Macrophages Characterization in an Injured Bone Tissue
Abstract
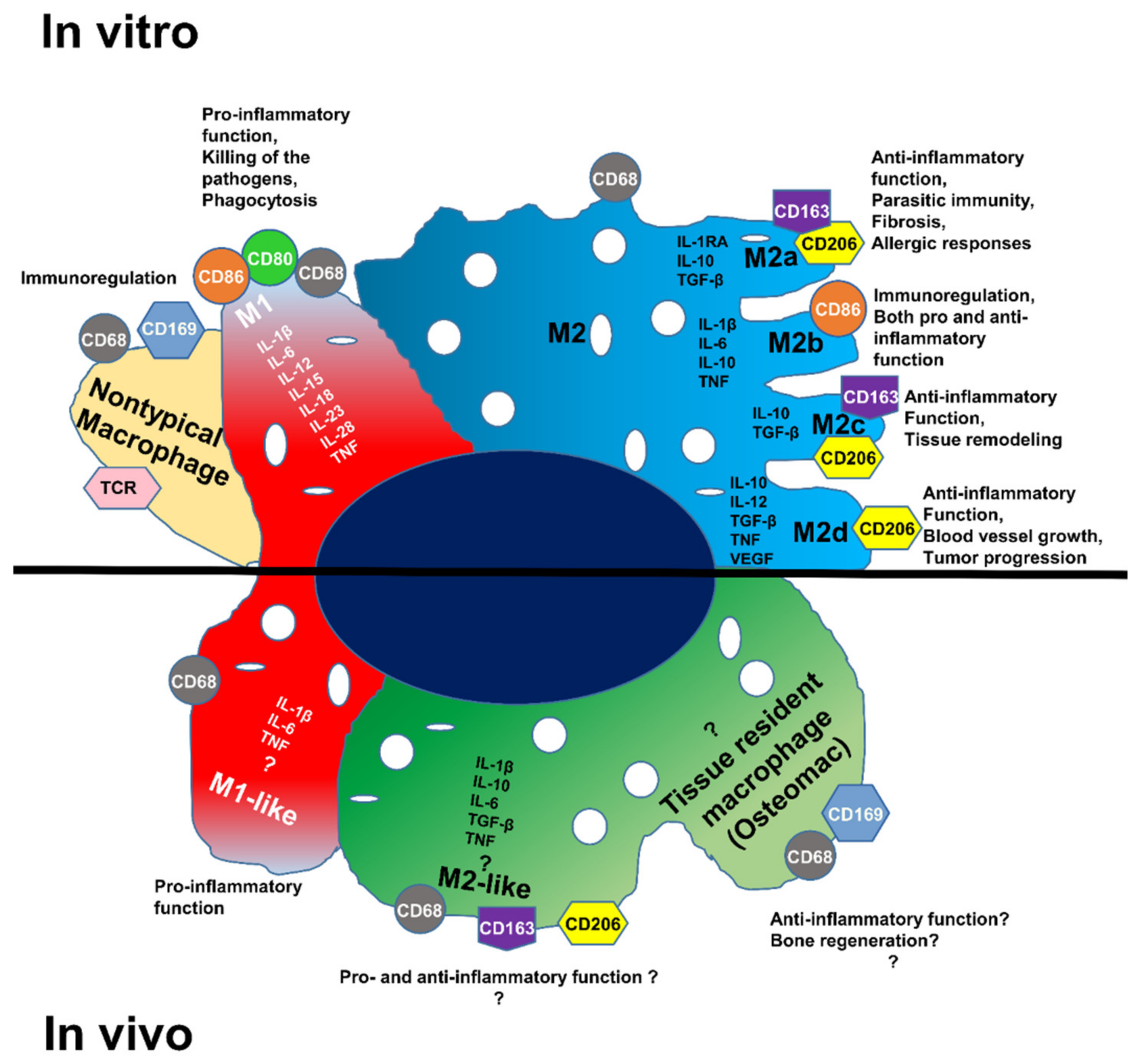
:1. Introduction
2. Materials and Methods
2.1. Rat Animal Model
2.2. Embedding and Cryosectioning of the Entire Femur (Bone and Muscle Together) of the Rat
2.3. Histological Staining
2.4. Immunofluorescence
2.5. RT-qPCR
2.6. In Situ DIG Hybridization
2.7. In Situ HCR Hybridization
2.8. Microspectroscopical Analysis
2.9. Quantification of HIS by ImageJ
3. Results
3.1. Detection of Immune Cell Accumulation around Biomaterial
3.2. Detection of Macrophages in Other Regions
3.3. Detection of Macrophages Based on Their Autofluorescence Feature
3.4. Investigation of Macrophage Fluorescence by Microspectroscopic Analysis
3.5. Detection of IL-1β and IL-6 Cytokines in the Operated Femur
3.6. Identification of Macrophage Phenotype Using Both I DIG and In Situ HCR Techniques
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pajarinen, J.; Lin, T.; Gibon, E.; Kohno, Y.; Maruyama, M.; Nathan, K.; Lu, L.; Yao, Z.; Goodman, S.B. Mesenchymal Stem Cell-Macrophage Crosstalk and Bone Healing. Biomaterials 2019, 196, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, J.; Xie, X.; Gu, F.; Sui, Z.; Zhang, K.; Yu, T. Macrophage-Osteoclast Associations: Origin, Polarization, and Subgroups. Front. Immunol. 2021, 12, 778078. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.K. Osteoclasts and Microgravity. Life 2020, 10, 207. [Google Scholar] [CrossRef] [PubMed]
- Gibon, E.; Lu, L.; Goodman, S.B. Aging, Inflammation, Stem Cells, and Bone Healing. Stem. Cell Res. Ther. 2016, 7, 44. [Google Scholar] [CrossRef]
- Weitzmann, M.N. Bone and the Immune System. Toxicol. Pathol. 2017, 45, 911–924. [Google Scholar] [CrossRef]
- Horwood, N.J. Macrophage Polarization and Bone Formation: A Review. Clin. Rev. Allergy Immunol. 2016, 51, 79–86. [Google Scholar] [CrossRef]
- Kaur, S.; Raggatt, L.J.; Batoon, L.; Hume, D.A.; Levesque, J.-P.; Pettit, A.R. Role of Bone Marrow Macrophages in Controlling Homeostasis and Repair in Bone and Bone Marrow Niches. Semin. Cell Dev. Biol. 2017, 61, 12–21. [Google Scholar] [CrossRef]
- Vi, L.; Baht, G.S.; Whetstone, H.; Ng, A.; Wei, Q.; Poon, R.; Mylvaganam, S.; Grynpas, M.; Alman, B.A. Macrophages Promote Osteoblastic Differentiation In-Vivo: Implications in Fracture Repair and Bone Homeostasis. J. Bone Miner. Res. 2015, 30, 1090–1102. [Google Scholar] [CrossRef]
- Heideveld, E.; Horcas-Lopez, M.; Lopez-Yrigoyen, M.; Forrester, L.M.; Cassetta, L.; Pollard, J.W. Methods for Macrophage Differentiation and in Vitro Generation of Human Tumor Associated-like Macrophages. Methods Enzym. 2020, 632, 113–131. [Google Scholar] [CrossRef]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage Activation and Polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Graff, J.W.; Dickson, A.M.; Clay, G.; McCaffrey, A.P.; Wilson, M.E. Identifying Functional MicroRNAs in Macrophages with Polarized Phenotypes. J. Biol. Chem. 2012, 287, 21816–21825. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Abdelaziz, M.H.; Abdelwahab, S.F.; Wan, J.; Cai, W.; Huixuan, W.; Jianjun, C.; Kumar, K.D.; Vasudevan, A.; Sadek, A.; Su, Z.; et al. Alternatively Activated Macrophages; a Double-Edged Sword in Allergic Asthma. J. Transl. Med. 2020, 18, 58. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef]
