Modulating Microglia/Macrophage Activation by CDNF Promotes Transplantation of Fetal Ventral Mesencephalic Graft Survival and Function in a Hemiparkinsonian Rat Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Hemiparkinsonian Rat Model
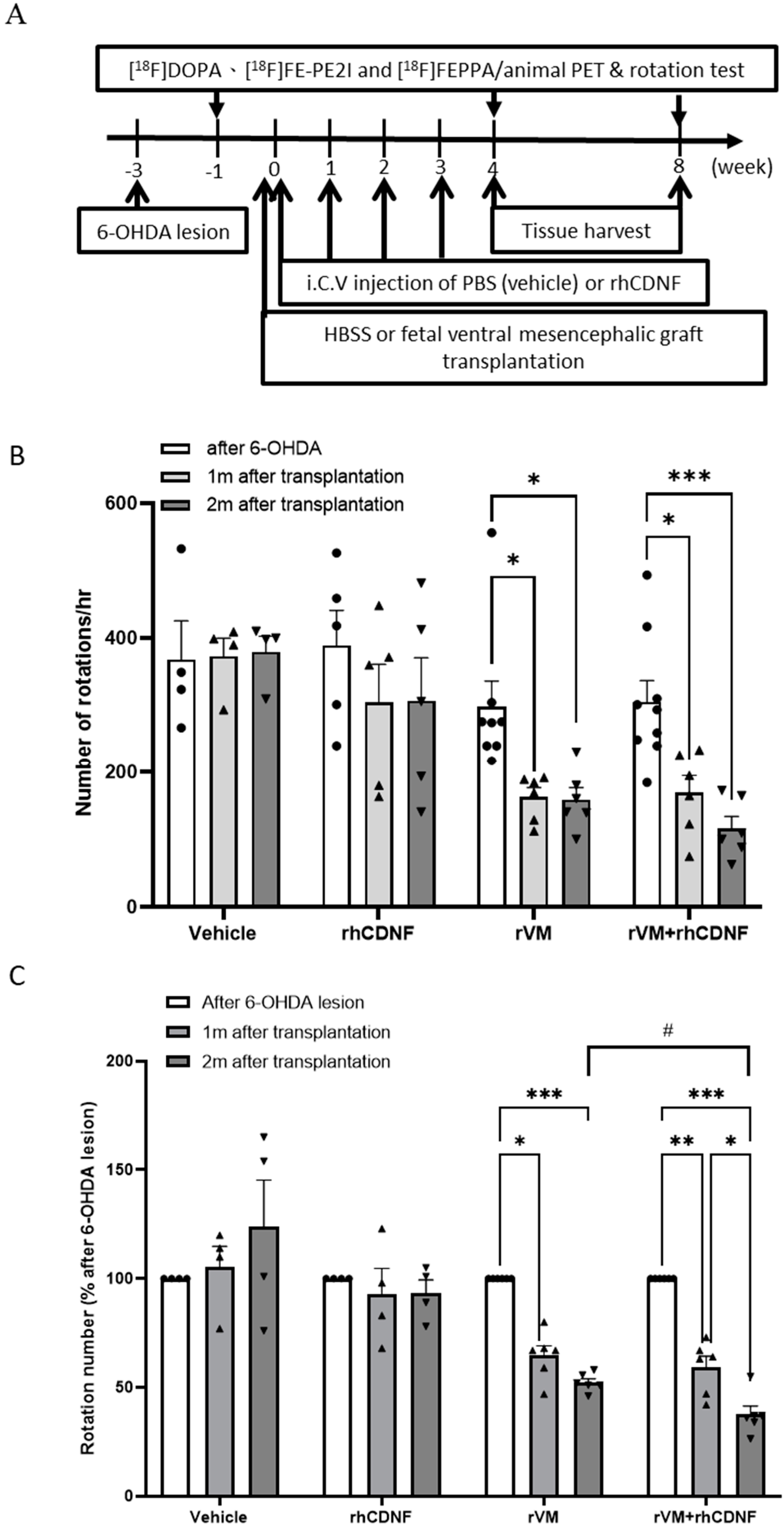
2.3. Behavioral Test
2.4. Production of Recombination CDNF Protein
2.5. Mesencephalic Tissue Preparation and Transplantation
2.6. Radiopharmaceuticals
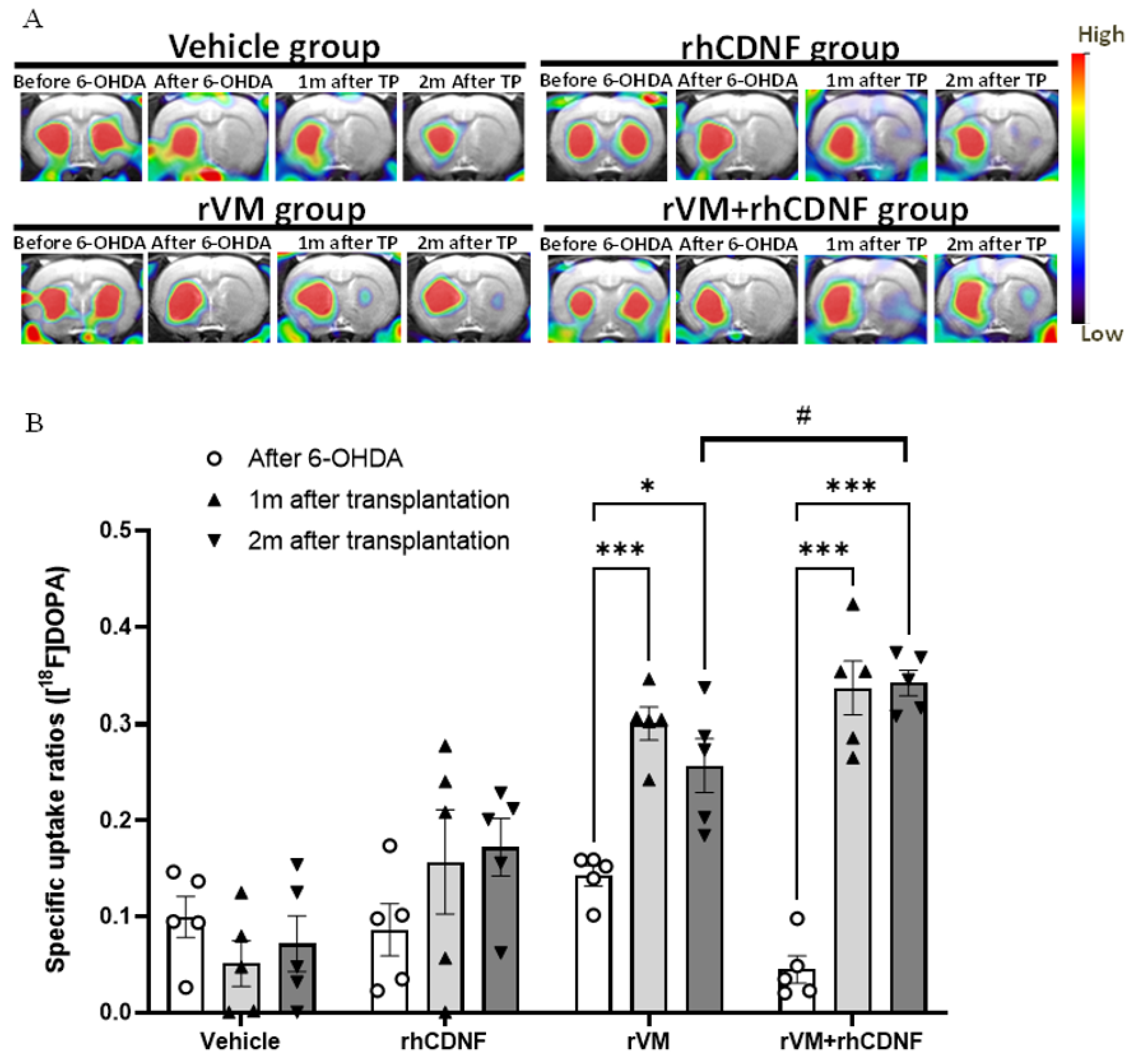
2.7. Small-Animal PET Imaging
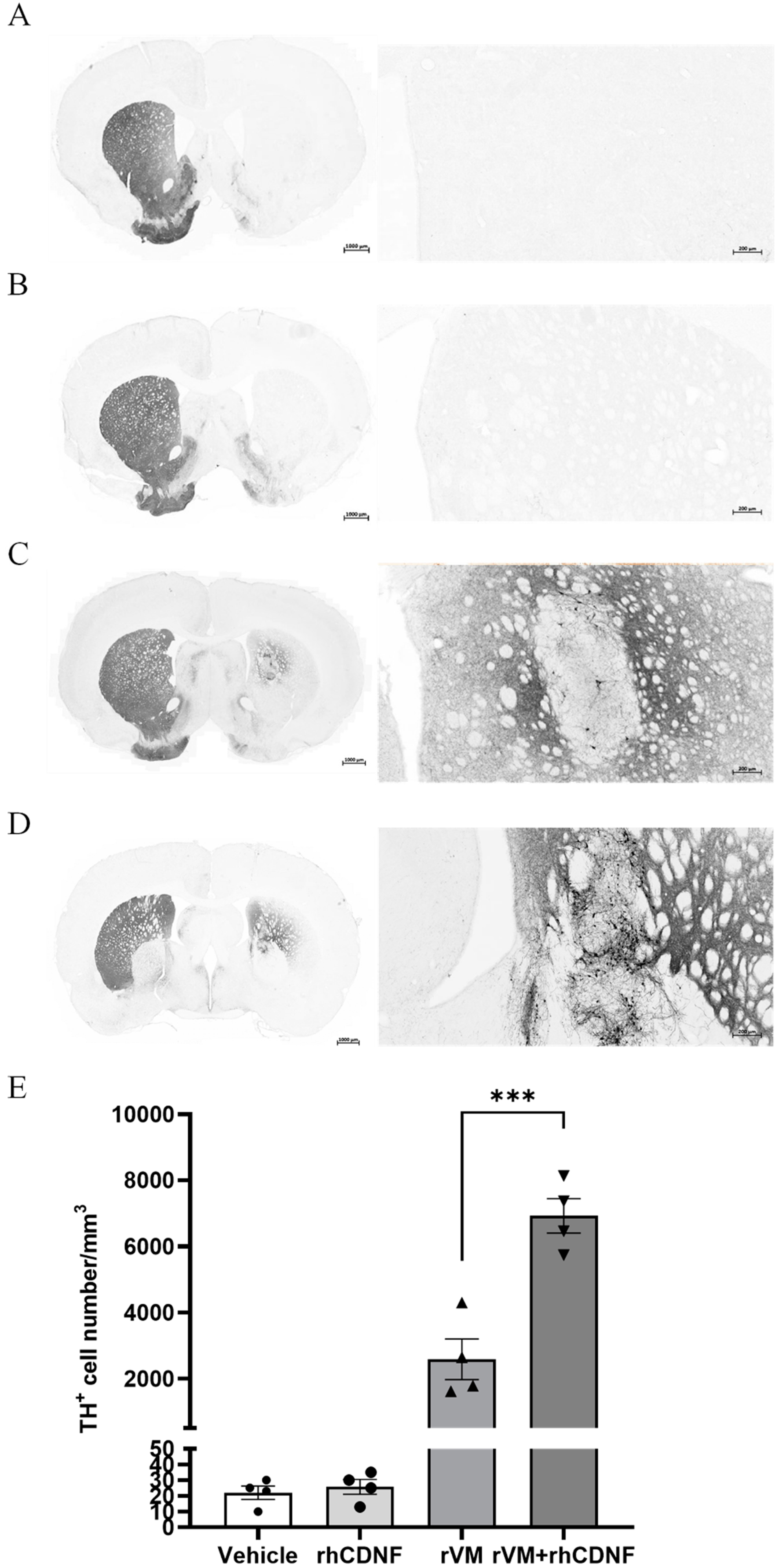
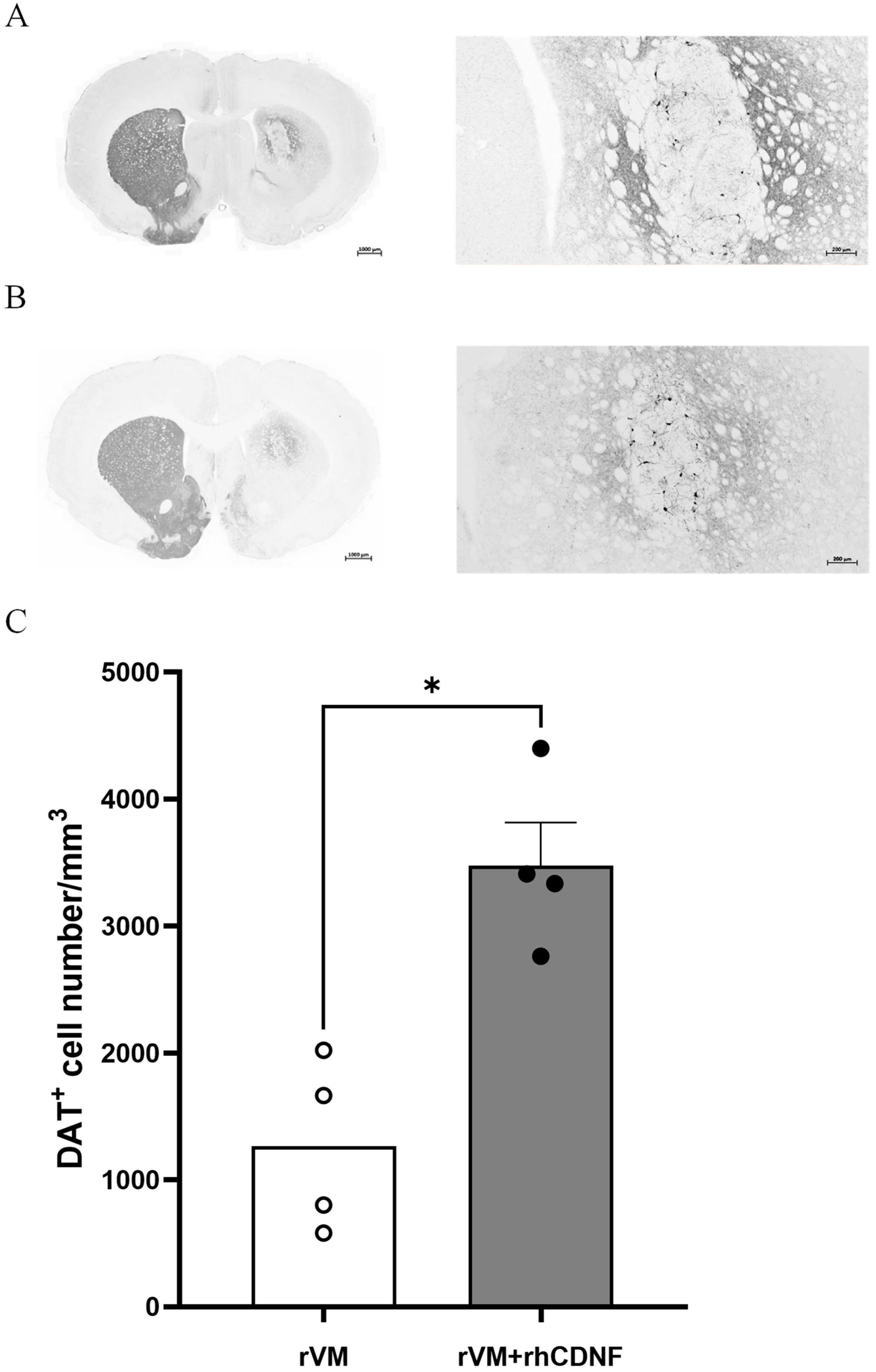
2.8. Histology, Immunohistochemistry, and Image Acquisition
2.9. Cell Culture and Treatment
2.10. Immunofluorescence Staining and Analysis of Imaging Data
2.11. Western Blot Analysis
2.12. Fractal Analysis Using FracLac for ImageJ
2.13. Assessment of Cytokines
2.14. RNA Preparation and RNA Sequencing
2.15. Bioinformatics
2.16. Statistical Analysis
3. Results
3.1. CDNF Boosts the Effect of rVM Transplantation on Functional Recovery in Hemiparkinsonian Rats
3.2. CDNF Improved the Functional Result of DA Allografts in the Striatum of Hemiparkinsonian Rats at 2 Months after Allotransplantation
3.3. CDNF Promotes Maturation of DA Allografts in the Striatum of Hemiparkinsonian Rats at 2 Months after Transplantation
3.4. CDNF Enhances the Survival and Maturation of DA Neurons in the rVM Grafted Striatum
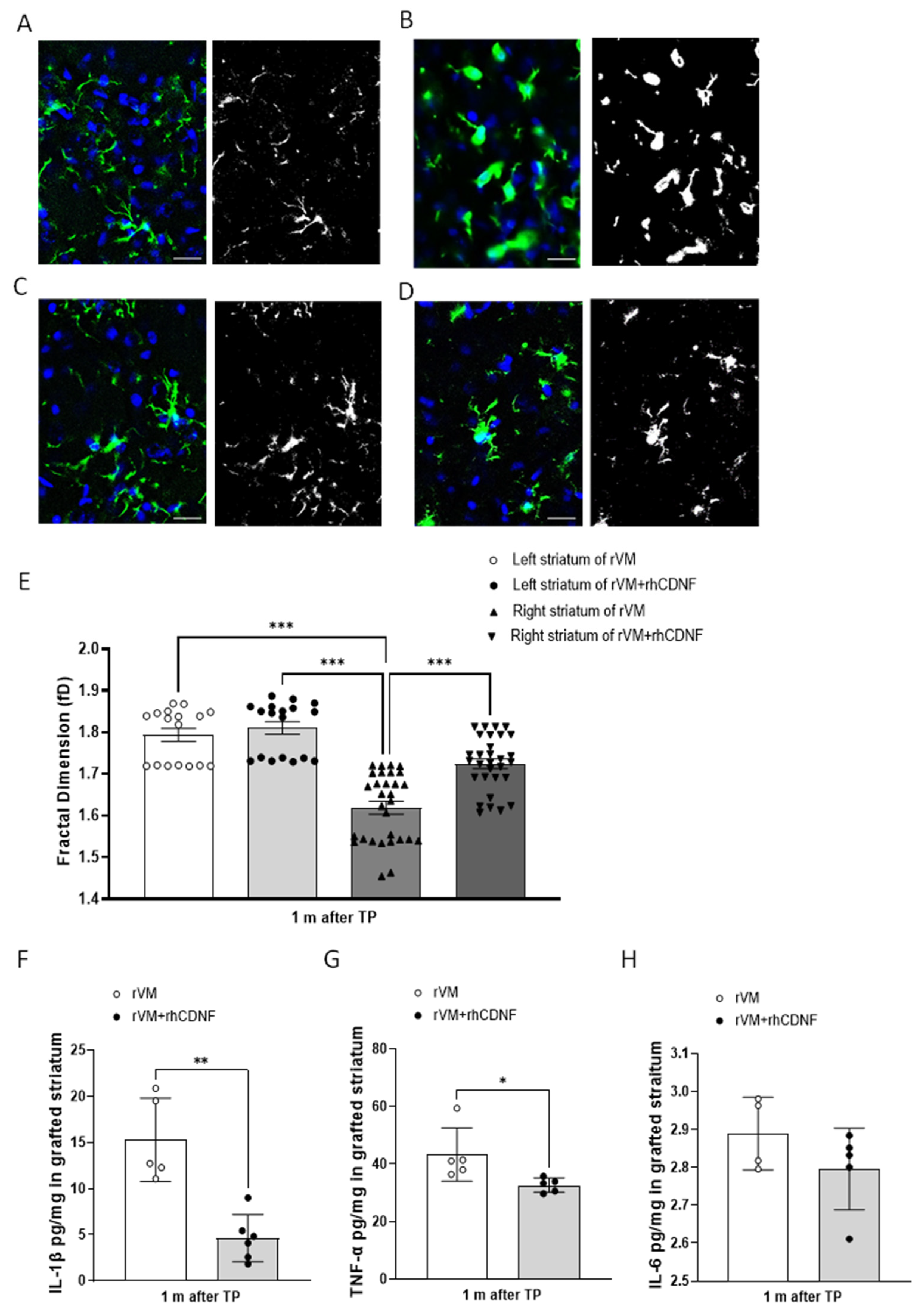
3.5. CDNF Attenuates Neuroinflamm. in Grafted Striatum after Allotransplantation
3.6. CDNF Modulates the Phenotype and Morphology of 6-OHDA-Stimulated BV2 Microglial Cell Lines
3.7. CDNF Treatment Alters Transcriptional Responses and Canonical Pathways in 6-OHDA-Stimulated BV2 Microglial Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 6-OHDA | 6-Hydroxydopamine hydrobromide |
| AAALAC International | Association for Assessment and Accreditation of Laboratory Animal Care International |
| AKT | Protein kinase B (PKB) |
| CDNF | Cerebral dopamine neurotrophic factor |
| DEGs | Differentially expressed genes |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| ER | Endoplasmic reticulum |
| FBS | Fetal bovine serum |
| GO | Gene ontology |
| IACUC | Institutional Animal Care and Use Committee |
| Iba1 | Ionized calcium binding adaptor molecule 1 |
| IPA | Ingenuity pathway analysis |
| ir | Immunoreactive |
| LPS | Lipopolysaccharide |
| PET | Positron emission tomography |
| PI3Ks | phosphoinositide 3 kinases |
| PIK3 | Phosphatidylinositol 3 kinase |
| RLE | Relative log expression |
| rVM tissue | Rat ventral mesencephalic tissue |
| TMM | Trimmed mean of M-values |
References
