Dietary Improvement during Lactation Normalizes miR-26a, miR-222 and miR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. miRNA Expression Profiling by Real-Time qPCR in the Mammary Gland
2.3. mRNA Expression Profiling by Real-Time qPCR in the Mammary Gland
2.4. miRNA Target Prediction and Pathway Analyses
2.5. miRNA Expression Profiling by Real-Time qPCR in Milk
2.6. Statistical Analysis
3. Results
3.1. Summary of Previously Described Phenotypic Traits of Dams
3.2. miRNA Profile in the Mammary Gland
3.3. Expression of Validated Target Genes in the Mammary Gland
3.4. In Silico Prediction of Altered Pathways in the Mammary Gland
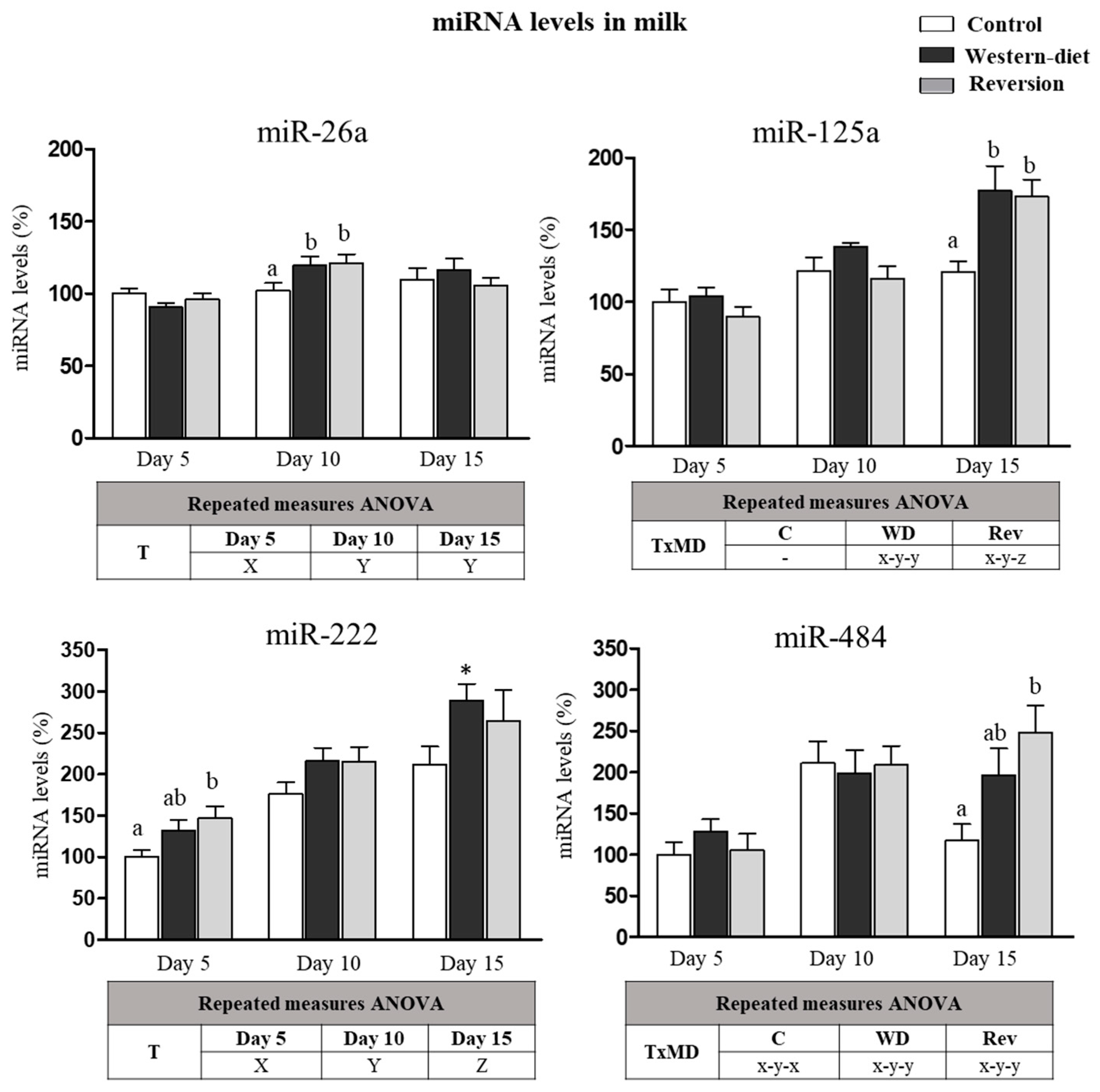
3.5. miRNA in Milk at Different Time Points of Lactation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Lönnerdal, B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173, S4–S9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef]
- Zamanillo, R.; Sánchez, J.; Serra, F.; Palou, A. Breast Milk Supply of MicroRNA Associated with Leptin and Adiponectin Is Affected by Maternal Overweight/Obesity and Influences Infancy BMI. Nutrients 2019, 11, 2589. [Google Scholar] [CrossRef] [Green Version]
- Xi, Y.; Jiang, X.; Li, R.; Chen, M.; Song, W.; Li, X. The levels of human milk microRNAs and their association with maternal weight characteristics. Eur. J. Clin. Nutr. 2016, 70, 445–449. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in Breastmilk and the Lactating Breast: Potential Immunoprotectors and Developmental Regulators for the Infant and the Mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [Green Version]
- McNeill, E.M.; Hirschi, K.D. Roles of Regulatory RNAs in Nutritional Control. Annu. Rev. Nutr. 2020, 40, 77–104. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.H.; Kaur, S.; Nielsen, L.B.; Størling, J.; Yarani, R.; Roursgaard, M.; Mathiesen, E.R.; Damm, P.; Svare, J.; Mortensen, H.B.; et al. Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers with Type 1 Diabetes Contain Aberrant Levels of microRNAs. Front. Immunol. 2019, 10, 2543. [Google Scholar] [CrossRef]
- Avril-Sassen, S.; Goldstein, L.D.; Stingl, J.; Blenkiron, C.; Le Quesne, J.; Spiteri, I.; Karagavriilidou, K.; Watson, C.J.; Tavaré, S.; Miska, E.A.; et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genom. 2009, 10, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human Milk Cells and Lipids Conserve Numerous Known and Novel miRNAs, Some of Which Are Differentially Expressed during Lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, D.J.; Travers, M.T.; Barber, M.C.; Binart, N.; Kelly, P.A. Diet-induced obesity impairs mammary development and lactogenesis in murine mammary gland. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E1179–E1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, L.L.; Grayson, B.E.; Yadav, E.; Seeley, R.J.; Horseman, N.D. High fat diet alters lactation outcomes: Possible involvement of inflammatory and serotonergic pathways. PLoS ONE 2012, 7, e32598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomar, C.A.; Serra, F.; Palou, A.; Sánchez, J. Lower miR-26a levels in breastmilk affect gene expression in adipose tissue of offspring. FASEB J. 2021, 35, e21924. [Google Scholar] [CrossRef]
