Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Methods and Strains
2.2. Analysis of Bacillus and C. difficile Spores in Fecal Samples
2.3. Screening of Spore Formers in Fecal Samples with Activity Against C. difficile
2.4. Purification and Identification of Heterogenous Lipopeptide Lytic Micelles (HELMs) from Bacillus Bacterial Culture
2.5. Synergy between RP-HPLC Separated Lipopeptides
2.6. HELM Bacteriolysis of C. difficile in Culture
2.7. DLS Analysis
2.8. RP-HPLC and MALDI-TOF Identification of HELMs in Small Intestinal (SI) Samples
2.9. Methanol (MeOH) Extraction of Intestinal Contents
2.10. Ex Vivo Analysis of Inhibitory Activity Against C. difficile
2.11. Synergy between HELMs and SI Extracts
2.12. Synergy between HELMs and DOC
2.13. In Vivo Studies
2.14. Preparation of Bacillus Test Material for In Vivo Studies
2.15. Murine C. difficile Colonization Susceptibility Study
2.16. Bacillus and C. difficile Colonization Cohort Study in Piglets
2.17. Hamster Clindamycin Colonization Model
2.18. Murine Microbiota Depletion Model
2.19. Statistics
2.20. Data Availability
3. Results
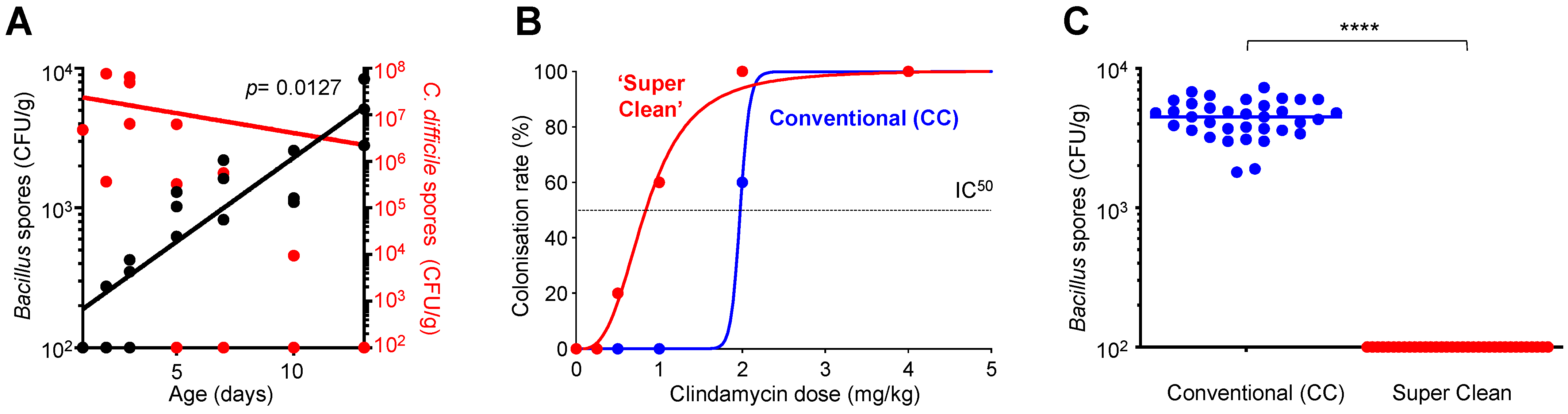
3.1. C. difficile Colonization Inversely Correlates to the Acquisition of Environmental Bacillus
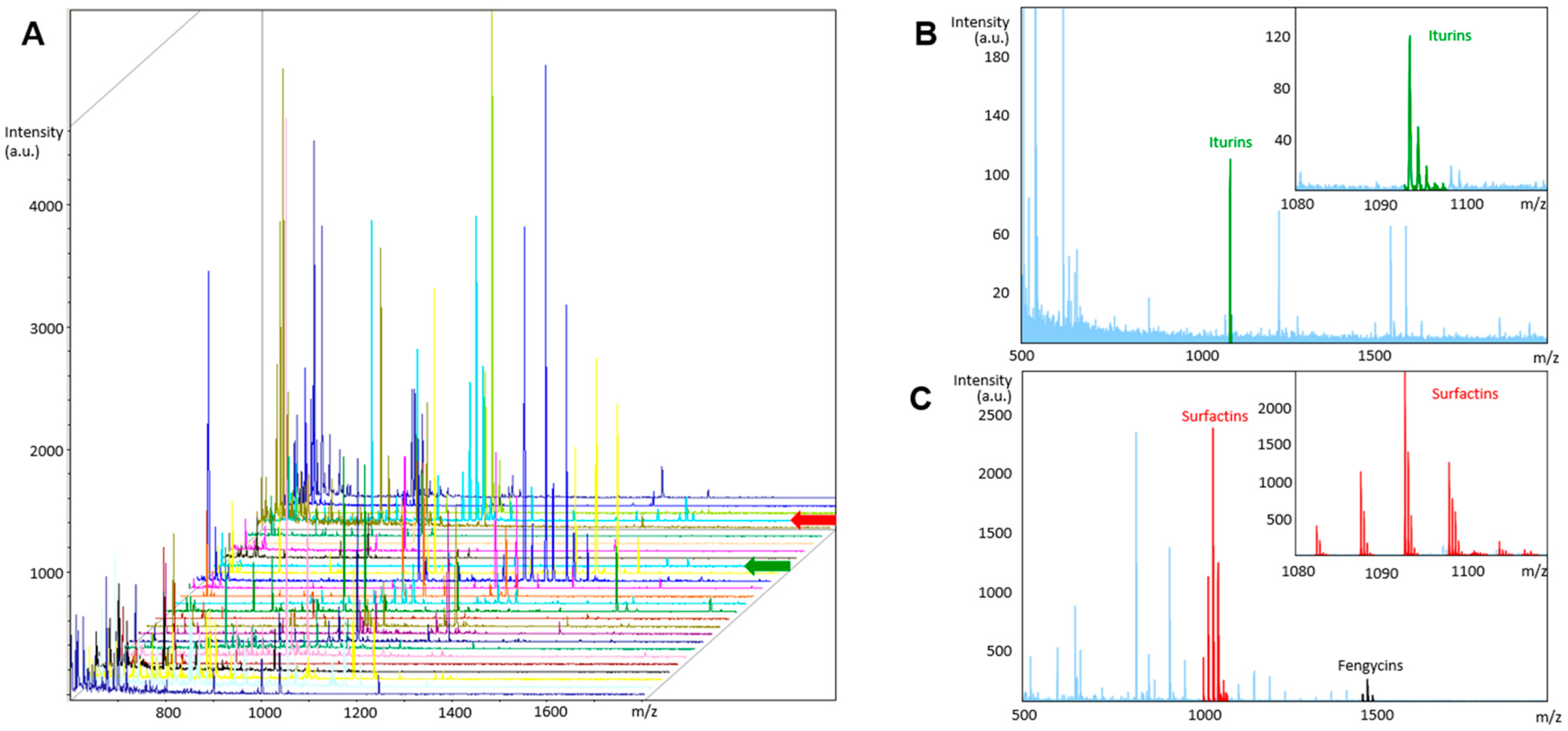
3.2. Environmentally Acquired Bacillus Have Inhibitory Activity towards C. difficile
3.3. HELMs in the SI Can Inhibit C. difficile
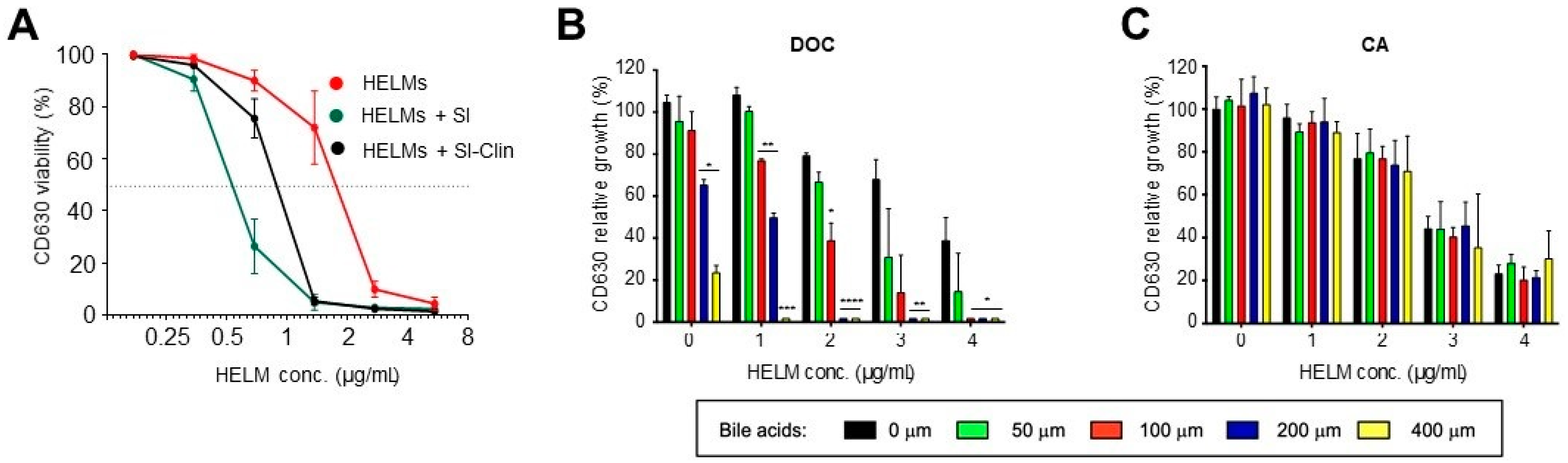
3.4. Synergistic Activity of HELMs
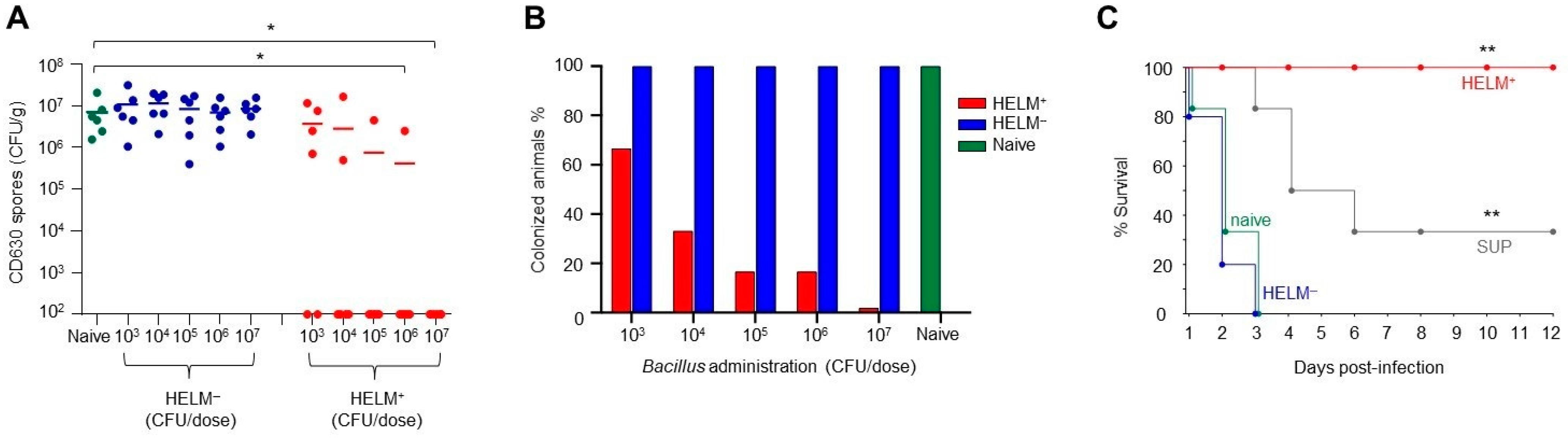
3.5. Exclusion of C. difficile Colonization by HELM-Producing Bacillus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depestel, D.D.; Aronoff, D.M. Epidemiology of Clostridium difficile infection. J. Pharm. Pract. 2013, 26, 464–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Smathers, S.A.; Prasad, P.; Leckerman, K.H.; Coffin, S.; Zaoutis, T. Epidemiological features of Clostridium difficile-associated disease among inpatients at children’s hospitals in the United States, 2001–2006. Pediatrics 2008, 122, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D.; et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology 2017, 152, 799–811.