- Arora, S.; Dev, K.; Agarwal, B.; Das, P.; Syed, M.A. Macrophages: Their Role, Activation and Polarization in Pulmonary Diseases. Immunobiology 2018, 223, 383–396. [Google Scholar] [CrossRef]
- Huang, X.; Xiu, H.; Zhang, S.; Zhang, G. The Role of Macrophages in the Pathogenesis of ALI/ARDS. Mediat. Inflamm. 2018, 2018, 1264913. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Van den Bossche, J.; O’Neill, L.A.; Menon, D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 2017, 38, 395–406. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage Biology in Development, Homeostasis and Disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Wang, L.-X.; Zhang, S.-X.; Wu, H.-J.; Rong, X.-L.; Guo, J. M2b Macrophage Polarization and Its Roles in Diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef]
- Minutti, C.M.; Knipper, J.A.; Allen, J.E.; Zaiss, D.M.W. Tissue-Specific Contribution of Macrophages to Wound Healing. Semin. Cell Dev. Biol. 2017, 61, 3–11. [Google Scholar] [CrossRef]
- Viniegra, A.; Goldberg, H.; Çil, Ç.; Fine, N.; Sheikh, Z.; Galli, M.; Freire, M.; Wang, Y.; Van Dyke, T.E.; Glogauer, M.; et al. Resolving Macrophages Counter Osteolysis by Anabolic Actions on Bone Cells. J. Dent. Res. 2018, 97, 1160–1169. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage Plasticity, Polarization, and Function in Health and Disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Selders, G.S.; Fetz, A.E.; Radic, M.Z.; Bowlin, G.L. An Overview of the Role of Neutrophils in Innate Immunity, Inflammation and Host-Biomaterial Integration. Regen. Biomater. 2017, 4, 55–68. [Google Scholar] [CrossRef]
- Mantovani, A. Wandering Pathways in the Regulation of Innate Immunity and Inflammation. J. Autoimmun. 2017, 85, 1–5. [Google Scholar] [CrossRef]
- Snyder, R.J.; Lantis, J.; Kirsner, R.S.; Shah, V.; Molyneaux, M.; Carter, M.J. Macrophages: A Review of Their Role in Wound Healing and Their Therapeutic Use. Wound Repair Regen. 2016, 24, 613–629. [Google Scholar] [CrossRef]
- Batoon, L.; Millard, S.M.; Wullschleger, M.E.; Preda, C.; Wu, A.C.-K.; Kaur, S.; Tseng, H.-W.; Hume, D.A.; Levesque, J.-P.; Raggatt, L.J.; et al. CD169+ Macrophages Are Critical for Osteoblast Maintenance and Promote Intramembranous and Endochondral Ossification during Bone Repair. Biomaterials 2019, 196, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Miron, R.J.; Bosshardt, D.D. OsteoMacs: Key Players around Bone Biomaterials. Biomaterials 2016, 82, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Tang, Q.; Zhong, Y.; Wei, Y.; He, L.; Wu, Y.; Wu, J.; Liao, J. Biomaterial-Based Strategies for Maxillofacial Tumour Therapy and Bone Defect Regeneration. Int. J. Oral. Sci. 2021, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Lebaudy, E.; Fournel, S.; Lavalle, P.; Vrana, N.E.; Gribova, V. Recent Advances in Antiinflammatory Material Design. Adv. Healthc. Mater. 2021, 10, e2001373. [Google Scholar] [CrossRef]