- Pomar, C.A.; Castro, H.; Picó, C.; Serra, F.; Palou, A.; Sánchez, J. Cafeteria Diet Consumption during Lactation in Rats, Rather than Obesity Per Se, alters miR-222, miR-200a, and miR-26a Levels in Milk. Mol. Nutr. Food Res. 2019, 63, e1800928. [Google Scholar] [CrossRef]
- Pomar, C.; Castillo, P.; Palou, M.; Palou, A.; Picó, C. Implementation of a healthy diet to lactating rats attenuates the early detrimental programming effects in the offspring born to obese dams. Putative relationship with milk hormone levels. J. Nutr. Biochem. 2022; proof in printing. [Google Scholar]
- Zarrinpar, A.; Gupta, S.; Maurya, M.R.; Subramaniam, S.; Loomba, R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: A prospective study. Gut 2016, 65, 1546–1554. [Google Scholar] [CrossRef] [Green Version]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef]
- Lin, H.; Tas, E.; Børsheim, E.; Mercer, K.E. Circulating miRNA Signatures Associated with Insulin Resistance in Adolescents with Obesity. Diabetes Metab. Syndr. Obes. 2020, 13, 4929–4939. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Badosa, G.; Bonmatí, A.; Ortega, F.J.; Mercader, J.M.; Guindo-Martínez, M.; Torrents, D.; Prats-Puig, A.; Martinez-Calcerrada, J.M.; de Zegher, F.; Ibáñez, L.; et al. Dysregulation of Placental miRNA in Maternal Obesity Is Associated With Pre- and Postnatal Growth. J. Clin. Endocrinol. Metab. 2017, 102, 2584–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalyfa, A.; Kheirandish-Gozal, L.; Bhattacharjee, R.; Khalyfa, A.A.; Gozal, D. Circulating microRNAs as Potential Biomarkers of Endothelial Dysfunction in Obese Children. Chest 2016, 149, 786–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulsmans, M.; Sinnaeve, P.; Van der Schueren, B.; Mathieu, C.; Janssens, S.; Holvoet, P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J. Clin. Endocrinol. Metab. 2012, 97, E1213–E1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Yi, S.; Yong, D.; Shaozhuang, L.; Guangyong, Z.; Sanyuan, H. miR-320 mediates diabetes amelioration after duodenal-jejunal bypass via targeting adipoR1. Surg. Obes. Relat. Dis. 2018, 14, 960–971. [Google Scholar] [CrossRef]
- Oses, M.; Margareto Sanchez, J.; Portillo, M.P.; Aguilera, C.M.; Labayen, I. Circulating miRNAs as Biomarkers of Obesity and Obesity-Associated Comorbidities in Children and Adolescents: A Systematic Review. Nutrients 2019, 11, 2890. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, L.B.; Wang, C.; Sørensen, K.; Bang-Berthelsen, C.H.; Hansen, L.; Andersen, M.L.; Hougaard, P.; Juul, A.; Zhang, C.Y.; Pociot, F.; et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: Evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp. Diabetes Res. 2012, 2012, 896362. [Google Scholar] [CrossRef] [Green Version]