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miezeiewski, M.; Schnaufer, T.; Muravsky, M.; Wang, S.; Caro-Aguilar, I.; Secore, S.; Thiriot, D.S.; Hsu, C.; Rogers, I.; DeSantis, T.; et al. An in vitro culture model to study the dynamics of colonic microbiota in Syrian golden hamsters and their susceptibility to infection with Clostridium difficile. ISME J. 2015, 9, 321–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jangi, S.; Lamont, J.T. Asymptomatic colonization by Clostridium difficile in infants: Implications for disease in later life. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 2–7. [Google Scholar] [CrossRef]
- Daquigan, N.; Seekatz, A.M.; Greathouse, K.L.; Young, V.B.; White, J.R. High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 2017, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, C.; Levenez, F.; Fouqueray, C.; Dore, J.; Collignon, A.; Lepage, P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J. Clin. Microbiol. 2011, 49, 858–865. [Google Scholar] [CrossRef] [Green Version]
- Ringel-Kulka, T.; Cheng, J.; Ringel, Y.; Salojarvi, J.; Carroll, I.; Palva, A.; de Vos, W.M.; Satokari, R. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PLoS ONE 2013, 8, e64315. [Google Scholar] [CrossRef]
- Li, Y.; Faden, H.S.; Zhu, L. The Response of the Gut Microbiota to Dietary Changes in the First Two Years of Life. Front. Pharmacol. 2020, 11, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winston, J.A.; Theriot, C.M. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016, 41, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopman, N.E.; Keessen, E.C.; Harmanus, C.; Sanders, I.M.; van Leengoed, L.A.; Kuijper, E.J.; Lipman, L.J.A. Acquisition of Clostridium difficile by piglets. Vet. Microbiol. 2011, 149, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Grzeskowiak, L.M.; Pieper, R.; Huynh, H.A.; Cutting, S.M.; Vahjen, W.; Zentek, J. Impact of early-life events on the susceptibility to Clostridium difficile colonisation and infection in the offspring of the pig. Gut Microbes 2019, 10, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weese, J.S.; Wakeford, T.; Reid-Smith, R.; Rousseau, J.; Friendship, R. Longitudinal investigation of Clostridium difficile shedding in piglets. Anaerobe 2010, 16, 501–504. [Google Scholar] [CrossRef]
- Mulder, I.E.; Schmidt, B.; Stokes, C.R.; Lewis, M.; Bailey, M.; Aminov, R.I.; Prosser, J.I.; Gill, B.P.; Pluske, J.R.; Mayer, C.D.; et al. Environmentally-acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biol. 2009, 7, 79. [Google Scholar] [CrossRef] [Green Version]
- Yue, S.; Li, Z.; Hu, F.; Picimbon, J.F. Curing piglets from diarrhea and preparation of a healthy microbiome with Bacillus treatment for industrial animal breeding. Sci. Rep. 2020, 10, 19476. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef]
