Identification of LOC101927355 as a Novel Biomarker for Preeclampsia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset and Differential Expression Analysis
2.2. Sample Collection
2.3. RNA Extraction
2.4. CDNA Synthesis and qPCR
2.5. Prediction of Subcellular Localization and Identification of MiRNAs’ Binding Sites for LOC101927355
2.6. Statistical Analysis
3. Results
3.1. Differentially Expressed Genes’ Identification in PE Samples
3.2. Validation of LncRNAs in the Term Placentas of Preeclamptic and Control Patients
3.3. Determination of LncRNAs in Maternal Plasma at Delivery
3.4. Subcellular Localization of LncRNA LOC101927355
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The American College of Obstetricians and Gynecologists (ACOG). Gestational Hypertension and Preeclampsia: ACOG Practice Bulletin, Number 222. Obstet. Gynecol. 2020, 135, e237–e260. [Google Scholar] [CrossRef] [PubMed]
- Steegers, E.A.P.; Von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Pre-eclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Gyselaers, W. Preeclampsia Is a Syndrome with a Cascade of Pathophysiologic Events. J. Clin. Med. 2020, 9, 2245. [Google Scholar] [CrossRef]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta—Gene Regul. Mech. 2016, 1859, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Marinescu, M.; Lazar, A.; Marta, M.M.; Cozma, A.; Catana, C. Non-Coding RNAs: Prevention, Diagnosis, and Treatment in Myocardial Ischemia–Reperfusion Injury. Int. J. Mol. Sci. 2022, 23, 2728. [Google Scholar] [CrossRef]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Non-Coding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, eaah7111. [Google Scholar] [CrossRef] [Green Version]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.