- Witherel, C.E.; Abebayehu, D.; Barker, T.H.; Spiller, K.L. Macrophage and Fibroblast Interactions in Biomaterial-Mediated Fibrosis. Adv. Healthc. Mater. 2019, 8, e1801451. [Google Scholar] [CrossRef]
- Sadtler, K.; Sommerfeld, S.D.; Wolf, M.T.; Wang, X.; Majumdar, S.; Chung, L.; Kelkar, D.S.; Pandey, A.; Elisseeff, J.H. Proteomic Composition and Immunomodulatory Properties of Urinary Bladder Matrix Scaffolds in Homeostasis and Injury. Semin. Immunol. 2017, 29, 14–23. [Google Scholar] [CrossRef]
- Klopfleisch, R. Macrophage Reaction against Biomaterials in the Mouse Model-Phenotypes, Functions and Markers. Acta Biomater. 2016, 43, 3–13. [Google Scholar] [CrossRef]
- Boersema, G.S.A.; Grotenhuis, N.; Bayon, Y.; Lange, J.F.; Bastiaansen-Jenniskens, Y.M. The Effect of Biomaterials Used for Tissue Regeneration Purposes on Polarization of Macrophages. Biores. Open Access 2016, 5, 6–14. [Google Scholar] [CrossRef]
- Ogle, M.E.; Segar, C.E.; Sridhar, S.; Botchwey, E.A. Monocytes and Macrophages in Tissue Repair: Implications for Immunoregenerative Biomaterial Design. Exp. Biol. Med. 2016, 241, 1084–1097. [Google Scholar] [CrossRef]
- Huffman, L.K.; Harris, J.G.; Suk, M. Using the Bi-Masquelet Technique and Reamer-Irrigator-Aspirator for Post-Traumatic Foot Reconstruction. Foot Ankle Int. 2009, 30, 895–899. [Google Scholar] [CrossRef]
- Masquelet, A.C. Free Vascularized Corticoperiosteal Grafts. Plast. Reconstr. Surg. 1991, 88, 1106. [Google Scholar] [CrossRef]
- Masquelet, A.C. Induced Membrane Technique: Pearls and Pitfalls. J. Orthop. Trauma 2017, 31 (Suppl. 5), S36–S38. [Google Scholar] [CrossRef]
- Masquelet, A.C. The Induced Membrane Technique. Orthop. Traumatol. Surg. Res. 2020, 106, 785–787. [Google Scholar] [CrossRef]
- Muñoz, J.; Akhavan, N.S.; Mullins, A.P.; Arjmandi, B.H. Macrophage Polarization and Osteoporosis: A Review. Nutrients 2020, 12, 2999. [Google Scholar] [CrossRef]
- Eggold, J.T.; Rankin, E.B. Erythropoiesis, EPO, Macrophages, and Bone. Bone 2019, 119, 36–41. [Google Scholar] [CrossRef]
- Michalski, M.N.; McCauley, L.K. Macrophages and Skeletal Health. Pharmacol. Ther. 2017, 174, 43–54. [Google Scholar] [CrossRef]
- Nikovics, K.; Favier, A.-L. Macrophage Identification in Situ. Biomedicines 2021, 9, 1393. [Google Scholar] [CrossRef]
- Nikovics, K.; Morin, H.; Riccobono, D.; Bendahmane, A.; Favier, A. Hybridization-chain-reaction Is a Relevant Method for in Situ Detection of M2d-like Macrophages in a Mini-pig Model. FASEB J. 2020, 34, 15675–15686. [Google Scholar] [CrossRef]