- Tam, N.K.; Uyen, N.Q.; Hong, H.A.; Duc le, H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andric, S.; Meyer, T.; Ongena, M. Bacillus Responses to Plant-Associated Fungal and Bacterial Communities. Front. Microbiol. 2020, 11, 1350. [Google Scholar] [CrossRef]
- Lopez, D.; Vlamakis, H.; Losick, R.; Kolter, R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 2009, 74, 609–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakar, K.U.; Duan, Y.P.; Nawaz, Z.; Sun, G.; Almoneafy, A.A.; Hassan, M.A.; Elshakh, A.; Li, B.; Xie, G.L. A novel rhizobacterium Bk7 for biological control of brown sheath rot of rice caused by Pseudomonas fuscovaginae and its mode of action. Eur. J. Plant Pathol. 2014, 138, 819–834. [Google Scholar] [CrossRef]
- Ferreira, W.T.; Hong, H.A.; Hess, M.; Adams, J.R.G.; Wood, H.; Bakun, K.; Tan, S.; Baccigalupi, L.; Ferrari, E.; Brisson, A.; et al. Micellar Antibiotics of Bacillus. Pharmaceutics 2021, 13, 1296. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martin, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the fecal microbiota: An optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef]
- Jenior, M.L.; Leslie, J.L.; Young, V.B.; Schloss, P.D. Clostridium difficile Alters the Structure and Metabolism of Distinct Cecal Microbiomes during Initial Infection To Promote Sustained Colonization. MSphere 2018, 3, e00261-18. [Google Scholar] [CrossRef] [Green Version]
- Filippidou, S.; Junier, T.; Wunderlin, T.; Lo, C.C.; Li, P.E.; Chain, P.S.; Junier, P. Under-detection of endospore-forming Firmicutes in metagenomic data. Comput. Struct. Biotechnol. J. 2015, 13, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Phister, T.G.; O’Sullivan, D.J.; McKay, L.L. Identification of bacilysin, chlorotetaine, and iturin a produced by Bacillus sp. strain CS93 isolated from pozol, a Mexican fermented maize dough. Appl. Environ. Microbiol. 2004, 70, 631–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molecular Biological Methods for Bacillus; Harwood, C.R.; Cutting, S.M. (Eds.) John Wiley & Sons Ltd.: Chichester, UK, 1990. [Google Scholar]
- Phetcharaburanin, J.; Hong, H.A.; Colenutt, C.; Bianconi, I.; Sempere, L.; Permpoonpattana, P.; Smith, K.; Dembek, M.; Tan, S.; Brisson, M.C.; et al. The spore-associated protein BclA1 affects the susceptibility of animals to colonization and infection by Clostridium difficile. Mol. Microbiol. 2014, 92, 1025–1038. [Google Scholar] [CrossRef]