- Dantas da Costa E Silva, M.E.; Polina, E.R.; Crispim, D.; Sbruzzi, R.C.; Lavinsky, D.; Mallmann, F.; Martinelli, N.C.; Canani, L.H.; Dos Santos, K.G. Plasma levels of miR-29b and miR-200b in type 2 diabetic retinopathy. J. Cell. Mol. Med. 2019, 23, 1280–1287. [Google Scholar] [CrossRef]
- Costé, É.; Rouleux-Bonnin, F. The crucial choice of reference genes: Identification of miR-191-5p for normalization of miRNAs expression in bone marrow mesenchymal stromal cell and HS27a/HS5 cell lines. Sci. Rep. 2020, 10, 17728. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Wang, H.; Luo, J.; Zhang, T.; Tian, H.; Ma, Y.; Xu, H.; Yao, D.; Loor, J.J. MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biol. 2016, 13, 500–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Xu, Y.; Zhang, P.; Zhao, X.; Gan, M.; Li, Q.; Ma, J.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Affects Adipocytes Proliferation, Differentiation and Fatty Acid Composition of Porcine Intramuscular Fat. Int. J. Mol. Sci. 2018, 19, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Du, J.; Zhang, P.; Zhao, X.; Li, Q.; Jiang, A.; Jiang, D.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Mediates 3T3-L1 Preadipocyte Proliferation and Differentiation. Molecules 2018, 23, 317. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Luan, S.; Xiao, X.; Lin, L.; Zhao, X.; Liu, X. Silenced microRNA-222 suppresses inflammatory response in gestational diabetes mellitus mice by promoting CXCR4. Life Sci. 2021, 266, 118850. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Shuang, T.; Gao, Y.; Lu, F.; Zhang, J.; He, W.; Qu, L.; Chen, B.; Hao, Q. Targeted delivery of exosomal miR-484 reprograms tumor vasculature for chemotherapy sensitization. Cancer Lett. 2022, 530, 45–58. [Google Scholar] [CrossRef]
- Ivanova, E.; Le Guillou, S.; Hue-Beauvais, C.; Le Provost, F. Epigenetics: New Insights into Mammary Gland Biology. Genes 2021, 12, 231. [Google Scholar] [CrossRef]
- Hue-Beauvais, C.; Faulconnier, Y.; Charlier, M.; Leroux, C. Nutritional Regulation of Mammary Gland Development and Milk Synthesis in Animal Models and Dairy Species. Genes 2021, 12, 523. [Google Scholar] [CrossRef]
- Pomar, C.A.; van Nes, R.; Sánchez, J.; Picó, C.; Keijer, J.; Palou, A. Maternal consumption of a cafeteria diet during lactation in rats leads the offspring to a thin-outside-fat-inside phenotype. Int. J. Obes. 2017, 41, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Lou, F.; Jove, R.; et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J. Clin. Investig. 2015, 125, 2497–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frørup, C.; Mirza, A.H.; Yarani, R.; Nielsen, L.B.; Mathiesen, E.R.; Damm, P.; Svare, J.; Engelbrekt, C.; Størling, J.; Johannesen, J.; et al. Plasma Exosome-Enriched Extracellular Vesicles From Lactating Mothers with Type 1 Diabetes Contain Aberrant Levels of miRNAs During the Postpartum Period. Front. Immunol. 