- Lewis, P.A. James Parkinson: The man behind the shaking palsy. J. Parkinsons Dis. 2012, 2, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Zhu, Y.; Tong, C.; Snyder, J.A.; Lipton, J.W.; Carvey, P.M. Progressive dopamine neuron loss following supra-nigral lipopolysaccharide (LPS) infusion into rats exposed to LPS prenatally. Exp. Neurol. 2006, 199, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Carvey, P.M.; Punati, A.; Newman, M.B. Progressive dopamine neuron loss in Parkinson’s disease: The multiple hit hypothesis. Cell Transplant. 2006, 15, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Bastide, M.F.; Meissner, W.G.; Picconi, B.; Fasano, S.; Fernagut, P.-O.; Feyder, M.; Francardo, V.; Alcacer, C.; Ding, Y.; Brambilla, R. Pathophysiology of L-dopa-induced motor and non-motor complications in Parkinson’s disease. Prog. Neurobiol. 2015, 132, 96–168. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Fernández, R.; Sossi, V.; Huang, Z.; Furtado, S.; Lu, J.-Q.; Calne, D.B.; Ruth, T.J.; Stoessl, A.J. Levodopa-induced changes in synaptic dopamine levels increase with progression of Parkinson’s disease: Implications for dyskinesias. Brain 2004, 127, 2747–2754. [Google Scholar] [CrossRef]
- Lindgren, H.S.; Andersson, D.R.; Lagerkvist, S.; Nissbrandt, H.; Cenci, M.A. l-DOPA-induced dopamine efflux in the striatum and the substantia nigra in a rat model of Parkinson’s disease: Temporal and quantitative relationship to the expression of dyskinesia. J. Neurochem. 2010, 112, 1465–1476. [Google Scholar] [CrossRef]
- Jenner, P. Molecular mechanisms of L-DOPA-induced dyskinesia. Nat. Rev. Neurosci. 2008, 9, 665–677. [Google Scholar] [CrossRef]
- Politis, M.; Lindvall, O. Clinical application of stem cell therapy in Parkinson’s disease. BMC Med. 2012, 10, 1. [Google Scholar] [CrossRef]
- Latchney, S.E.; Eisch, A.J. Therapeutic application of neural stem cells and adult neurogenesis for neurodegenerative disorders: Regeneration and beyond. Eur. J. Neurodegener. Dis. 2012, 1, 335–351. [Google Scholar]
- Yamagishi, H.; Shigematsu, K. Perspectives on Stem Cell-Based Regenerative Medicine with a Particular Emphasis on Mesenchymal Stem Cell Therapy. JMA J. 2022, 5, 36–43. [Google Scholar]
- Grinand, L.; Takahashi, J. Automated measurement of fluorescence signals reveals a significant increase of the graft-derived neurite extension in neonates compared to aged rats. Regen. Ther. 2022, 19, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Freeman, T.B.; Olanow, C.W.; Hauser, R.A.; Nauert, G.M.; Smith, D.A.; Borlongan, C.V.; Sanberg, P.R.; Holt, D.A.; Kordower, J.H.; Vingerhoets, F.J.; et al. Bilateral fetal nigral transplantation into the postcommissural putamen in Parkinson’s disease. Ann. Neurol. 1995, 38, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Kordower, J.H.; Freeman, T.B.; Chen, E.Y.; Mufson, E.J.; Sanberg, P.R.; Hauser, R.A.; Snow, B.; Olanow, C.W. Fetal nigral grafts survive and mediate clinical benefit in a patient with Parkinson’s disease. Mov. Disord. 1998, 13, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Kordower, J.H.; Goetz, C.G.; Chu, Y.; Halliday, G.M.; Nicholson, D.A.; Musial, T.F.; Marmion, D.J.; Stoessl, A.J.; Sossi, V.; Freeman, T.B.; et al. Robust graft survival and normalized dopaminergic innervation do not obligate recovery in a Parkinson disease patient. Ann. Neurol. 2017, 81, 46–57. [Google Scholar] [CrossRef]
