Uncertainty Quantification in Segmenting Tuberculosis-Consistent Findings in Frontal Chest X-rays
Abstract
:1. Introduction
- We conduct extensive empirical evaluations to select an appropriate loss function to improve model performance.
- The proposed method quantifies uncertainty in predictions using the MCD method and identifies the optimal number of MC samples required to stabilize model performance.
- We evaluate, quantify, and compare various uncertainties in model representations and arrive at an optimal uncertainty threshold using these uncertainty metrics. The predictions exceeding this threshold could be referred to an expert to ensure reliable outcomes.
- To the best of our knowledge, this is the first study that uses fine-grained annotations of TB-consistent regions for model training and evaluation.
2. Materials and Methods
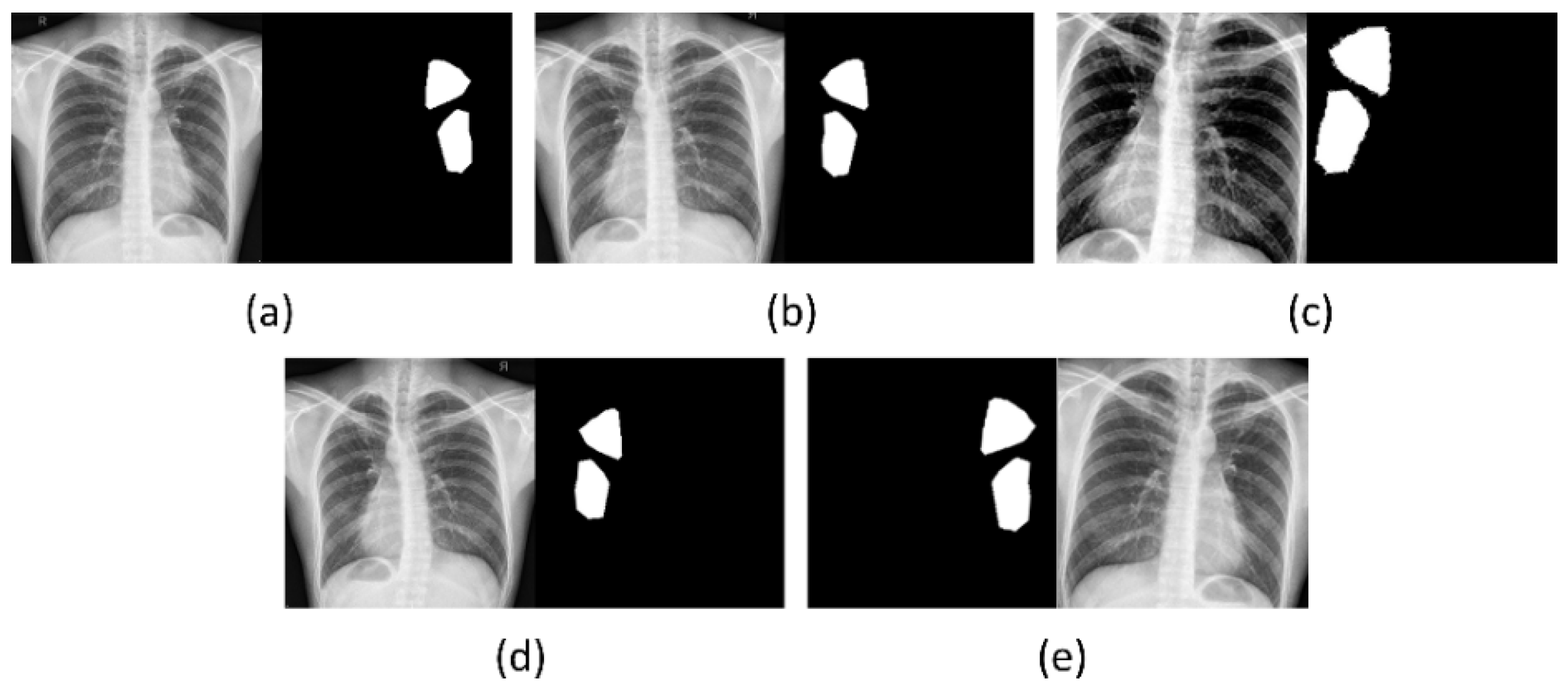
2.1. Data Collection and Preprocessing
2.2. Statistical Analysis
2.3. TB-Consistent Region Segmentation
2.3.1. Model Architecture
2.3.2. Loss Functions
2.3.3. Uncertainty Quantification
3. Results
3.1. Segmentation Performance Achieved with the Proposed Loss Functions
3.2. Uncertainty Quantification
3.3. Identifying the Optimal Uncertainty Threshold
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. World Health Organization Global Tuberculosis Report; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Sivaramakrishnan, R.; Antani, S.; Candemir, S.; Xue, Z.; Abuya, J.; Sivaramakrishnan, R.; Antani, S.; Candemir, S.; Xue, Z.; Abuya, J.; et al. Comparing Deep Learning Models for Population Screening Using Chest Radiography. In Proceedings of the SPIE Medical Imaging, Houston, TX, USA, 10–15 February 2018; p. 105751E. [Google Scholar]
- Jaeger, S.; Candemir, S.; Antani, S.; Wang, Y.-X.J.; Lu, P.-X.; Thoma, G. Two Public Chest X-Ray Datasets for Computer-Aided Screening of Pulmonary Diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [CrossRef]
- Rajaraman, S.; Antani, S.K. Modality-Specific Deep Learning Model Ensembles Toward Improving TB Detection in Chest Radiographs. IEEE Access 2020, 8, 27318–27326. [Google Scholar] [CrossRef]
- Balabanova, Y.; Coker, R.; Fedorin, I.; Zakharova, S.; Plavinskij, S.; Krukov, N.; Atun, R.; Drobniewski, F. Variability in Interpretation of Chest Radiographs among Russian Clinicians and Implications for Screening Programmes: Observational Study. BMJ 2005, 331, 379–382. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, A.; Goyal, A.; Guleria, R.; Gupta, A. Chest Tuberculosis: Radiological Review and Imaging Recommendations. Indian J. Radiol. Imaging 2015, 25, 213. [Google Scholar] [CrossRef]