-C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, L.J.; Peñailillo, R.; Sánchez, M.; Acuña-Gallardo, S.; Mönckeberg, M.; Ong, J.; Choolani, M.; Illanes, S.E.; Nardocci, G. The role of long non-coding rnas in trophoblast regulation in preeclampsia and intrauterine growth restriction. Genes 2021, 12, 970. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Meng, T. Long Noncoding RNA in Preeclampsia: Transcriptional Noise or Innovative Indicators? Biomed Res. Int. 2019, 2019, 5437621. [Google Scholar] [CrossRef] [PubMed]
- He, X.; He, Y.; Xi, B.; Zheng, J.; Zeng, X.; Cai, Q.; OuYang, Y.; Wang, C.; Zhou, X.; Huang, H.; et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS ONE 2013, 8, e81437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, J.; Zhao, W.; Lv, H.; Li, W.; Chen, Z.; Zhang, C. Transcriptomic Profiling in Human Decidua of Severe Preeclampsia Detected by RNA Sequencing. J. Cell. Biochem. 2018, 119, 607–615. [Google Scholar] [CrossRef]
- Awamleh, Z.; Gloor, G.B.; Han, V.K.M. Placental microRNAs in pregnancies with early onset intrauterine growth restriction and preeclampsia: Potential impact on gene expression and pathophysiology. BMC Med. Genom. 2019, 12, 91. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Wu, Y.; Lu, C.; Zhang, F.; Wu, F.-X.; Li, M. DeepLncLoc: A deep learning framework for long non-coding RNA subcellular localization prediction based on subsequence embedding. Brief. Bioinform. 2022, 23, bbab360. [Google Scholar] [CrossRef]