2021, 12, 744509. [Google Scholar] [CrossRef] [PubMed]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Prattichizzo, F.; Di Silvestre, D.; Perna, F.; La Sala, L.; Ceriello, A.; Mozzillo, E.; Fattorusso, V.; et al. Blood Co-Circulating Extracellular microRNAs and Immune Cell Subsets Associate with Type 1 Diabetes Severity. Int. J. Mol. Sci. 2020, 21, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, S.; Mo, Z.; Sun, S.; Yin, K.; Lv, Y. Emerging role of Insig-1 in lipid metabolism and lipid disorders. Clin. Chim. Acta 2020, 508, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Qiu, L.; Teng, X.; Zhang, Y.; Miao, Y. Effect of INSIG1 on the milk fat synthesis of buffalo mammary epithelial cells. J. Dairy Res. 2020, 87, 349–355. [Google Scholar] [CrossRef]
- Goodrich, D.W.; Wang, N.P.; Qian, Y.W.; Lee, E.Y.; Lee, W.H. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 1991, 67, 293–302. [Google Scholar] [CrossRef]
- Ahmed, F.W.; Bakhashab, S.; Bastaman, I.T.; Crossland, R.E.; Glanville, M.; Weaver, J.U. Anti-Angiogenic miR-222, miR-195, and miR-21a Plasma Levels in T1DM Are Improved by Metformin Therapy, Thus Elucidating Its Cardioprotective Effect: The MERIT Study. Int. J. Mol. Sci. 2018, 19, 3242. [Google Scholar] [CrossRef] [Green Version]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Rovira, O.; Guerra, E.; Esteve, E.; Xifra, G.; Martínez, C.; Ricart, W.; Rieusset, J.; et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014, 37, 1375–1383. [Google Scholar] [CrossRef] [Green Version]
- Coleman, C.B.; Lightell, D.J.; Moss, S.C.; Bates, M.; Parrino, P.E.; Woods, T.C. Elevation of miR-221 and -222 in the internal mammary arteries of diabetic subjects and normalization with metformin. Mol. Cell Endocrinol. 2013, 374, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Villard, A.; Marchand, L.; Thivolet, C.; Rome, S. Diagnostic Value of Cell-free Circulating MicroRNAs for Obesity and Type 2 Diabetes: A Meta-analysis. J. Mol. Biomark. Diagn. 2015, 6, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERα expression in estrogen-induced insulin resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filardi, T.; Catanzaro, G.; Grieco, G.E.; Splendiani, E.; Trocchianesi, S.; Santangelo, C.; Brunelli, R.; Guarino, E.; Sebastiani, G.; Dotta, F.; et al. Identification and Validation of miR-222-3p and miR-409-3p as Plasma Biomarkers in Gestational Diabetes Mellitus Sharing Validated Target Genes Involved in Metabolic Homeostasis. Int. J. Mol. Sci. 2022, 23, 4276. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Igata, M.; Kondo, T.; Kitano, S.; Takaki, Y.; Hanatani, S.; Sakaguchi, M.; Goto, R.; Senokuchi, T.; Kawashima, J.; et al. Identification of microRNA that represses IRS-1 expression in liver. PLoS ONE 2018, 13, e0191553. [Google Scholar] [CrossRef]
- Fan, L.; Shan, A.; Su, Y.; Cheng, Y.; Ji, H.; Yang, Q.; Lei, Y.; Liu, B.; Wang, W.; Ning, G.; et al. MiR-221/222 Inhibit Insulin Production of Pancreatic β-Cells in Mice. Endocrinology 2020, 161, bqz027. [Google Scholar] [CrossRef]