- Nikovics, K.; Castellarin, C.; Holy, X.; Durand, M.; Morin, H.; Bendahmane, A.; Favier, A. In Situ Gene Expression in Native Cryofixed Bone Tissue. Biomedicines 2022, 10, 484–498. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Beck, V.A.; Pierce, N.A. Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano 2014, 8, 4284–4294. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Calvert, C.R.; Husain, N.; Huss, D.; Barsi, J.C.; Deverman, B.E.; Hunter, R.C.; Kato, M.; Lee, S.M.; Abelin, A.C.T.; et al. Mapping a Multiplexed Zoo of MRNA Expression. Development 2016, 143, 3632–3637. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Schwarzkopf, M.; Fornace, M.E.; Acharya, A.; Artavanis, G.; Stegmaier, J.; Cunha, A.; Pierce, N.A. Third-Generation in Situ Hybridization Chain Reaction: Multiplexed, Quantitative, Sensitive, Versatile, Robust. Development 2018, 145, dev165753. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Schwarzkopf, M.; Pierce, N.A. Multiplexed Quantitative in Situ Hybridization with Subcellular or Single-Molecule Resolution Within Whole-Mount Vertebrate Embryos: QHCR and DHCR Imaging (v3.0). Methods Mol. Biol. 2020, 2148, 159–178. [Google Scholar] [CrossRef]
- Song, S.H.; Kim, J.E.; Lee, Y.J.; Kwak, M.H.; Sung, G.Y.; Kwon, S.H.; Son, H.J.; Lee, H.S.; Jung, Y.J.; Hwang, D.Y. Cellulose Film Regenerated from Styela Clava Tunics Have Biodegradability, Toxicity and Biocompatibility in the Skin of SD Rats. J. Mater. Sci. Mater. Med. 2014, 25, 1519–1530. [Google Scholar] [CrossRef]
- Betjes, M.G.; Haks, M.C.; Tuk, C.W.; Beelen, R.H. Monoclonal Antibody EBM11 (Anti-CD68) Discriminates between Dendritic Cells and Macrophages after Short-Term Culture. Immunobiology 1991, 183, 79–87. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/Macrosialin: Not Just a Histochemical Marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef]
- Kosmac, K.; Peck, B.D.; Walton, R.G.; Mula, J.; Kern, P.A.; Bamman, M.M.; Dennis, R.A.; Jacobs, C.A.; Lattermann, C.; Johnson, D.L.; et al. Immunohistochemical Identification of Human Skeletal Muscle Macrophages. Bio-Protoc. 2018, 8, e2883. [Google Scholar] [CrossRef]
- Tidball, J.G.; Villalta, S.A. Regulatory Interactions between Muscle and the Immune System during Muscle Regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef]
- Fujimori, M.; Kimura, Y.; Ueshima, E.; Dupuy, D.E.; Adusumilli, P.S.; Solomon, S.B.; Srimathveeravalli, G. Lung Ablation with Irreversible Electroporation Promotes Immune Cell Infiltration by Sparing Extracellular Matrix Proteins and Vasculature: Implications for Immunotherapy. Bioelectricity 2021, 3, 204–214. [Google Scholar] [CrossRef]
- Zhang, X.; Thompkins-Johns, A.; Ziober, A.; Zhang, P.J.; Furth, E.E. Hepatic Macrophage Types Cluster with Disease Etiology in Chronic Liver Disease and Differ Compared to Normal Liver: Implications for Their Biologic and Diagnostic Role. Int. J. Surg. Pathol. 2022. [Google Scholar] [CrossRef]