- Ahmad, A.; Lin, H.; Shatabda, S. Locate-R: Subcellular localization of long non-coding RNAs using nucleotide compositions. Genomics 2020, 112, 2583–2589. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Pan, X.; Shen, H. Bin LncLocator 2.0: A cell-line-specific subcellular localization predictor for long non-coding RNAs with interpretable deep learning. Bioinformatics 2021, 37, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Gao, L.; Guo, X.; Li, X.; Huang, X.; Wang, Y.; Xu, H.; He, R.; Jia, C.; Liang, F. lncSLdb: A resource for long non-coding RNA subcellular localization. Database 2018, 2018, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef]
- Song, X.; Luo, X.; Gao, Q.; Wang, Y.; Gao, Q.; Long, W. Dysregulation of LncRNAs in Placenta and Pathogenesis of Preeclampsia. Curr. Drug Targets 2017, 18, 1165–1170. [Google Scholar] [CrossRef]
- Long, W.; Rui, C.; Song, X.; Dai, X.; Xue, X.; Lu, Y.; Shen, R.; Li, J.; Li, J.; Ding, H. Distinct expression profiles of lncRNAs between early-onset preeclampsia and preterm controls. Clin. Chim. Acta 2016, 463, 193–199. [Google Scholar] [CrossRef]
- Chen, F.; Wang, N.; Tan, H.Y.; Guo, W.; Zhang, C.; Feng, Y. The functional roles of exosomes-derived long non-coding RNA in human cancer. Cancer Biol. Ther. 2019, 20, 583–592. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Xie, X.J.; Zhao, L.; Chen, W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008, 582, 1919–1927. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.M.; Lu, J.H.; Chen, W.Y.; Gu, A.Q. Up-regulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int. J. Clin. Exp. Med. 2015, 8, 11824–11830. [Google Scholar] [PubMed]