- Schumacher, J.M.; Ellias, S.A.; Palmer, E.P.; Kott, H.S.; Dinsmore, J.; Dempsey, P.K.; Fischman, A.J.; Thomas, C.; Feldman, R.G.; Kassissieh, S.; et al. Transplantation of embryonic porcine mesencephalic tissue in patients with PD. Neurology 2000, 54, 1042–1050. [Google Scholar] [CrossRef]
- Camp, D.M.; Loeffler, D.A.; Farrah, D.M.; Borneman, J.N.; LeWitt, P.A. Cellular immune response to intrastriatally implanted allogeneic bone marrow stromal cells in a rat model of Parkinson’s disease. J. Neuroinflamm. 2009, 6, 17. [Google Scholar] [CrossRef]
- Shamekh, R.; Newcomb, J.; Mallery, J.; Cassady, C.J.; Saporta, S.; Cameron, D.F.; Sanberg, P.R.; Willing, A.E. Survival of rat or mouse ventral mesencephalon neurons after cotransplantation with rat sertoli cells in the mouse striatum. Cell Transplant. 2005, 14, 551–564. [Google Scholar] [CrossRef]
- Mori, D.N.; Kreisel, D.; Fullerton, J.N.; Gilroy, D.W.; Goldstein, D.R. Inflammatory triggers of acute rejection of organ allografts. Immunol. Rev. 2014, 258, 132–144. [Google Scholar] [CrossRef]
- Adler, A.F.; Cardoso, T.; Nolbrant, S.; Mattsson, B.; Hoban, D.B.; Jarl, U.; Wahlestedt, J.N.; Grealish, S.; Bjorklund, A.; Parmar, M. hESC-Derived Dopaminergic Transplants Integrate into Basal Ganglia Circuitry in a Preclinical Model of Parkinson’s Disease. Cell Rep. 2019, 28, 3462–3473.e5. [Google Scholar] [CrossRef]
- Steinbeck, J.A.; Studer, L. Moving stem cells to the clinic: Potential and limitations for brain repair. Neuron 2015, 86, 187–206. [Google Scholar] [CrossRef]
- Tansey, M.G.; McCoy, M.K.; Frank-Cannon, T.C. Neuroinflammatory mechanisms in Parkinson’s disease: Potential environmental triggers, pathways, and targets for early therapeutic intervention. Exp. Neurol. 2007, 208, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Rabaneda-Lombarte, N.; Vidal-Taboada, J.M.; Valente, T.; Ezquerra, M.; Fernandez-Santiago, R.; Marti, M.J.; Compta, Y.; Saura, J.; Sola, C. Altered expression of the immunoregulatory ligand-receptor pair CD200-CD200R1 in the brain of Parkinson’s disease patients. NPJ Parkinsons Dis. 2022, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- London, A.; Cohen, M.; Schwartz, M. Microglia and monocyte-derived macrophages: Functionally distinct populations that act in concert in CNS plasticity and repair. Front. Cell Neurosci. 2013, 7, 34. [Google Scholar] [CrossRef]
- Forbes, L.H.; Miron, V.E. Monocytes in central nervous system remyelination. Glia 2022, 70, 797–807. [Google Scholar] [CrossRef]
- Jha, M.K.; Lee, W.H.; Suk, K. Functional polarization of neuroglia: Implications in neuroinflammation and neurological disorders. Biochem. Pharmacol. 2016, 103, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Leak, R.K.; Shi, Y.; Suenaga, J.; Gao, Y.; Zheng, P.; Chen, J. Microglial and macrophage polarization-new prospects for brain repair. Nat. Rev. Neurol. 2015, 11, 56–64. [Google Scholar] [CrossRef]
- Garcia-Revilla, J.; Alonso-Bellido, I.M.; Burguillos, M.A.; Herrera, A.J.; Espinosa-Oliva, A.M.; Ruiz, R.; Cruz-Hernandez, L.; Garcia-Dominguez, I.; Roca-Ceballos, M.A.; Santiago, M.; et al. Reformulating Pro-Oxidant Microglia in Neurodegeneration. J. Clin. Med. 2019, 8, 1719. [Google Scholar] [CrossRef]