- Vidaković, M.; Grdović, N.; Dinić, S.; Mihailović, M.; Uskoković, A.; Arambašić Jovanović, J. The Importance of the CXCL12/CXCR4 Axis in Therapeutic Approaches to Diabetes Mellitus Attenuation. Front. Immunol. 2015, 6, 403. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Kim, J.; Yoon, J.H.; Ghim, J.; Yea, K.; Song, P.; Park, S.; Lee, A.; Hong, C.P.; Jang, M.S.; et al. CXCL12 secreted from adipose tissue recruits macrophages and induces insulin resistance in mice. Diabetologia 2014, 57, 1456–1465. [Google Scholar] [CrossRef]
- Gongol, B.; Marin, T.; Zhang, J.; Wang, S.C.; Sun, W.; He, M.; Chen, S.; Chen, L.; Li, J.; Liu, J.H.; et al. Shear stress regulation of miR-93 and miR-484 maturation through nucleolin. Proc. Natl. Acad. Sci. USA 2019, 116, 12974–12979. [Google Scholar] [CrossRef] [Green Version]
- Marzano, F.; Faienza, M.F.; Caratozzolo, M.F.; Brunetti, G.; Chiara, M.; Horner, D.S.; Annese, A.; D’Erchia, A.M.; Consiglio, A.; Pesole, G.; et al. Pilot study on circulating miRNA signature in children with obesity born small for gestational age and appropriate for gestational age. Pediatr. Obes. 2018, 13, 803–811. [Google Scholar] [CrossRef]
- Wu, L.E.; Meoli, C.C.; Mangiafico, S.P.; Fazakerley, D.J.; Cogger, V.C.; Mohamad, M.; Pant, H.; Kang, M.J.; Powter, E.; Burchfield, J.G.; et al. Systemic VEGF-A neutralization ameliorates diet-induced metabolic dysfunction. Diabetes 2014, 63, 2656–2667. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, S.; Maeno, N.; Matsuo, K.; Nakajima, T.; Kitajima, I.; Saito, H.; Maruyama, I. Human lactiferous mammary gland cells produce vascular endothelial growth factor (VEGF) and express the VEGF receptors, Flt-1 AND KDR/Flk-1. Cytokine 2002, 18, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Pepper, M.S.; Baetens, D.; Mandriota, S.J.; Di Sanza, C.; Oikemus, S.; Lane, T.F.; Soriano, J.V.; Montesano, R.; Iruela-Arispe, M.L. Regulation of VEGF and VEGF receptor expression in the rodent mammary gland during pregnancy, lactation, and involution. Dev. Dyn. 2000, 218, 507–524. [Google Scholar] [CrossRef]
- Rossiter, H.; Barresi, C.; Ghannadan, M.; Gruber, F.; Mildner, M.; Födinger, D.; Tschachler, E. Inactivation of VEGF in mammary gland epithelium severely compromises mammary gland development and function. FASEB J. 2007, 21, 3994–4004. [Google Scholar] [CrossRef]
- Simionescu, N.; Niculescu, L.S.; Sanda, G.M.; Margina, D.; Sima, A.V. Analysis of circulating microRNAs that are specifically increased in hyperlipidemic and/or hyperglycemic sera. Mol. Biol. Rep. 2014, 41, 5765–5773. [Google Scholar] [CrossRef]