- Liu, J.; Luo, C.; Zhang, C.; Cai, Q.; Lin, J.; Zhu, T.; Huang, X. Up-regulated lncRNA UCA1 inhibits trophoblast cell invasion and proliferation by downregulating JAK2. J. Cell. Physiol. 2020, 235, 7410–7419. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Li, J.; Mao, B. Urothelial cancer associated 1 (UCA1) regulates trophoblast viability, proliferation, and migration via modulating the UCA1/miR-455/RUNX2 signaling pathway. Acta Biochim. Biophys. Sin. 2020, 52, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Jin, F.; Hu, J.; Zhu, Z.; Tian, F.; Tao, M.; Teng, Y. Urothelial carcinoma associated 1 promotes trophoblast invasion by regulating MMP9. Cell Biosci. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, H.; Wu, B.; Zhang, C.; Yu, H.; Li, X.; Wang, Q.; Shi, X.; Fan, C.; Wang, D.; et al. Long noncoding rna linc00551 suppresses glycolysis and tumor progression by regulating c-myc-mediated pkm2 expression in lung adenocarcinoma. Onco. Targets. Ther. 2020, 13, 11459–11470. [Google Scholar] [CrossRef]
- Peng, X.; Zhou, Y.; Chen, Y.; Tang, L.; Wang, G.; Jiang, H.; Wang, X.; Tao, Y.; Zhuang, W. Reduced LINC00551 expression promotes proliferation and invasion of esophageal squamous cancer by increase in HSP27 phosphorylation. J. Cell. Physiol. 2021, 236, 1418–1431. [Google Scholar] [CrossRef]