- Glass, C.K.; Saijo, K.; Winner, B.; Marchetto, M.C.; Gage, F.H. Mechanisms underlying inflammation in neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef]
- Praet, J.; Santermans, E.; Daans, J.; Le Blon, D.; Hoornaert, C.; Goossens, H.; Hens, N.; Van der Linden, A.; Berneman, Z.; Ponsaerts, P. Early Inflammatory Responses Following Cell Grafting in the CNS Trigger Activation of the Subventricular Zone: A Proposed Model of Sequential Cellular Events. Cell Transplant. 2015, 24, 1481–1492. [Google Scholar] [CrossRef]
- Lindholm, P.; Voutilainen, M.H.; Lauren, J.; Peranen, J.; Leppanen, V.M.; Andressoo, J.O.; Lindahl, M.; Janhunen, S.; Kalkkinen, N.; Timmusk, T.; et al. Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature 2007, 448, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, P.; Saarma, M. Novel CDNF/MANF family of neurotrophic factors. Dev. Neurobiol. 2010, 70, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Jntti, M.; Harvey, B.K. Trophic activities of endoplasmic reticulum proteins CDNF and MANF. Cell Tissue Res. 2020, 382, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Tseng, K.Y.; Danilova, T.; Domanskyi, A.; Saarma, M.; Lindahl, M.; Airavaara, M. MANF Is Essential for Neurite Extension and Neuronal Migration in the Developing Cortex. eNeuro 2017, 4, 0214-17. [Google Scholar] [CrossRef] [PubMed]
- Tseng, K.Y.; Anttila, J.E.; Khodosevich, K.; Tuominen, R.K.; Lindahl, M.; Domanskyi, A.; Airavaara, M. MANF Promotes Differentiation and Migration of Neural Progenitor Cells with Potential Neural Regenerative Effects in Stroke. Mol. Ther. 2018, 26, 238–255. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Victor, P.; Jasper, H.; Neves, J. Trophic Factors in Inflammation and Regeneration: The Role of MANF and CDNF. Front. Physiol. 2018, 9, 1629. [Google Scholar] [CrossRef]
- Jhao, Y.T.; Chiu, C.H.; Chen, C.F.; Chou, T.K.; Lin, Y.W.; Ju, Y.T.; Wu, S.C.; Yan, R.F.; Shiue, C.Y.; Chueh, S.H.; et al. The Effect of Sertoli Cells on Xenotransplantation and Allotransplantation of Ventral Mesencephalic Tissue in a Rat Model of Parkinson’s Disease. Cells 2019, 8, 1420. [Google Scholar] [CrossRef]
- Truong, L.; Allbutt, H.; Kassiou, M.; Henderson, J.M. Developing a preclinical model of Parkinson’s disease: A study of behaviour in rats with graded 6-OHDA lesions. Behav. Brain Res. 2006, 169, 1–9. [Google Scholar] [CrossRef]
- Ghadery, C.; Koshimori, Y.; Coakeley, S.; Harris, M.; Rusjan, P.; Kim, J.; Houle, S.; Strafella, A.P. Microglial activation in Parkinson’s disease using [(18)F]-FEPPA. J. Neuroinflamm. 2017, 14, 8. [Google Scholar] [CrossRef]