- Pasa, F.; Golkov, V.; Pfeiffer, F.; Cremers, D.; Pfeiffer, D. Efficient Deep Network Architectures for Fast Chest X-ray Tuberculosis Screening and Visualization. Sci. Rep. 2019, 9, 6268. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.H.; Acharya, U.R.; Tan, C.; Abraham, K.T.; Lim, C.M. Computer-Assisted Diagnosis of Tuberculosis: A First Order Statistical Approach to Chest Radiograph. J. Med. Syst. 2011, 36, 2751–2759. [Google Scholar] [CrossRef]
- Stirenko, S.; Kochura, Y.; Alienin, O.; Rokovyi, O.; Gordienko, Y.; Gang, P.; Zeng, W. Chest X-Ray Analysis of Tuberculosis by Deep Learning with Segmentation and Augmentation. In Proceedings of the 2018 IEEE 38th International Conference on Electronics and Nanotechnology, Kyiv, Ukraine, 24–26 April 2018. [Google Scholar]
- Rajaraman, S.; Folio, L.R.; Dimperio, J.; Alderson, P.O.; Antani, S.K. Improved Semantic Segmentation of Tuberculosis—Consistent Findings in Chest X-Rays Using Augmented Training of Modality-Specific u-Net Models with Weak Localizations. Diagnostics 2021, 11, 616. [Google Scholar] [CrossRef]
- Jadon, S. A Survey of Loss Functions for Semantic Segmentation. In Proceedings of the IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Viña del Mar, Viña del Mar, Chile, 27–29 October 2020. [Google Scholar] [CrossRef]
- Couso, I.; Sánchez, L. Machine Learning Models, Epistemic Set-Valued Data and Generalized Loss Functions: An Encompassing Approach. Inf. Sci. 2016, 358–359, 129–150. Inf. Sci. 2016, 358–359, 129–150. [Google Scholar] [CrossRef]
- Abraham, N.; Khan, N.M. A Novel Focal Tversky Loss Function with Improved Attention U-Net for Lesion Segmentation. In Proceedings of the International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019. [Google Scholar]
- Liu, Y.; Wu, Y.H.; Ban, Y.; Wang, H.; Cheng, M.M. Rethinking Computer-Aided Tuberculosis Diagnosis. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Loquercio, A.; Segu, M.; Scaramuzza, D. A General Framework for Uncertainty Estimation in Deep Learning. IEEE Robot. Autom. Lett. 2020, 5, 3153–3160. [Google Scholar] [CrossRef] [Green Version]
- Asgharnezhad, H.; Shamsi, A.; Alizadehsani, R.; Khosravi, A.; Nahavandi, S.; Sani, Z.A.; Srinivasan, D.; Islam, S.M.S. Objective Evaluation of Deep Uncertainty Predictions for COVID-19 Detection. Sci. Rep. 2022, 12, 815. [Google Scholar] [CrossRef]