- Sun, M.; Geng, D.; Li, S.; Chen, Z.; Zhao, W. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol. Chem. 2018, 399, 387–395. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, R. LNCRNA PART1 promotes breast cancer cell progression by directly targeting miR-4516. Cancer Manag. Res. 2020, 12, 7753–7760. [Google Scholar] [CrossRef]
- Li, B.; Lou, G.; Zhang, J.; Cao, N.; Yu, X. Repression of lncRNA PART1 attenuates ovarian cancer cell viability, migration and invasion through the miR-503-5p/FOXK1 axis. BMC Cancer 2022, 22, 124. [Google Scholar] [CrossRef]
- Cruickshank, B.M.; Wasson, M.C.D.; Brown, J.M.; Fernando, W.; Venkatesh, J.; Walker, O.L.; Morales-quintanilla, F.; Dahn, M.L.; Dean, C.A.; Vidovic, D.; et al. Lncrna part1 promotes proliferation and migration, is associated with cancer stem cells, and alters the mirna landscape in triple-negative breast cancer. Cancers 2021, 13, 2644. [Google Scholar] [CrossRef]
- Ruan, Z.; Zhao, D. Long intergenic noncoding RNA LINC00284 knockdown reduces angiogenesis in ovarian cancer cells via up-regulation of MEST through NF-κB1. FASEB J. 2019, 33, 12047–12059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Yang, S.; Zhao, W. Long non-coding RNA nrad1 and linc00152 are highly expressed and associated with prognosis in patients with hepatocellular carcinoma. Onco. Targets. Ther. 2020, 13, 10409–10416. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2020, 27, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Wei, G.; Zhang, L.; Xu, Z. Signature microRNAs and long noncoding RNAs in laryngeal cancer recurrence identified using a competing endogenous RNA network. Mol. Med. Rep. 2019, 19, 4806–4818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Qin, X. MicroRNA-708 targeting ZNF549 regulates colon adenocarcinoma development through PI3K/AKt pathway. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
