar] [CrossRef]
- Okabe, M.; Tsukahara, Y.; Tanaka, M.; Suzuki, K.; Saito, S.; Kamiya, Y.; Tsujimura, T.; Nakamura, K.; Miyajima, A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 2009, 136, 1951–1960. [Google Scholar] [CrossRef]
- van Driel, P.B.A.A.; Boonstra, M.C.; Prevoo, H.A.J.M.; Van De Giessen, M.; Snoeks, T.J.A.; Tummers, Q.R.J.G.; Keereweer, S.; Cordfunke, R.A.; Fish, A.; Van Eendenburg, J.D.H.; et al. EpCAM as multi-tumour target for near-infrared fluorescence guided surgery. BMC Cancer 2016, 16, 1–14. [Google Scholar] [CrossRef]
- Liao, M.Y.; Lai, J.K.; Kuo, M.Y.P.; Lu, R.M.; Lin, C.W.; Cheng, P.C.; Liang, K.-H.; Wu, H.-C. An anti-EpCAM antibody EpAb2-6 for the treatment of colon cancer. Oncotarget 2015, 6, 24947–24968. [Google Scholar] [CrossRef]
- El Khoury, F.; Corcos, L.; Durand, S.; Simon, B.; Le Jossic-Corcos, C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int. J. Oncol. 2016, 49, 2558–2568. [Google Scholar] [CrossRef]
- Manuscript, A. Cell cycle dependent variation of a CD133 epitope in human embryonic stem cell, colon cancer and melanoma cell lines. Cancer Res. 2009, 68, 7882–7886. [Google Scholar] [CrossRef]
- Aguirre-Ghiso, J.A. Translating the Science of Cancer Dormancy to the Clinic. Cancer Res. 2021, 81, 4673–4675. [Google Scholar] [CrossRef]
- Manjili, M.H. Tumor dormancy and relapse: From a natural by-product of evolution to a disease state Masoud. Cancer Res. 2017, 77, 2564–2569. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Liang, X.; Zhan, Y.; Wang, Z.; Xu, J.; Qiu, Y.; Wang, J.; Cao, Y.; Le, V.-M.; Ly, H.T.; et al. Targeting CD133 reverses drug-resistance via the AKT/NF-κB/MDR1 pathway in colorectal cancer. Br. J. Cancer 2020, 122, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon Cancer Stem Cells Dictate Tumor Growth and Resist Cell Death by Production of Interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liu, X.; Wang, H.; Liu, X.; Zhang, Q.; Li, K.; Chen, Y.; Zhu, Q.; Shen, Y.; Sui, M. A nanotherapeutic strategy to overcome chemoresistance to irinotecan/7-ethyl-10-hydroxy-camptothecin in colorectal cancer. Acta Biomater. 2022, 137, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Jaeger, C.; Rashid, R.; Munir, R.; Zaidi, N. Cancer cell lipid class homeostasis is altered under nutrient-deprivation but stable under hypoxia. BMC Cancer 2019, 19, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Liu, H.-G.; Dong, S.-Y.; Yang, F.; Wang, Q.-X.; Guo, G.-L.; Pan, Y.-F.; Zhang, X.-H. Upregulation of CD44v6 contributes to acquired chemoresistance via the modulation of autophagy in colon cancer SW480 cells. Tumor Biol. 2016, 37, 8811–8824. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Muto, Y.; Ichida, K.; Fukui, T.; Takayama, Y.; Kakizawa, N.; Kato, T.; Hasegawa, F.; Watanabe, F.; Kaneda, Y.; et al. Morphological response contributes to patient selection for rescue liver resection in chemotherapy patients with initially un-resectable colorectal liver metastasis. Oncol. Lett. 2017, 14, 1491–1499. [Google Scholar] [CrossRef][Green Version]