- Hong, H.A.; Khaneja, R.; Tam, N.M.; Cazzato, A.; Tan, S.; Urdaci, M.; Brisson, A.; Gasbarrini, A.; Barnes, I.; Cutting, S.M. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 2009, 160, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Munoz, M.; Rios-Chaparro, D.I.; Herrera, G.; Soto-De Leon, S.C.; Birchenall, C.; Pinilla, D.; Pardo-Oviedo, J.M.; Josa, D.F.; Patarroyo, M.A.; Ramírez, J.D. New Insights into Clostridium difficile (CD) Infection in Latin America: Novel Description of Toxigenic Profiles of Diarrhea-Associated to CD in Bogota, Colombia. Front. Microbiol. 2018, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Bae, K.S. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 2000, 78, 123–127. [Google Scholar] [CrossRef]
- Hoa, N.T.; Baccigalupi, L.; Huxham, A.; Smertenko, A.; Van, P.H.; Ammendola, S.; Ricca, E.; Cutting, S.M. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 2000, 66, 5241–5247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.A.; Hitri, K.; Hosseini, S.; Kotowicz, N.; Bryan, D.; Mawas, F.; Wilkinson, A.J.; van Broekhoven, A.; Kearsey, J.; Cutting, S.M. Mucosal Antibodies to the C Terminus of Toxin A Prevent Colonization of Clostridium difficile. Infect. Immun. 2017, 85, e01060-16. [Google Scholar] [CrossRef] [Green Version]
- Jump, R.L. Clostridium difficile infection in older adults. Aging Health 2013, 9, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adimpong, D.B.; Sorensen, K.I.; Thorsen, L.; Stuer-Lauridsen, B.; Abdelgadir, W.S.; Nielsen, D.S.; Derkx, P.M.; Jespersen, L. Antimicrobial susceptibility of Bacillus strains isolated from primary starters for African traditional bread production and characterization of the bacitracin operon and bacitracin biosynthesis. Appl. Environ. Microbiol. 2012, 78, 7903–7914. [Google Scholar] [CrossRef] [Green Version]
- Hryckowian, A.J.; Van Treuren, W.; Smits, S.A.; Davis, N.M.; Gardner, J.O.; Bouley, D.M.; Sonnenburg, J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 2018, 3, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapp, C.; Jung, G.; Katzer, W.; Loeffler, W. Chlorotetain from Bacillus subtilis, an antifungal dipeptide with an unusual chlorine-containing amino acid. Angew. Chem. Int. Ed. Engl. 1988, 27, 1733–1734. [Google Scholar] [CrossRef]