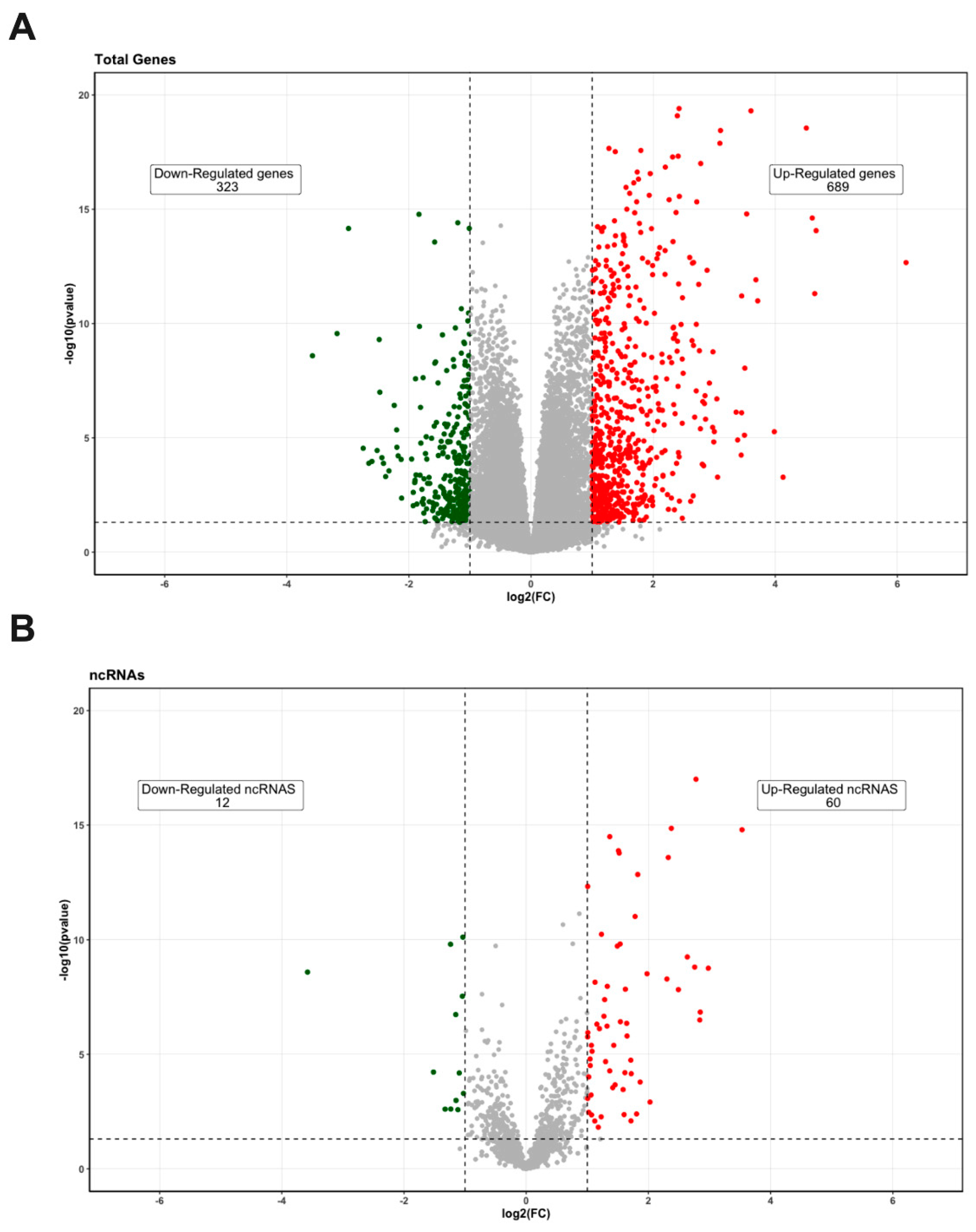
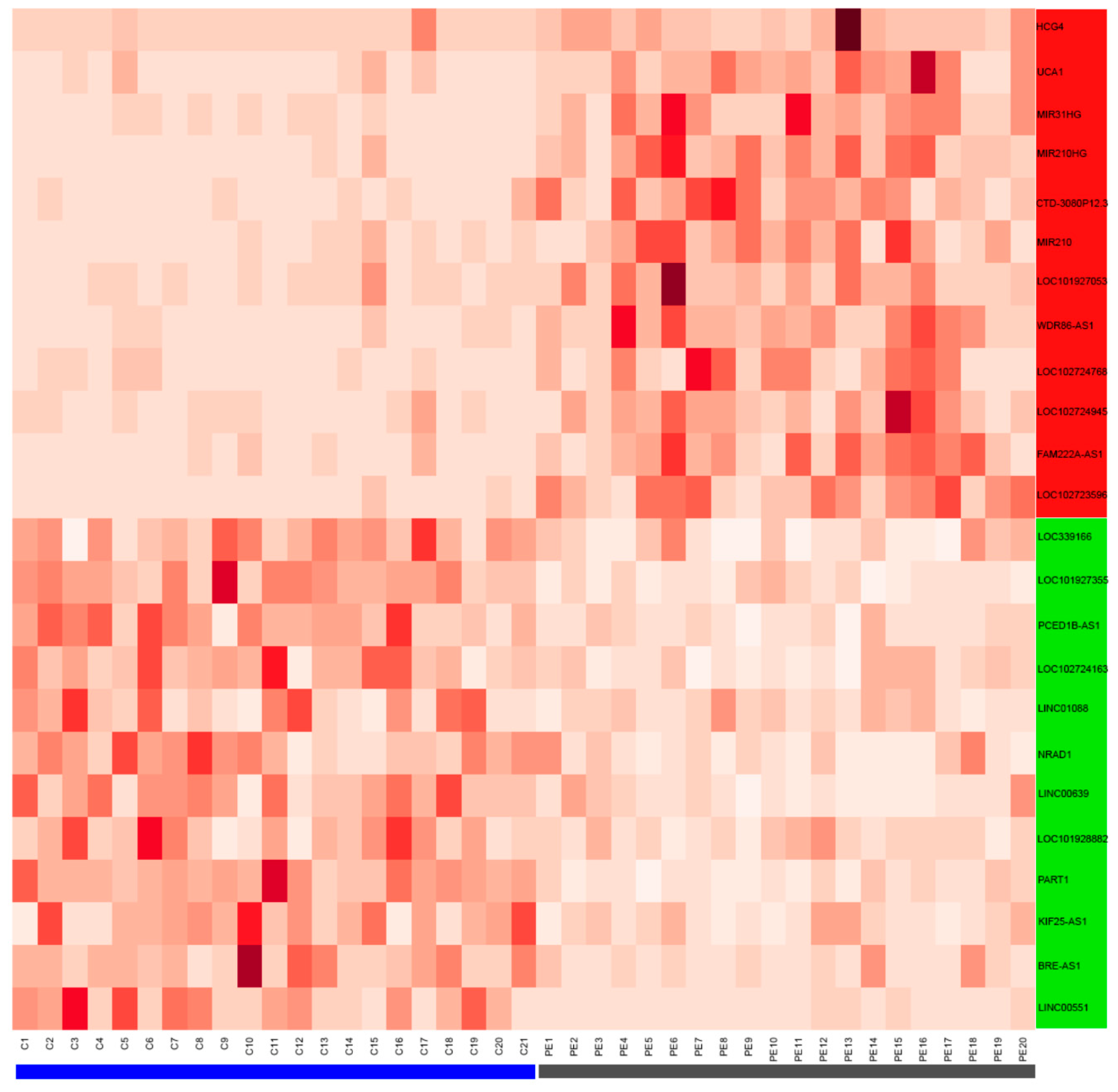
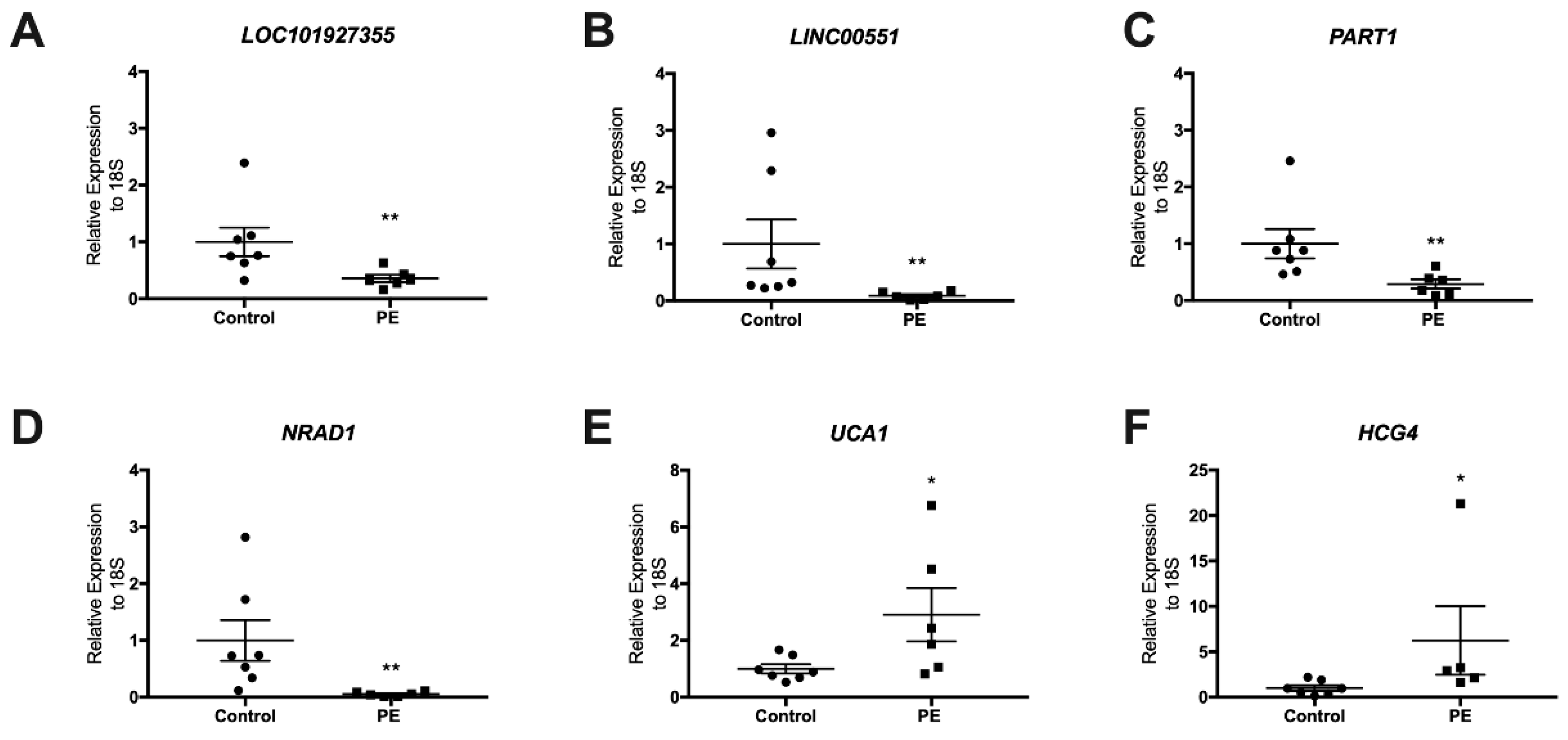

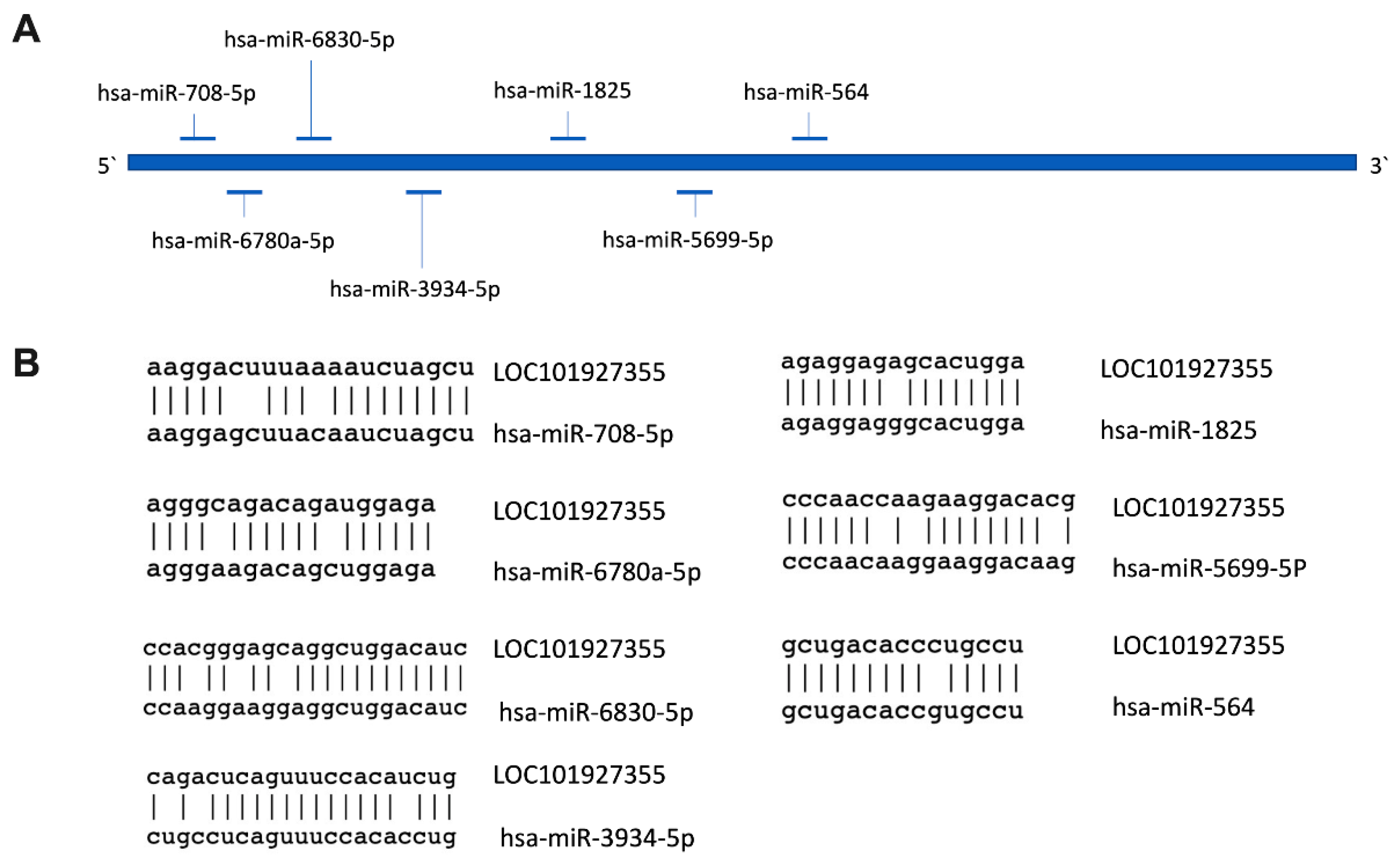
| Characteristics | Control (n = 7) | Preeclampsia (n = 7) | p-Value |
|---|---|---|---|
| Mean ± SD | Mean ± SD | ||
| Age (years) | 34.8 ± 6.3 | 29.6 ± 4.0 | 0.1358 |
| BMI (kg/m2) | 31.5 ± 2.2 | 30.6 ± 3.2 | 0.9825 |
| Systolic blood pressure | 113.8 ± 10.2 | 153.4 ± 27.9 | 0.0006 |
| Diastolic blood pressure | 66.2 ± 7.6 | 88.3 ± 14.3 | 0.0017 |
| Gestational age at delivery | 38.4 ± 1.0 | 33.14 ± 4.4 | 0.0006 |
| Birth weight | 3551.0 ± 442.7 | 1953.6 ± 768.4 | 0.0006 |
| Gene | Sequence |
|---|---|
| 18S Fw | GCCGCTAGAGGTGAAATTCTTGGA |
| 18S Rev | ATCGCCGGTCGGCATCGTTTAT |
| U6 Fw | CTCGCTTCGGCAGCACA |
| U6 Rev | AACGCTTCACGAATTTGCGT |
| LOC101927355 Fw | CTCTGACTCTGTATTTCAGGAAGC |
| LOC101927355 Rev | TTGTGGTAAAGGGAGATAGGAAGG |
| LINC00551 Fw | GGATTTGGAAGAACAAACGGG |
| LINC00551 Rev | GGTCAAATACTCTGGTAGCTCC |
| PART1 Fw | GTGATCTGGGGAAAACGCA |
| PART1 Rev | GGGAATCGGTTGTGAGTAGG |
| NRAD1 Fw | ATGTGAGTGATCAGTAACACC |
| NRAD1 Rev | GAACCACGAAGACAAGGAT |
| UCA1 Fw | GGCCCTCATTCCGTGAAGAG |
| UCA1 Rev | CTCCACCGTAAGAGTTACCCGA |
| HCG4 Fw | CCAGGGAGAAACCCTCGGAAT |
| HCG4 Rev | AAACCCTGTCTCTACACCTCCATT |
| Predictor | Nucleus | Cytoplasm | Cytosol | Exome | Ribosome | Predicted Location |