- Rasbridge, S.A.; Gillett, C.E.; Seymour, A.M.; Patel, K.; Richards, M.A.; Rubens, R.D.; Millis, R. The effects of chemotherapy on morphology, cellular proliferation, apoptosis and oncoprotein expression in primary breast carcinoma. Br. J. Cancer 1994, 70, 335–341. [Google Scholar] [CrossRef]
- Liu, Z.; Qiu, M.; Tang, Q.-L.; Liu, M.; Lang, N.; Bi, F. Establishment and biological characteristics of oxaliplatin-resistant human colon cancer cell lines. Chin. J. Cancer 2010, 29, 661–667. [Google Scholar] [CrossRef]
- Hua, Y.; Zhu, Y.; Zhang, J.; Zhu, Z.; Ning, Z.; Chen, H.; Liu, L.; Chen, Z.; Meng, Z. MiR-122 targets X-linked inhibitor of apoptosis protein to sensitize oxaliplatin-resistant colorectal cancer cells to oxaliplatin-mediated cytotoxicity. Cell Physiol. Biochem. 2018, 51, 2148–2159. [Google Scholar] [CrossRef]
- Rodríguez-Fanjul, V.; Guerrero-López, R.; Fernández-Varas, B.; Perona, R.; Sastre-Perona, A.; Sastre, L. Comparison of Colorectal Cancer Stem Cells and Oxaliplatin-Resistant Cells Unveils Functional Similarities. Cells 2022, 11, 511. [Google Scholar] [CrossRef] [PubMed]




| Antibody | Dilution | Antibody | Dilution |
|---|---|---|---|
| anti-BCRP1 | 1:11 | anti-CD36 | 1:50 |
| anti-CD133 | 1:50 | anti-CD44V6 | 1:50 |
| anti-AC133 | 1:50 | anti-TROP2 | 1:100 |
| anti-EPCAM | 1:50 | anti-LGR5 | 1:50 |
| anti-CD34 | 1:50 | anti-CD166 | 1:11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Díez, M.; Alegría-Aravena, N.; López-Montes, M.; Quiroz-Troncoso, J.; González-Martos, R.; Menéndez-Rey, A.; Sánchez-Sánchez, J.L.; Pastor, J.M.; Ramírez-Castillejo, C. Implication of Different Tumor Biomarkers in Drug Resistance and Invasiveness in Primary and Metastatic Colorectal Cancer Cell Lines. Biomedicines 2022, 10, 1083. https://doi.org/10.3390/biomedicines10051083
Sánchez-Díez M, Alegría-Aravena N, López-Montes M, Quiroz-Troncoso J, González-Martos R, Menéndez-Rey A, Sánchez-Sánchez JL, Pastor JM, Ramírez-Castillejo C. Implication of Different Tumor Biomarkers in Drug Resistance and Invasiveness in Primary and Metastatic Colorectal Cancer Cell Lines. Biomedicines. 2022; 10(5):1083. https://doi.org/10.3390/biomedicines10051083
Chicago/Turabian StyleSánchez-Díez, Marta, Nicolás Alegría-Aravena, Marta López-Montes, Josefa Quiroz-Troncoso, Raquel González-Martos, Adrián Menéndez-Rey, José Luis Sánchez-Sánchez, Juan Manuel Pastor, and Carmen Ramírez-Castillejo. 2022. "Implication of Different Tumor Biomarkers in Drug Resistance and Invasiveness in Primary and Metastatic Colorectal Cancer Cell Lines" Biomedicines 10, no. 5: 1083. https://doi.org/10.3390/biomedicines10051083
APA StyleSánchez-Díez, M., Alegría-Aravena, N., López-Montes, M., Quiroz-Troncoso, J., González-Martos, R., Menéndez-Rey, A., Sánchez-Sánchez, J. L., Pastor, J. M., & Ramírez-Castillejo, C. (2022). Implication of Different Tumor Biomarkers in Drug Resistance and Invasiveness in Primary and Metastatic Colorectal Cancer Cell Lines. Biomedicines, 10(5), 1083. https://doi.org/10.3390/biomedicines10051083









