Self-Organization of the Retina during Eye Development, Retinal Regeneration In Vivo, and in Retinal 3D Organoids In Vitro
Abstract
:1. Introduction
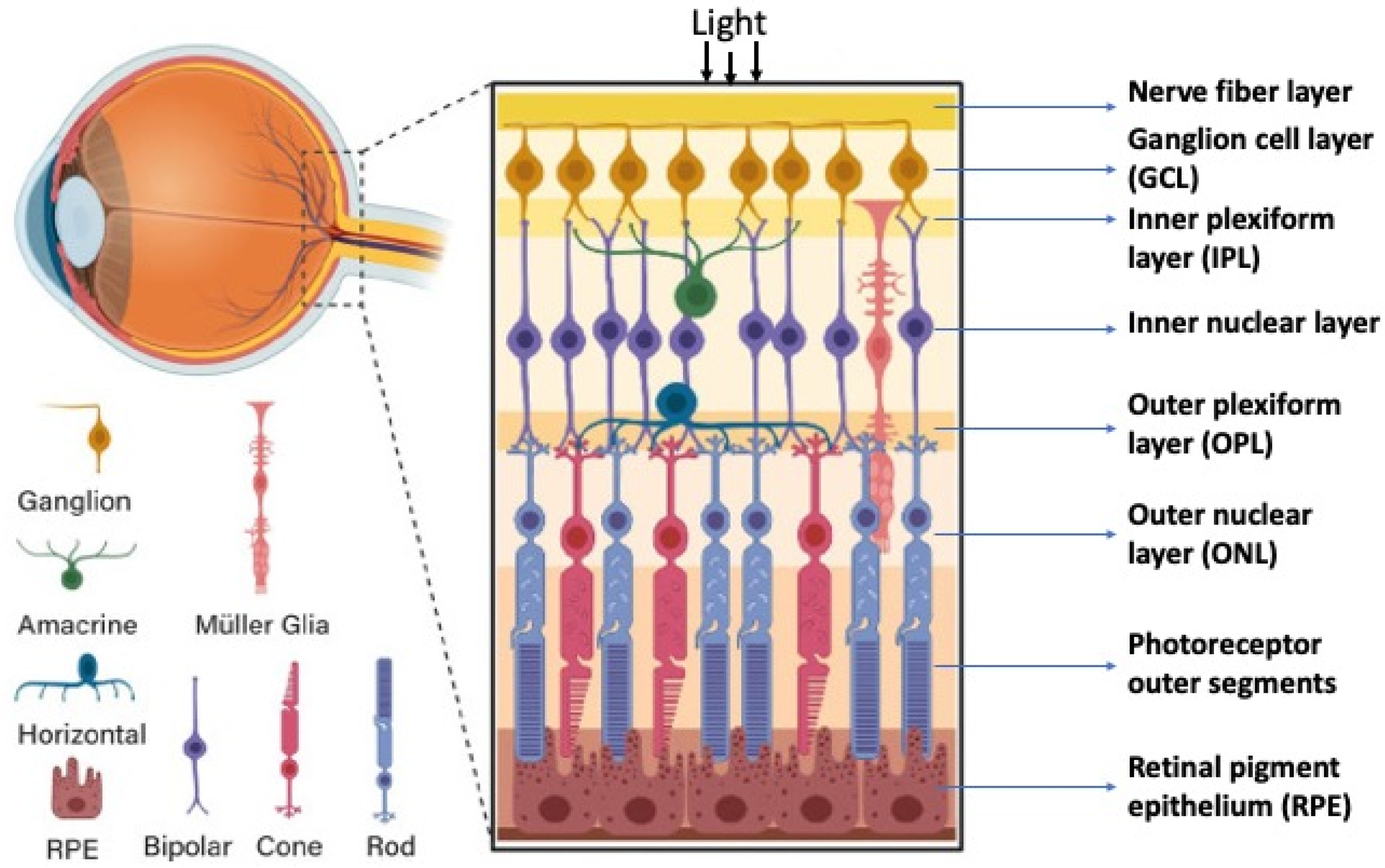
2. Retinal Structure and Function in Adult Vertebrates
3. Embryonic Retina Self-Organization In Vivo
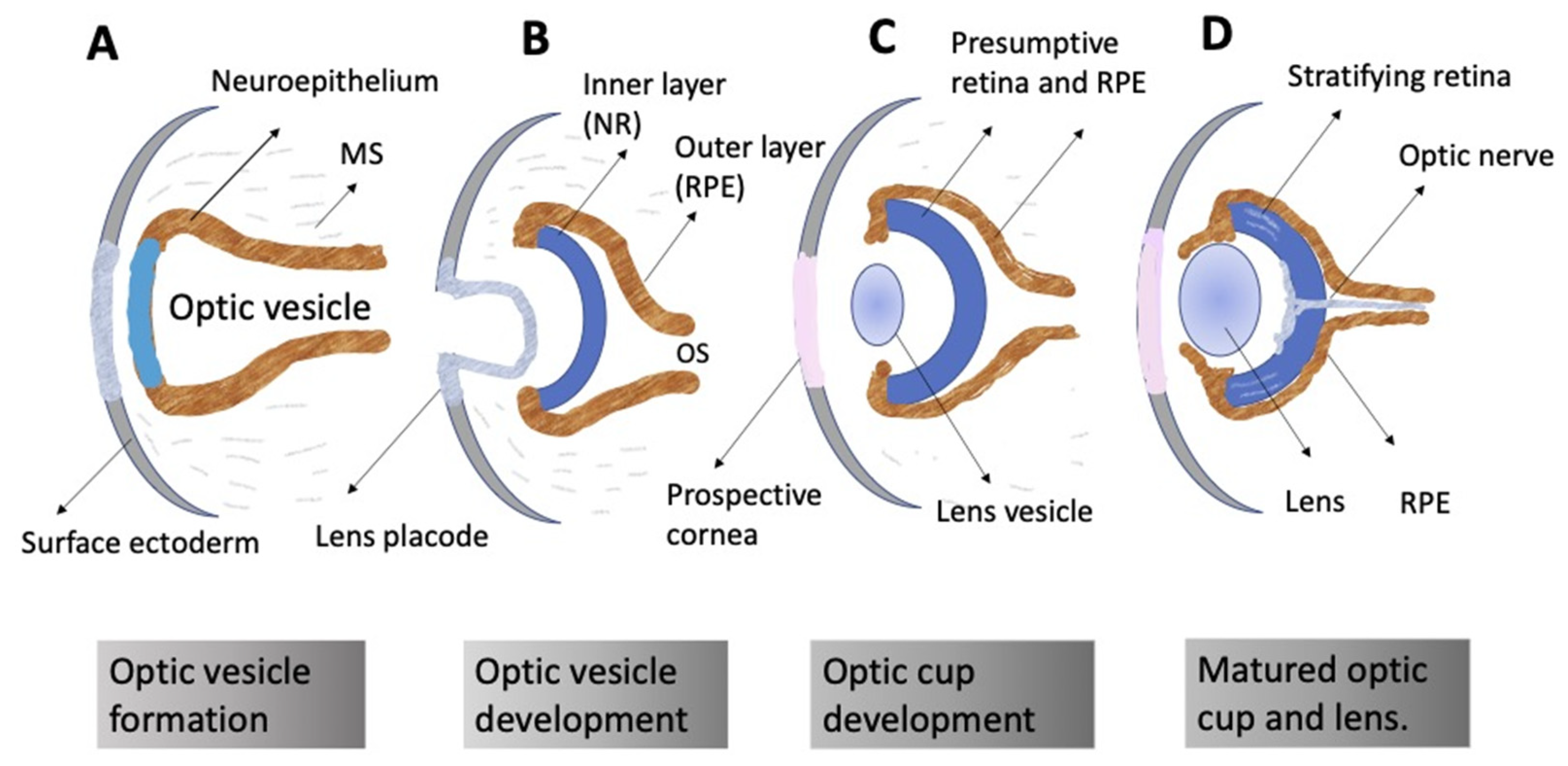
3.1. General Concepts
3.2. Regulation of Eye and Retina Development by Extrinsic Factors in Vertebrates In Vivo
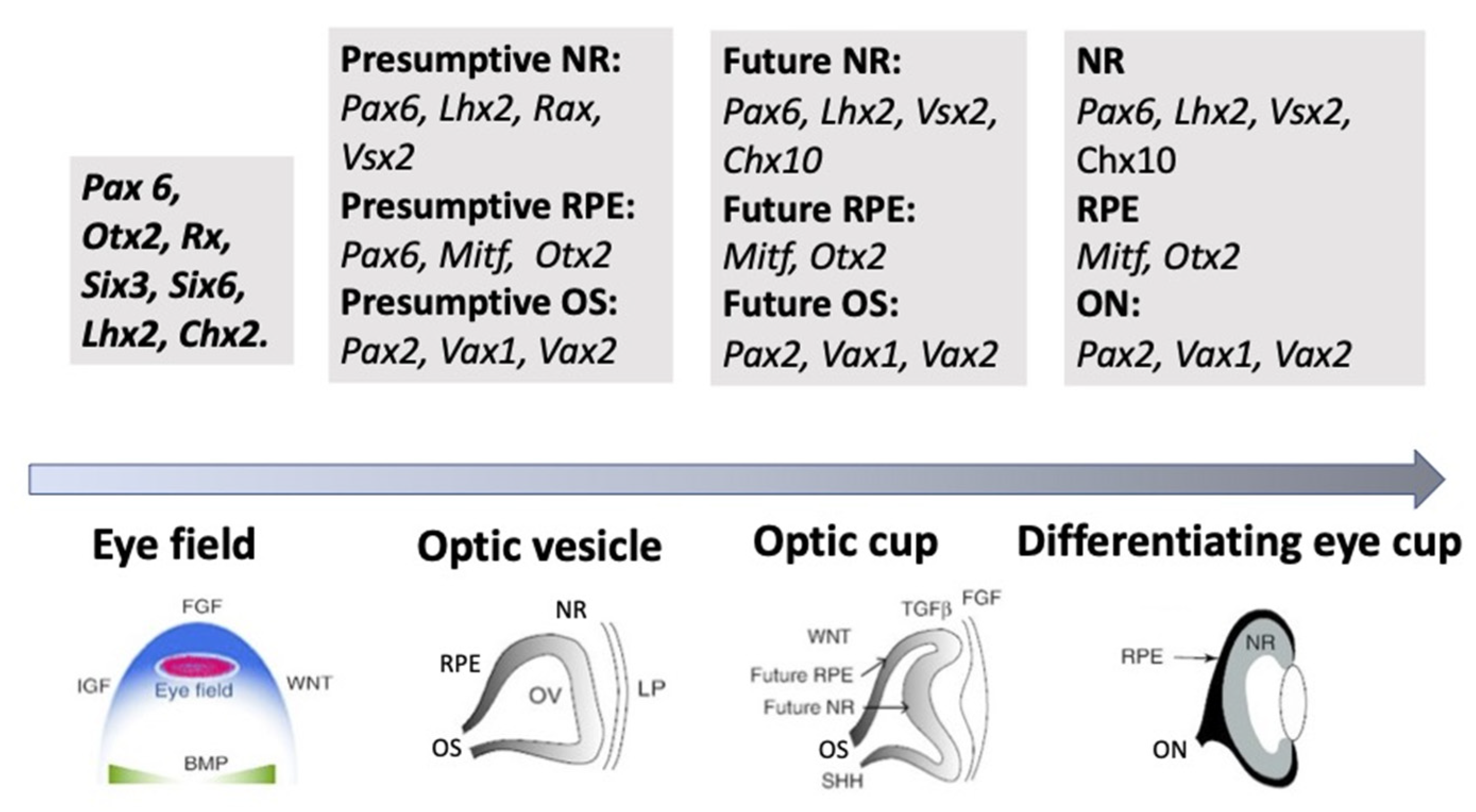
3.3. Regulation of Eye and Retina Development by Intrinsic Factors in Vertebrates In Vivo
3.4. Morphogenetic Factors of Retinal Self-Organization In Vivo
4. Self-Organization of the Retina during Regeneration in Mature Amphibians
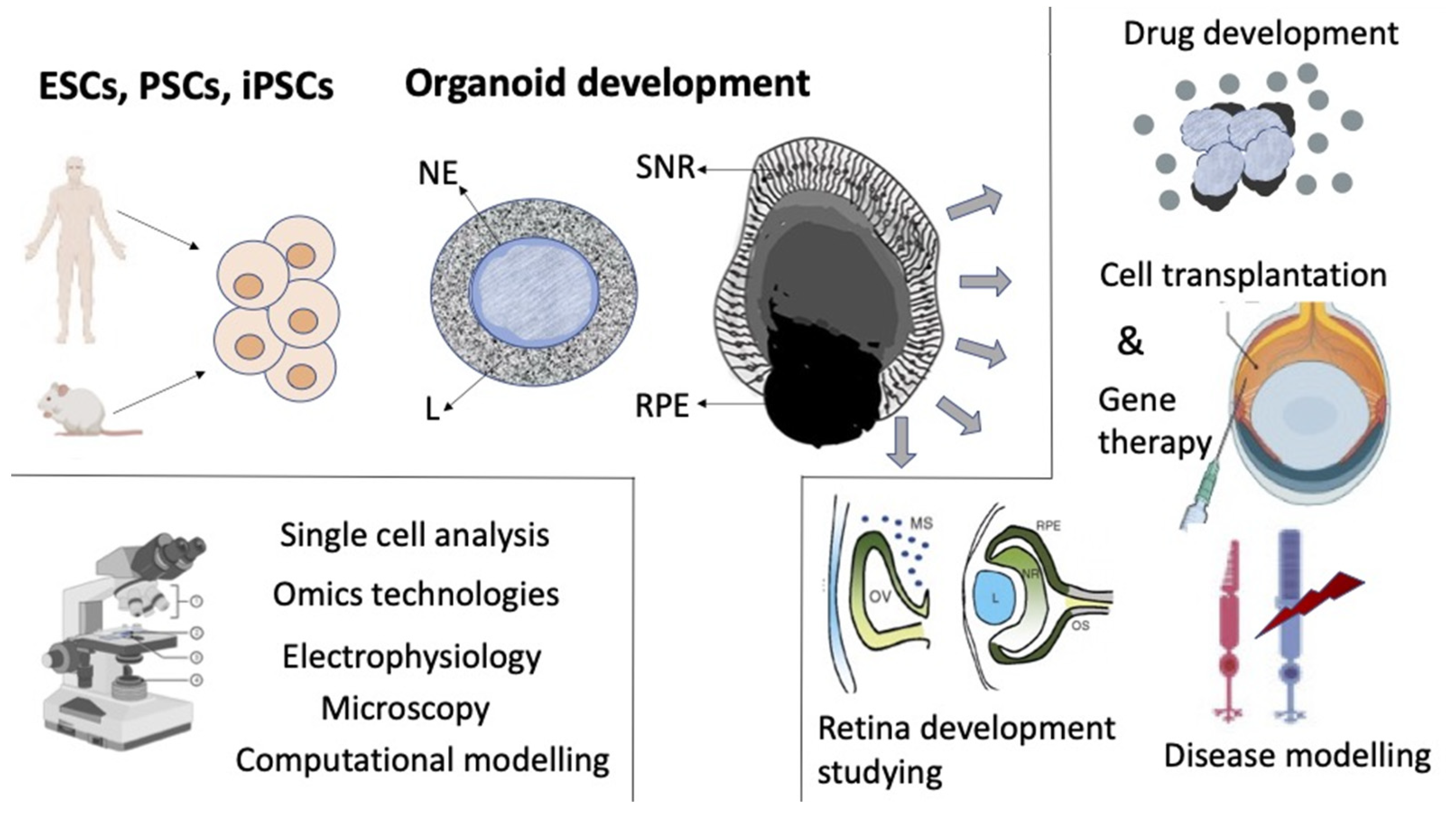
5. Retinal Self-Organization In Vitro