- Mnif, I.; Ghribi, D. Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers 2015, 104, 129–147. [Google Scholar] [CrossRef]
- Sheikh, M.; Khanam, A.J.; Matto, R.; Din, K. Comparative study of the micellar and antimicrobial activity of Gemini conventional surfactants in pure and mixed micelles. J. Surfactants Deterg. 2013, 16, 503–508. [Google Scholar] [CrossRef]
- Granum, P.E.; Lund, T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Ferreira, W.T.; Hosseini, S.; Anwar, S.; Hitri, K.; Wilkinson, A.J.; Vahjen, W.; Zentek, J.; Soloviev, M.; Cutting, S.M. The Spore Coat Protein CotE Facilitates Host Colonization by Clostridium difficile. J. Infect. Dis. 2017, 216, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. MSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Coote, P.J. Bactericidal synergy of lysostaphin in combination with antimicrobial peptides. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1015–1021. [Google Scholar] [CrossRef]
- Rivardo, F.; Martinotti, M.G.; Turner, R.J.; Ceri, H. Synergistic effect of lipopeptide biosurfactant with antibiotics against Escherichia coli CFT073 biofilm. Int. J. Antimicrob. Agents 2011, 37, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Lichtenberger, L.M.; Ajami, N.; Dial, E.J.; Jiang, Z.D.; DuPont, H.L. Bile acids improve the antimicrobial effect of rifaximin. Antimicrob. Agents Chemother. 2010, 54, 3618–3624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleksic Sabo, V.; Skoric, D.; Jovanovic-Santa, S.; Nikolic, I.; Janos, C.; Knezevic, P. Synergistic activity of bile salts and their derivatives in combination with conventional antimicrobial agents against Acinetobacter baumannii. J. Ethnopharmacol. 2021, 264, 113266. [Google Scholar] [CrossRef]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile Acid 7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2019, 26, 27–34.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enright, E.F.; Joyce, S.A.; Gahan, C.G.; Griffin, B.T. Impact of Gut Microbiota-Mediated Bile Acid Metabolism on the Solubilization Capacity of Bile Salt Micelles and Drug Solubility. Mol. Pharm. 2017, 14, 1251–1263. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Wang, X.; Zhang, G.; Guo, B.; Hou, X.; Ran, J.; Zhang, Q.; Li, C.; Zhao, X.; et al. Mechanism of Asbt (Slc10a2)-related bile acid malabsorption in diarrhea after pelvic radiation. Int. J. Radiat. Biol. 2020, 96, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Matsumoto, H.; Kennedy, S.; Newberry, E.P.; Moritz, W.; DeBosch, B.J.; Moley, K.H.; Rubin, D.C.; Warner, B.W.; Kau, A.L.; et al. Impaired Chylomicron Assembly Modifies Hepatic Metabolism through Bile Acid-Dependent and Transmissible Microbial Adaptations. Hepatology 2019, 70, 1168–1184. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalova, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018, 9, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.A.; To, E.; Fakhry, S.; Baccigalupi, L.; Ricca, E.; Cutting, S.M. Defining the natural habitat of Bacillus spore-formers. Res. Microbiol. 2009, 160, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Heyndrickx, M. The Importance of Endospore-Forming Bacteria Originating from Soil for Contamination of Industrial Food Processing. Appl. Environ. Soil Sci. 2011, 2011, 561975. [Google Scholar] [CrossRef] [Green Version]
- Nybroe, O.; Sorensen, J. Production of Cyclic Lipopeptides by Fluorescent Pseudomonads. In Pseudomonas; Ramons, J.-L., Ed.; Kluwer Academic/Plenum: New York, NY, USA, 2004; pp. 147–172. [Google Scholar]
- Hofemeister, J.; Conrad, B.; Adler, B.; Hofemeister, B.; Feesche, J.; Kucheryava, N.; Steinborn, G.; Franke, P.; Grammel, N.; Zwintscher, A.; et al. Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol. Genet. Genom. 2004, 272, 363–378. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [Green Version]
- Shakerifard, P.; Gancel, F.; Jacques, P.; Faille, C. Effect of different Bacillus subtilis lipopeptides on surface hydrophobicity and adhesion of Bacillus cereus 98/4 spores to stainless steel and Teflon. Biofouling 2009, 25, 533–541. [Google Scholar] [CrossRef]
- Ghribi, D.; Ellouze-Chaabouni, S. Enhancement of Bacillus subtilis Lipopeptide Biosurfactants Production through Optimization of Medium Composition and Adequate Control of Aeration. Biotechnol. Res. Int. 2011, 2011, 653654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocin-Antimicrobial Synergy: A Medical and Food Perspective. Front. Microbiol. 2017, 8, 1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorbara, M.T.; Pamer, E.G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 2019, 12, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaCourse, K.D.; Peterson, S.B.; Kulasekara, H.D.; Radey, M.C.; Kim, J.; Mougous, J.D. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat. Microbiol. 2018, 3, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Vinarov, Z.; Katev, V.; Radeva, D.; Tcholakova, S.; Denkov, N.D. Micellar solubilization of poorly water-soluble drugs: Effect of surfactant and solubilizate molecular structure. Drug Dev. Ind. Pharm. 2018, 44, 677–686. [Google Scholar] [CrossRef]