- Yeung, M.; Rundo, L.; Nan, Y.; Sala, E.; Schönlieb, C.-B.; Yang, G. Calibrating the Dice Loss to Handle Neural Network Overconfidence for Biomedical Image Segmentation. arXiv 2021, arXiv:2111.00528. [Google Scholar]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gros, C.; Lemay, A.; Cohen-Adad, J. SoftSeg: Advantages of Soft versus Binary Training for Image Segmentation. Med. Image Anal. 2021, 71, 102038. [Google Scholar] [CrossRef]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamzadeh, M.; Fieguth, P.; Cao, X.; Khosravi, A.; Acharya, U.R.; et al. A Review of Uncertainty Quantification in Deep Learning: Techniques, Applications and Challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Kwon, Y.; Won, J.-H.; Kim, B.J.; Paik, M.C. Uncertainty Quantification Using Bayesian Neural Networks in Classification: Application to Ischemic Stroke Lesion Segmentation. Comput. Stat. Data Anal. 2020, 142, 106816. [Google Scholar] [CrossRef]
- Dechesne, C.; Lassalle, P.; Lefèvre, S. Bayesian U-Net: Estimating Uncertainty in Semantic Segmentation of Earth Observation Images. Remote Sens. 2021, 13, 3836. [Google Scholar] [CrossRef]
- Gal, Y.; Hron, J.; Kendall, A. Concrete Dropout. In Proceedings of the 31st International Conference on Neural Information Processing SystemsDecember (NIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 3584–3593. [Google Scholar]
- Zhang, G.; Dang, H.; Xu, Y. Epistemic and Aleatoric Uncertainties Reduction with Rotation Variation for Medical Image Segmentation with ConvNets. SN Appl. Sci. 2022, 4, 56. [Google Scholar] [CrossRef]
- Petschnigg, C.; Spitzner, M.; Weitzendorf, L. From a Point Cloud to a Simulation Model—Bayesian 3D Modelling. Entropy 2021, 23, 301. [Google Scholar] [CrossRef]
- Bloice, M.D.; Roth, P.M.; Holzinger, A. Biomedical Image Augmentation Using Augmentor. Bioinformatics 2019, 35, 4522–4524. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Statistics Notes: How to Obtain the P Value from a Confidence Interval. BMJ 2011, 343, d2304. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Yeung, M.; Yang, G.; Sala, E.; Schönlieb, C.-B.; Rundo, L. Incorporating Boundary Uncertainty into Loss Functions for Biomedical Image Segmentation. arXiv 2021, arXiv:arXiv:2111.00533. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar] [CrossRef] [Green Version]
- Seedat, N. MCU-Net: A Framework towards Uncertainty Representations for Decision Support System Patient Referrals in Healthcare Contexts. arXiv 2020, arXiv:arXiv:2007.03995. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zamzmi, G.; Rajaraman, S.; Sachdev, V.; Antani, S. Trilateral Attention Network for Real-Time Cardiac Region Segmentation. IEEE Access 2021, 9, 118205–118214. [Google Scholar] [CrossRef] [PubMed]