- Kuo, T.T.; Wang, V.; Wu, J.S.; Chen, Y.H.; Tseng, K.Y. Post-stroke Delivery of Valproic Acid Promotes Functional Recovery and Differentially Modifies Responses of Peri-Infarct Microglia. Front. Mol. Neurosci. 2021, 14, 639145. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Chang, H.-F.; Tsai, M.-J.; Chen, J.-S.; Wang, M.-J. 6-Mercaptopurine attenuates tumor necrosis factor-α production in microglia through Nur77-mediated transrepression and PI3K/Akt/mTOR signaling-mediated translational regulation. J. Neuroinflamm. 2016, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, Y.; Han, X.; Xing, Q.; Zhao, L. Dexmedetomidine Regulates 6-hydroxydopamine-Induced Microglial Polarization. Neurochem. Res. 2017, 42, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cheng, L.; Liu, Y.; Zhang, W.; Maharjan, S.; Cui, Z.; Wang, X.; Tang, D.; Nie, L. Mechanisms of anti-inflammatory property of conserved dopamine neurotrophic factor: Inhibition of JNK signaling in lipopolysaccharide-induced microglia. J. Mol. Neurosci. 2014, 52, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Karperien, A.; Ahammer, H.; Jelinek, H.F. Quantitating the subtleties of microglial morphology with fractal analysis. Front. Cell Neurosci. 2013, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Morrison, H.; Young, K.; Qureshi, M.; Rowe, R.K.; Lifshitz, J. Quantitative microglia analyses reveal diverse morphologic responses in the rat cortex after diffuse brain injury. Sci. Rep. 2017, 7, 13211. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierlinski, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dong, C.; Li, P.; Ren, Z.; Wang, H.; Yu, F.; Ning, C.; Liu, K.; Wei, W.; Huang, R.; et al. Identification of candidate genes associated with porcine meat color traits by genome-wide transcriptome analysis. Sci. Rep. 2016, 6, 35224. [Google Scholar] [CrossRef] [PubMed]
- Maza, E. In Papyro Comparison of TMM (edgeR), RLE (DESeq2), and MRN Normalization Methods for a Simple Two-Conditions-Without-Replicates RNA-Seq Experimental Design. Front. Genet. 2016, 7, 164. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Das, A.; Kim, S.H.; Arifuzzaman, S.; Yoon, T.; Chai, J.C.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflamm. 2016, 13, 182. [Google Scholar] [CrossRef]
- Das, A.; Chai, J.C.; Kim, S.H.; Park, K.S.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Dual RNA sequencing reveals the expression of unique transcriptomic signatures in lipopolysaccharide-induced BV-2 microglial cells. PLoS ONE 2015, 10, e0121117. [Google Scholar] [CrossRef]
- Inaji, M.; Yoshizaki, T.; Okauchi, T.; Maeda, J.; Nagai, Y.; Nariai, T.; Ohno, K.; Ando, K.; Okano, H.; Obayashi, S.; et al. In vivo PET measurements with [11C]PE2I to evaluate fetal mesencephalic transplantations to unilateral 6-OHDA-lesioned rats. Cell Transplant. 2005, 14, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Rusjan, P.M.; Wilson, A.A.; Bloomfield, P.M.; Vitcu, I.; Meyer, J.H.; Houle, S.; Mizrahi, R. Quantitation of translocator protein binding in human brain with the novel radioligand [18F]-FEPPA and positron emission tomography. J. Cereb. Blood Flow Metab. 2011, 31, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.J.; Chen, C.F.; Huang, Y.S.; Chiu, C.H.; Wu, S.C.; Lin, C.Y.; Chueh, S.H.; Cheng, C.Y.; Ma, K.H. Olfactory ensheathing cells improve the survival of porcine neural xenografts in a Parkinsonian rat model. Xenotransplantation 2020, 27, e12569. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.; Zhu, J.; Sousa-Victor, P.; Konjikusic, M.; Riley, R.; Chew, S.; Qi, Y.; Jasper, H.; Lamba, D.A. Immune modulation by MANF promotes tissue repair and regenerative success in the retina. Science 2016, 353, aaf3646. [Google Scholar] [CrossRef]