- Oh, T.; Do, D.T.; Vo, H.V.; Kwon, H.-I.; Lee, S.-C.; Kim, M.H.; Nguyen, D.T.T.; Le, Q.T.V.; Tran, T.M.; Nguyen, T.T.; et al. The Isolation and Replication of African Swine Fever Virus in Primary Renal-Derived Swine Macrophages. Front. Vet. Sci. 2021, 8, 645456. [Google Scholar] [CrossRef] [PubMed]
- Fauch, L.; Palander, A.; Dekker, H.; Schulten, E.A.; Koistinen, A.; Kullaa, A.; Keinänen, M. Narrowband-Autofluorescence Imaging for Bone Analysis. Biomed. Opt. Express 2019, 10, 2367–2382. [Google Scholar] [CrossRef] [PubMed]
- Capasso, L.; D’Anastasio, R.; Guarnieri, S.; Viciano, J.; Mariggiò, M. Bone Natural Autofluorescence and Confocal Laser Scanning Microscopy: Preliminary Results of a Novel Useful Tool to Distinguish between Forensic and Ancient Human Skeletal Remains. Forensic. Sci. Int. 2017, 272, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Monici, M. Cell and Tissue Autofluorescence Research and Diagnostic Applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [CrossRef] [PubMed]
- Vedeswari In Vivo Autofluorescence Characteristics of Pre- and Post-Treated Oral Submucous Fibrosis: A Pilot Study-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/19884705/ (accessed on 17 February 2021).
- Weiss, G.; Schaible, U.E. Macrophage Defense Mechanisms against Intracellular Bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Mukherjee, R. Cytokine-Mediated Communication: A Quantitative Appraisal of Immune Complexity. Nat. Rev. Immunol. 2019, 19, 205–217. [Google Scholar] [CrossRef]
- Chu, Y.-H.; Hardin, H.; Zhang, R.; Guo, Z.; Lloyd, R.V. In Situ Hybridization: Introduction to Techniques, Applications and Pitfalls in the Performance and Interpretation of Assays. Semin. Diagn. Pathol. 2019, 36, 336–341. [Google Scholar] [CrossRef]
- Herrmann, M.; Engelke, K.; Ebert, R.; Müller-Deubert, S.; Rudert, M.; Ziouti, F.; Jundt, F.; Felsenberg, D.; Jakob, F. Interactions between Muscle and Bone-Where Physics Meets Biology. Biomolecules 2020, 10, 432. [Google Scholar] [CrossRef]
- Sandberg, O.H.; Tätting, L.; Bernhardsson, M.E.; Aspenberg, P. Temporal Role of Macrophages in Cancellous Bone Healing. Bone 2017, 101, 129–133. [Google Scholar] [CrossRef]
- Jablonská, E.; Horkavcová, D.; Rohanová, D.; Brauer, D.S. A Review of in Vitro Cell Culture Testing Methods for Bioactive Glasses and Other Biomaterials for Hard Tissue Regeneration. J. Mater. Chem. B 2020, 8, 10941–10953. [Google Scholar] [CrossRef]
- Bonnardel, J.; Guilliams, M. Developmental Control of Macrophage Function. Curr. Opin. Immunol. 2018, 50, 64–74. [Google Scholar] [CrossRef]
- Eme-Scolan, E.; Dando, S.J. Tools and Approaches for Studying Microglia In Vivo. Front. Immunol. 2020, 11, 583647. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage Plasticity and Polarization: In Vivo Veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Loi, F.; Córdova, L.A.; Pajarinen, J.; Lin, T.; Yao, Z.; Goodman, S.B. Inflammation, Fracture and Bone Repair. Bone 2016, 86, 119–130. [Google Scholar] [CrossRef]