- Studer, N.; Desharnais, L.; Beutler, M.; Brugiroux, S.; Terrazos, M.A.; Menin, L.; Schürch, C.M.; McCoy, K.D.; Kuehne, S.A.; Minton, N.P.; et al. Functional Intestinal Bile Acid 7alpha-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front. Cell Infect. Microbiol. 2016, 6, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribet, D.; Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef]
- Blum, W.E.H.; Zechmeister-Boltenstern, S.; Keiblinger, K.M. Does Soil Contribute to the Human Gut Microbiome? Microorganisms 2019, 7, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, I.; Stegen, J.C.; Maldonado-Gomez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The gut microbiota of rural papua new guineans: Composition, diversity patterns, and ecological processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, J.L.; Sonnenburg, E.D. Vulnerability of the industrialized microbiota. Science 2019, 366, eaaw9255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Mäkelä, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Zhang, H.; Bai, Z.; Zhang, A.; Bai, F.; Luo, X.; Hou, Y.; Ding, X.; Sun, B.; Sun, X.; et al. Exposure to soil, house dust and decaying plants increases gut microbial diversity and decreases serum immunoglobulin E levels in BALB/c mice. Environ. Microbiol. 2016, 18, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Baddour, L.M.; Huskins, W.C.; Kammer, P.P.; Faubion, W.A.; Zinsmeister, A.R.; Harmsen, W.S.; Pardi, D.S. The epidemiology of Clostridium difficile infection in children: A population-based study. Clin. Infect. Dis. 2013, 56, 1401–1406. [Google Scholar] [CrossRef] [Green Version]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Browne, H.P.; Viciani, E.; Forster, S.C.; Clare, S.; Harcourt, K.; Stares, M.D.; Dougan, G.; Fairley, D.J.; Roberts, P.; et al. Adaptation of host transmission cycle during Clostridium difficile speciation. Nat. Genet. 2019, 51, 1315–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, J.; Robinson, C.; Danhof, H.; Knetsch, C.W.; van Leeuwen, H.C.; Lawley, T.D.; Auchtung, J.M.; Britton, R.A. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 2018, 553, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Granato, E.T.; Meiller-Legrand, T.A.; Foster, K.R. The Evolution and Ecology of Bacterial Warfare. Curr. Biol. 2019, 29, R521–R537. [Google Scholar] [CrossRef]