- Matlik, K.; Anttila, J.E.; Kuan-Yin, T.; Smolander, O.P.; Pakarinen, E.; Lehtonen, L.; Abo-Ramadan, U.; Lindholm, P.; Zheng, C.; Harvey, B.; et al. Poststroke delivery of MANF promotes functional recovery in rats. Sci. Adv. 2018, 4, eaap8957. [Google Scholar] [CrossRef]
- Rong, Z.; Wang, M.; Hu, Z.; Stradner, M.; Zhu, S.; Kong, H.; Yi, H.; Goldrath, A.; Yang, Y.G.; Xu, Y.; et al. An effective approach to prevent immune rejection of human ESC-derived allografts. Cell Stem Cell 2014, 14, 121–130. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Akhmetzyanova, E.R.; Mukhamedshina, Y.O.; Zhuravleva, M.N.; Galieva, L.R.; Kostennikov, A.A.; Garanina, E.E.; Rizvanov, A.A. Transplantation of Microglia in the Area of Spinal Cord Injury in an Acute Period Increases Tissue Sparing, but Not Functional Recovery. Front. Cell Neurosci. 2018, 12, 507. [Google Scholar] [CrossRef]
- Gao, J.; Grill, R.J.; Dunn, T.J.; Bedi, S.; Labastida, J.A.; Hetz, R.A.; Xue, H.; Thonhoff, J.R.; DeWitt, D.S.; Prough, D.S.; et al. Human Neural Stem Cell Transplantation-Mediated Alteration of Microglial/Macrophage Phenotypes after Traumatic Brain Injury. Cell Transplant. 2016, 25, 1863–1877. [Google Scholar] [CrossRef]
- Duan, W.M.; Widner, H.; Brundin, P. Temporal pattern of host responses against intrastriatal grafts of syngeneic, allogeneic or xenogeneic embryonic neuronal tissue in rats. Exp. Brain Res. 1995, 104, 227–242. [Google Scholar] [CrossRef]
- Lawrence, J.M.; Morris, R.J.; Wilson, D.J.; Raisman, G. Mechanisms of allograft rejection in the rat brain. Neuroscience 1990, 37, 431–462. [Google Scholar] [CrossRef]
- Piehl, F.; Lidman, O. Neuroinflammation in the rat—CNS cells and their role in the regulation of immune reactions. Immunol. Rev. 2001, 184, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Weiner, H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, M.; Danilova, T.; Palm, E.; Lindholm, P.; Voikar, V.; Hakonen, E.; Ustinov, J.; Andressoo, J.O.; Harvey, B.K.; Otonkoski, T.; et al. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep. 2014, 7, 366–375. [Google Scholar] [CrossRef]
- Lindahl, M.; Chalazonitis, A.; Palm, E.; Pakarinen, E.; Danilova, T.; Pham, T.D.; Setlik, W.; Rao, M.; Voikar, V.; Huotari, J.; et al. Cerebral dopamine neurotrophic factor-deficiency leads to degeneration of enteric neurons and altered brain dopamine neuronal function in mice. Neurobiol. Dis. 2020, 134, 104696. [Google Scholar] [CrossRef]
- Voutilainen, M.H.; De Lorenzo, F.; Stepanova, P.; Back, S.; Yu, L.Y.; Lindholm, P.; Porsti, E.; Saarma, M.; Mannisto, P.T.; Tuominen, R.K. Evidence for an Additive Neurorestorative Effect of Simultaneously Administered CDNF and GDNF in Hemiparkinsonian Rats: Implications for Different Mechanism of Action. eNeuro 2017, 4, 0117-16. [Google Scholar] [CrossRef]