- Sagar, A. Uncertainty Quantification Using Variational Inference for Biomedical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 44–51. [Google Scholar] [CrossRef]
- Tang, P.; Yang, P.; Nie, D.; Wu, X.; Zhou, J.; Wang, Y. Unified Medical Image Segmentation by Learning from Uncertainty in an End-to-End Manner. Knowl. Based Syst. 2022, 241, 108215. [Google Scholar] [CrossRef]









| Dataset | Train | Validation | Test |
|---|---|---|---|
| Shenzhen TB (n = 330) | 2231 | 66 | 33 |
| Loss | AUC | mAP | IOU | Dice |
|---|---|---|---|---|
| CE | 0.7763 | 0.5150 (0.3444, 0.6856) | 0.3334 | 0.5000 |
| CE + BU | 0.7704 | 0.5289 (0.3585, 0.6993) | 0.3511 | 0.5197 |
| Dice | 0.7961 | 0.5497 (0.3799, 0.7195) | 0.3653 | 0.5351 |
| Dice + BU | 0.7359 | 0.5162 (0.3456, 0.6868) | 0.3400 | 0.5074 |
| IOU | 0.7864 | 0.5248 (0.3544, 0.6952) | 0.3398 | 0.5073 |
| IOU + BU | 0.7477 | 0.5337 (0.3634, 0.7040) | 0.3563 | 0.5254 |
| Tversky | 0.8075 | 0.5366 (0.3664, 0.7068) | 0.3405 | 0.5080 |
| Tversky + BU | 0.8125 | 0.5368 (0.3666, 0.7070) | 0.3364 | 0.5034 |
| Focal Tversky | 0.8284 | 0.5400 (0.3699, 0.7101) | 0.3242 | 0.4896 |
| Focal Tversky + BU | 0.8281 | 0.5710 (0.4021, 0.7399) | 0.3723 | 0.5426 |
| Model | AUC | mAP | IOU | Dice |
|---|---|---|---|---|
| Focal Tversky + BU (Baseline) | 0.8281 | 0.5710 (0.4021,0.7399) | 0.3723 | 0.5426 |
| Monte–Carlo (30) Focal Tversky + BU | 0.8279 | 0.5721 (0.4032, 0.7410) | 0.3726 | 0.5430 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajaraman, S.; Zamzmi, G.; Yang, F.; Xue, Z.; Jaeger, S.; Antani, S.K. Uncertainty Quantification in Segmenting Tuberculosis-Consistent Findings in Frontal Chest X-rays. Biomedicines 2022, 10, 1323. https://doi.org/10.3390/biomedicines10061323
Rajaraman S, Zamzmi G, Yang F, Xue Z, Jaeger S, Antani SK. Uncertainty Quantification in Segmenting Tuberculosis-Consistent Findings in Frontal Chest X-rays. Biomedicines. 2022; 10(6):1323. https://doi.org/10.3390/biomedicines10061323
Chicago/Turabian StyleRajaraman, Sivaramakrishnan, Ghada Zamzmi, Feng Yang, Zhiyun Xue, Stefan Jaeger, and Sameer K. Antani. 2022. "Uncertainty Quantification in Segmenting Tuberculosis-Consistent Findings in Frontal Chest X-rays" Biomedicines 10, no. 6: 1323. https://doi.org/10.3390/biomedicines10061323
APA StyleRajaraman, S., Zamzmi, G., Yang, F., Xue, Z., Jaeger, S., & Antani, S. K. (2022). Uncertainty Quantification in Segmenting Tuberculosis-Consistent Findings in Frontal Chest X-rays. Biomedicines, 10(6), 1323. https://doi.org/10.3390/biomedicines10061323