|---|---|---|---|---|---|---|
| DeepLncLoc | 0.330 | 0.447 | 0.090 | 0.017 | 0.116 | Cytoplasm |
| Locate-R | 0.06 | 0.85 | - | 0 | 0.09 | Cytoplasm |
| lncLocator | 0.154 | 0.799 | 0.022 | 0.0087 | 0.0140 | Cytoplasm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peñailillo, R.; Monteiro, L.J.; Acuña-Gallardo, S.; García, F.; Velásquez, V.; Correa, P.; Díaz, P.; Valdebenito, P.P.; Navarro, C.; Romero, R.; et al. Identification of LOC101927355 as a Novel Biomarker for Preeclampsia. Biomedicines 2022, 10, 1253. https://doi.org/10.3390/biomedicines10061253
Peñailillo R, Monteiro LJ, Acuña-Gallardo S, García F, Velásquez V, Correa P, Díaz P, Valdebenito PP, Navarro C, Romero R, et al. Identification of LOC101927355 as a Novel Biomarker for Preeclampsia. Biomedicines. 2022; 10(6):1253. https://doi.org/10.3390/biomedicines10061253
Chicago/Turabian StylePeñailillo, Reyna, Lara J. Monteiro, Stephanie Acuña-Gallardo, Felipe García, Victoria Velásquez, Paula Correa, Pilar Díaz, Patricia P. Valdebenito, Cristina Navarro, Roberto Romero, and et al. 2022. "Identification of LOC101927355 as a Novel Biomarker for Preeclampsia" Biomedicines 10, no. 6: 1253. https://doi.org/10.3390/biomedicines10061253
APA StylePeñailillo, R., Monteiro, L. J., Acuña-Gallardo, S., García, F., Velásquez, V., Correa, P., Díaz, P., Valdebenito, P. P., Navarro, C., Romero, R., Sánchez, M., Illanes, S. E., & Nardocci, G. (2022). Identification of LOC101927355 as a Novel Biomarker for Preeclampsia. Biomedicines, 10(6), 1253. https://doi.org/10.3390/biomedicines10061253