- Nich, C.; Takakubo, Y.; Pajarinen, J.; Ainola, M.; Salem, A.; Sillat, T.; Rao, A.J.; Raska, M.; Tamaki, Y.; Takagi, M.; et al. Macrophages-Key Cells in the Response to Wear Debris from Joint Replacements. J. Biomed. Mater. Res. A 2013, 101, 3033–3045. [Google Scholar] [CrossRef]
- Claes, L.; Recknagel, S.; Ignatius, A. Fracture Healing under Healthy and Inflammatory Conditions. Nat. Rev. Rheumatol. 2012, 8, 133–143. [Google Scholar] [CrossRef]
- Bauman, J.G.; Wiegant, J.; Borst, P.; van Duijn, P. A New Method for Fluorescence Microscopical Localization of Specific DNA Sequences by in Situ Hybridization of Fluorochromelabelled RNA. Exp. Cell Res. 1980, 128, 485–490. [Google Scholar] [CrossRef]
- Bayani, J.; Squire, J.A. Fluorescence in Situ Hybridization (FISH). Curr. Protoc. Cell Biol. 2004, 23, 22–24. [Google Scholar] [CrossRef]
- Bayani, J.; Squire, J.A. Comparative Genomic Hybridization. Curr. Protoc. Cell Biol. 2005, 25, 22–26. [Google Scholar] [CrossRef]
- Pothos, A.; Plastira, K.; Plastiras, A.; Vlachodimitropoulos, D.; Goutas, N.; Angelopoulou, R. Comparison of Chromogenic in Situ Hybridisation with Fluorescence in Situ Hybridisation and Immunohistochemistry for the Assessment of Her-2/Neu Oncogene in Archival Material of Breast Carcinoma. Acta Histochem. Cytochem. 2008, 41, 59–64. [Google Scholar] [CrossRef]









| CD68 | CD206 | CD163 | Iba1 | |
|---|---|---|---|---|
| M1-like macrophages | + | − | − | + |
| M2-like macrophages | + | + | + | + |
| Satellite cells | − | + | − | − |
| M1-like Macrophages | M2-like Macrophages | |
|---|---|---|
| CD68/CD206 | 6.2% | 16.6% |
| Iba1/CD206 | 4.1% | 17.5% |
| CD163 | / | 21.6% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikovics, K.; Durand, M.; Castellarin, C.; Burger, J.; Sicherre, E.; Collombet, J.-M.; Oger, M.; Holy, X.; Favier, A.-L. Macrophages Characterization in an Injured Bone Tissue. Biomedicines 2022, 10, 1385. https://doi.org/10.3390/biomedicines10061385
Nikovics K, Durand M, Castellarin C, Burger J, Sicherre E, Collombet J-M, Oger M, Holy X, Favier A-L. Macrophages Characterization in an Injured Bone Tissue. Biomedicines. 2022; 10(6):1385. https://doi.org/10.3390/biomedicines10061385
Chicago/Turabian StyleNikovics, Krisztina, Marjorie Durand, Cédric Castellarin, Julien Burger, Emma Sicherre, Jean-Marc Collombet, Myriam Oger, Xavier Holy, and Anne-Laure Favier. 2022. "Macrophages Characterization in an Injured Bone Tissue" Biomedicines 10, no. 6: 1385. https://doi.org/10.3390/biomedicines10061385
APA StyleNikovics, K., Durand, M., Castellarin, C., Burger, J., Sicherre, E., Collombet, J. -M., Oger, M., Holy, X., & Favier, A. -L. (2022). Macrophages Characterization in an Injured Bone Tissue. Biomedicines, 10(6), 1385. https://doi.org/10.3390/biomedicines10061385