- La Ragione, R.M.; Woodward, M.J. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol. 2003, 94, 245–256. [Google Scholar] [CrossRef]
- Wust, J.; Sullivan, N.M.; Hardegger, U.; Wilkins, T.D. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol. 1982, 16, 1096–1101. [Google Scholar] [CrossRef]
- Fort, P.; Errington, J. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J. Gen. Microbiol. 1985, 131, 1091–1105. [Google Scholar] [CrossRef] [Green Version]
- Vehmaanpera, J. Transformation of Bacillus amyloliquefaciens protoplasts with plasmid DNA. FEMS Microbiol. Lett. 1988, 49, 101–105. [Google Scholar]
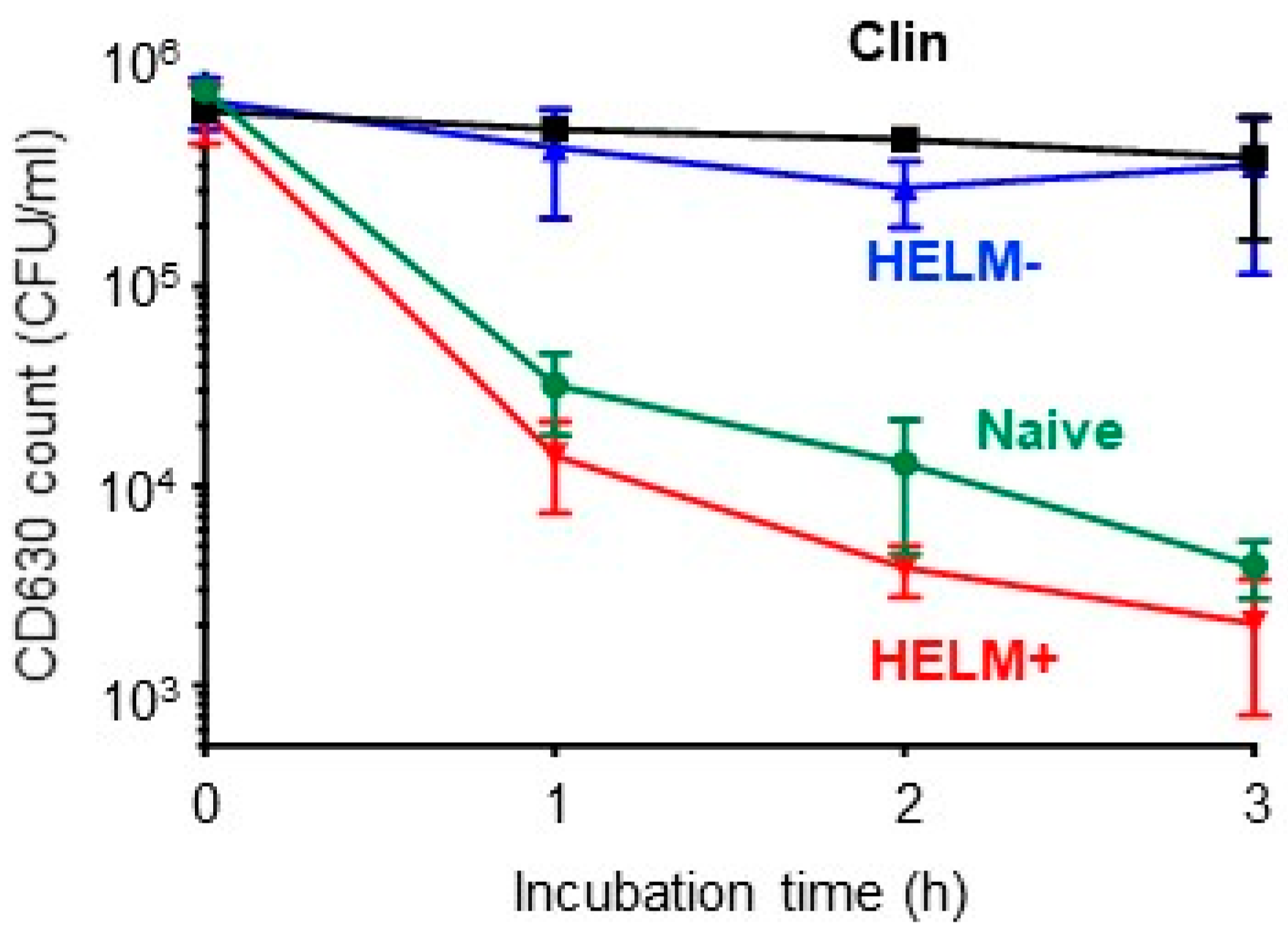
 ), HELM− (srfAA-) culture supernatant (
), HELM− (srfAA-) culture supernatant (  ), HELM+ (Bv277) SEC-fractionated ‘active’ fraction (
), HELM+ (Bv277) SEC-fractionated ‘active’ fraction (  ) and untreated (PBS) (
) and untreated (PBS) (  ).
).
 ), HELM− (srfAA-) culture supernatant (
), HELM− (srfAA-) culture supernatant (  ), HELM+ (Bv277) SEC-fractionated ‘active’ fraction (
), HELM+ (Bv277) SEC-fractionated ‘active’ fraction (  ) and untreated (PBS) (
) and untreated (PBS) (  ).
).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, W.T.; Hong, H.A.; Adams, J.R.G.; Hess, M.; Kotowicz, N.K.; Tan, S.; Ferrari, E.; Brisson, A.; Zentek, J.; Soloviev, M.; et al. Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance. Biomedicines 2022, 10, 930. https://doi.org/10.3390/biomedicines10050930
Ferreira WT, Hong HA, Adams JRG, Hess M, Kotowicz NK, Tan S, Ferrari E, Brisson A, Zentek J, Soloviev M, et al. Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance. Biomedicines. 2022; 10(5):930. https://doi.org/10.3390/biomedicines10050930
Chicago/Turabian StyleFerreira, William T., Huynh A. Hong, James R. G. Adams, Mateusz Hess, Natalia K. Kotowicz, Sisareuth Tan, Enrico Ferrari, Alain Brisson, Jurgen Zentek, Mikhail Soloviev, and et al. 2022. "Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance" Biomedicines 10, no. 5: 930. https://doi.org/10.3390/biomedicines10050930
APA StyleFerreira, W. T., Hong, H. A., Adams, J. R. G., Hess, M., Kotowicz, N. K., Tan, S., Ferrari, E., Brisson, A., Zentek, J., Soloviev, M., & Cutting, S. M. (2022). Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance. Biomedicines, 10(5), 930. https://doi.org/10.3390/biomedicines10050930