- Nadella, R.; Voutilainen, M.H.; Saarma, M.; Gonzalez-Barrios, J.A.; Leon-Chavez, B.A.; Jimenez, J.M.; Jimenez, S.H.; Escobedo, L.; Martinez-Fong, D. Transient transfection of human CDNF gene reduces the 6-hydroxydopamine-induced neuroinflammation in the rat substantia nigra. J. Neuroinflamm. 2014, 11, 209. [Google Scholar] [CrossRef]
- Mei, J.; Niu, C. Effects of engineered conserved dopamine neurotrophic factor-expressing bone marrow stromal cells on dopaminergic neurons following 6-OHDA administrations. Mol. Med. Rep. 2015, 11, 1207–1213. [Google Scholar] [CrossRef]
- Rocha, S.M.; Cristovao, A.C.; Campos, F.L.; Fonseca, C.P.; Baltazar, G. Astrocyte-derived GDNF is a potent inhibitor of microglial activation. Neurobiol. Dis. 2012, 47, 407–415. [Google Scholar] [CrossRef]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Neuroinflammation and M2 microglia: The good, the bad, and the inflamed. J. Neuroinflamm. 2014, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; de Castro, F.; Del Rio-Hortega, J.; Rafael Iglesias-Rozas, J.; Garrosa, M.; Kettenmann, H. The “Big-Bang” for modern glial biology: Translation and comments on Pio del Rio-Hortega 1919 series of papers on microglia. Glia 2016, 64, 1801–1840. [Google Scholar] [CrossRef] [PubMed]
- Norris, G.T.; Kipnis, J. Immune cells and CNS physiology: Microglia and beyond. J. Exp. Med. 2019, 216, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.S.; Zhao, M.L.; Derico, L.; Choi, N.; Lee, S.C. Insulin-like growth factor 1 and 2 (IGF1, IGF2) expression in human microglia: Differential regulation by inflammatory mediators. J. Neuroinflamm. 2013, 10, 37. [Google Scholar] [CrossRef]
- Spadaro, O.; Camell, C.D.; Bosurgi, L.; Nguyen, K.Y.; Youm, Y.H.; Rothlin, C.V.; Dixit, V.D. IGF1 Shapes Macrophage Activation in Response to Immunometabolic Challenge. Cell Rep. 2017, 19, 225–234. [Google Scholar] [CrossRef]
- Tarassishin, L.; Suh, H.S.; Lee, S.C. Interferon regulatory factor 3 plays an anti-inflammatory role in microglia by activating the PI3K/Akt pathway. J. Neuroinflamm. 2011, 8, 187. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, X.; Dai, X.; Lu, S.; Gui, B.; Jin, W.; Zhang, S.; Zhang, S.; Qian, Y. Lithium ameliorates lipopolysaccharide-induced microglial activation via inhibition of toll-like receptor 4 expression by activating the PI3K/Akt/FoxO1 pathway. J. Neuroinflamm. 2014, 11, 140. [Google Scholar] [CrossRef]
- Myers, K.V.; Amend, S.R.; Pienta, K.J. Targeting Tyro3, Axl and MerTK (TAM receptors): Implications for macrophages in the tumor microenvironment. Mol. Cancer 2019, 18, 94. [Google Scholar] [CrossRef]
- Han, C.Z.; Juncadella, I.J.; Kinchen, J.M.; Buckley, M.W.; Klibanov, A.L.; Dryden, K.; Onengut-Gumuscu, S.; Erdbrugger, U.; Turner, S.D.; Shim, Y.M.; et al. Macrophages redirect phagocytosis by non-professional phagocytes and influence inflammation. Nature 2016, 539, 570–574. [Google Scholar] [CrossRef]