- Brovkina, O.; Nikitin, A.; Khodyrev, D.; Shestakova, E.; Sklyanik, I.; Panevina, A.; Stafeev, I.; Menshikov, M.; Kobelyatskaya, A.; Yurasov, A.; et al. Role of MicroRNAs in the Regulation of Subcutaneous White Adipose Tissue in Individuals With Obesity and Without Type 2 Diabetes. Front. Endocrinol. 2019, 10, 840. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Wu, M.; Zhu, J.J.; Zhang, C.H.; Yao, D.W.; Luo, J.; Loor, J.J. Fatty acid elongase 6 plays a role in the synthesis of long-chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2017, 100, 4987–4995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Cruz, M.; Sánchez, R.; Sánchez, A.M.; Kelleher, S.L.; Sánchez-Muñoz, F.; Maldonado, J.; López-Alarcón, M. Participation of mammary gland in long-chain polyunsaturated fatty acid synthesis during pregnancy and lactation in rats. Biochim. Biophys. Acta 2011, 1811, 284–293. [Google Scholar] [CrossRef]



| Phenotypic Traits | Control | Western Diet | Reversion | ANOVA | |
|---|---|---|---|---|---|
| Prior to gestation | Body weight (g) | 223 ± 9 | 253 ± 7 | 252 ± 11 | - |
| Body fat (g) | 29.1 ± 2.1 a | 51.5 ± 4.7 b | 52.9 ± 6.6 b | MD | |
| Body fat (%) | 12.4 ± 0.8 a | 20.0 ± 1.3 b | 20.4 ± 1.7 b | MD | |
| Day 17 of gestation | Body weight (g) | 309 ± 9 | 322 ± 8 | 324 ± 14 | - |
| Day 21 of lactation | Body weight (g) | 304 ± 9 a | 283 ± 5 b | 311 ± 7 a | MD |
| Body fat (g) | 29.5 ± 2.4 a | 32.0 ± 2.2 a | 41.3 ± 3.9 b | MD | |
| Body fat (%) | 9.7 ± 0.8 a | 11.3 ± 0.7 ab | 13.2 ± 1 b | MD | |
| GO ID | GO Term | Corrected p-Value | % Associated Genes | No. Genes |
|---|---|---|---|---|
| WP:3888 | VEGFA-VEGFR2 signaling pathway | 0.0002 | 16.9 | 74 |
| WP:2853 | Endoderm differentiation | <0.0001 | 26.0 | 38 |
| WP:2380 | Brain-derived neurotrophic factor (BDNF) signaling pathway | <0.0001 | 25.0 | 36 |
| WP:481 | Insulin signaling | 0.0003 | 22.4 | 36 |
| WP:2857 | Mesodermal commitment pathway | 0.0004 | 22.3 | 35 |
| WP:673 | ErbB signaling pathway | 0.0001 | 28.6 | 26 |
| WP:4685 | Melanoma | 0.0002 | 30.9 | 21 |
| WP:3657 | Hematopoietic stem cell gene regulation by the GABP alpha/beta Complex | 0.0004 | 50.0 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomar, C.A.; Castillo, P.; Palou, A.; Palou, M.; Picó, C. Dietary Improvement during Lactation Normalizes miR-26a, miR-222 and miR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats. Biomedicines 2022, 10, 1292. https://doi.org/10.3390/biomedicines10061292
Pomar CA, Castillo P, Palou A, Palou M, Picó C. Dietary Improvement during Lactation Normalizes miR-26a, miR-222 and miR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats. Biomedicines. 2022; 10(6):1292. https://doi.org/10.3390/biomedicines10061292
Chicago/Turabian StylePomar, Catalina A., Pedro Castillo, Andreu Palou, Mariona Palou, and Catalina Picó. 2022. "Dietary Improvement during Lactation Normalizes miR-26a, miR-222 and miR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats" Biomedicines 10, no. 6: 1292. https://doi.org/10.3390/biomedicines10061292
APA StylePomar, C. A., Castillo, P., Palou, A., Palou, M., & Picó, C. (2022). Dietary Improvement during Lactation Normalizes miR-26a, miR-222 and miR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats. Biomedicines, 10(6), 1292. https://doi.org/10.3390/biomedicines10061292