- Healy, L.M.; Perron, G.; Won, S.Y.; Michell-Robinson, M.A.; Rezk, A.; Ludwin, S.K.; Moore, C.S.; Hall, J.A.; Bar-Or, A.; Antel, J.P. MerTK Is a Functional Regulator of Myelin Phagocytosis by Human Myeloid Cells. J. Immunol. 2016, 196, 3375–3384. [Google Scholar] [CrossRef]
- Nomura, K.; Vilalta, A.; Allendorf, D.H.; Hornik, T.C.; Brown, G.C. Activated Microglia Desialylate and Phagocytose Cells via Neuraminidase, Galectin-3, and Mer Tyrosine Kinase. J. Immunol. 2017, 198, 4792–4801. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Chen, X.; Lu, S.; Yang, D.; Li, W.; Li, F.; Wu, F.; Sun, M. Insulin-like growth factor 1 (IGF-1) promotes phagocytic activity of mouse BV-2 microglial cells via activating PI3K/AKT signaling pathway. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2021, 37, 199–204. [Google Scholar] [PubMed]
- Hoban, D.B.; Shrigley, S.; Mattsson, B.; Breger, L.S.; Jarl, U.; Cardoso, T.; Nelander Wahlestedt, J.; Luk, K.C.; Bjorklund, A.; Parmar, M. Impact of alpha-synuclein pathology on transplanted hESC-derived dopaminergic neurons in a humanized alpha-synuclein rat model of PD. Proc. Natl. Acad. Sci. USA 2020, 117, 15209–15220. [Google Scholar] [CrossRef] [PubMed]
- Albert, K.; Raymundo, D.P.; Panhelainen, A.; Eesmaa, A.; Shvachiy, L.; Araujo, G.R.; Chmielarz, P.; Yan, X.; Singh, A.; Cordeiro, Y.; et al. Cerebral dopamine neurotrophic factor reduces alpha-synuclein aggregation and propagation and alleviates behavioral alterations in vivo. Mol. Ther. 2021, 29, 2821–2840. [Google Scholar] [CrossRef]
- Lalancette-Hebert, M.; Swarup, V.; Beaulieu, J.M.; Bohacek, I.; Abdelhamid, E.; Weng, Y.C.; Sato, S.; Kriz, J. Galectin-3 is required for resident microglia activation and proliferation in response to ischemic injury. J. Neurosci. 2012, 32, 10383–10395. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, K.-Y.; Wu, J.-S.; Chen, Y.-H.; Airavaara, M.; Cheng, C.-Y.; Ma, K.-H. Modulating Microglia/Macrophage Activation by CDNF Promotes Transplantation of Fetal Ventral Mesencephalic Graft Survival and Function in a Hemiparkinsonian Rat Model. Biomedicines 2022, 10, 1446. https://doi.org/10.3390/biomedicines10061446
Tseng K-Y, Wu J-S, Chen Y-H, Airavaara M, Cheng C-Y, Ma K-H. Modulating Microglia/Macrophage Activation by CDNF Promotes Transplantation of Fetal Ventral Mesencephalic Graft Survival and Function in a Hemiparkinsonian Rat Model. Biomedicines. 2022; 10(6):1446. https://doi.org/10.3390/biomedicines10061446
Chicago/Turabian StyleTseng, Kuan-Yin, Jui-Sheng Wu, Yuan-Hao Chen, Mikko Airavaara, Cheng-Yi Cheng, and Kuo-Hsing Ma. 2022. "Modulating Microglia/Macrophage Activation by CDNF Promotes Transplantation of Fetal Ventral Mesencephalic Graft Survival and Function in a Hemiparkinsonian Rat Model" Biomedicines 10, no. 6: 1446. https://doi.org/10.3390/biomedicines10061446
APA StyleTseng, K.-Y., Wu, J.-S., Chen, Y.-H., Airavaara, M., Cheng, C.-Y., & Ma, K.-H. (2022). Modulating Microglia/Macrophage Activation by CDNF Promotes Transplantation of Fetal Ventral Mesencephalic Graft Survival and Function in a Hemiparkinsonian Rat Model. Biomedicines, 10(6), 1446. https://doi.org/10.3390/biomedicines10061446