5.1. Embryonic Retina Self-Organization in Reaggregation Culture
5.2. Retinal Self-Organization in the Process of Organoid Formation In Vitro
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gershenson, C. Guiding the self-organization of random Boolean networks. Theory Biosci. 2012, 131, 181–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas, F.; Mediano, P.A.M.; Ugarte, M.; Jensen, H.J. An Information-Theoretic Approach to Self-Organisation: Emergence of Complex Interdependencies in Coupled Dynamical Systems. Entropy 2018, 20, 793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios, E.R.; Razi, A.; Parr, T.; Kirchhoff, M.; Friston, K. On Markov blankets and hierarchical self-organisation. J. Theor. Biol. 2020, 486, 110089. [Google Scholar] [CrossRef] [PubMed]
- Camazine, S. Self-Organization in Biological Systems; Princeton University Press: Princeton, NJ, USA, 2003; p. 562. [Google Scholar]
- Crommelinck, M.; Feltz, B.; Goujon, P. Self-Organization and Emergence in Life Sciences; Springer: Dordrecht, The Netherlands, 2006; p. 360. [Google Scholar]
- Isaeva, V. Self-Organization in Biological Systems. Biol. Bull. 2012, 39, 110–118. [Google Scholar] [CrossRef]
- Werner, S.; Vu, H.T.-K.; Rink, J.C. Self-organization in development, regeneration and organoids. Curr. Opin. Cell Biol. 2016, 44, 102–109. [Google Scholar] [CrossRef]
- Gomez-Galvez, P.; Anbari, S.; Escudero, L.M.; Buceta, J. Mechanics and self-organization in tissue development. Semin. Cell Dev. Biol. 2021, 120, 147–159. [Google Scholar] [CrossRef]
- Kolb, H. Simple Anatomy of the Retina. In Webvision: The Organization of the Retina and Visual System [Internet]; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2005. [Google Scholar]
- Hoon, M.; Okawa, H.; Della Santina, L.; Wong, R.O. Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res. 2014, 42, 44–84. [Google Scholar] [CrossRef] [Green Version]
- Heavner, W.; Pevny, L. Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008391. [Google Scholar] [CrossRef] [Green Version]
- Graw, J. Eye development. Curr. Top. Dev. Biol. 2010, 90, 343–386. [Google Scholar] [CrossRef]
- Picker, A.; Cavodeassi, F.; Machate, A.; Bernauer, S.; Hans, S.; Abe, G.; Kawakami, K.; Wilson, S.W.; Brand, M. Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina. PLoS Biol. 2009, 7, e1000214. [Google Scholar] [CrossRef] [Green Version]
- Kwan, K.M.; Otsuna, H.; Kidokoro, H.; Carney, K.R.; Saijoh, Y.; Chien, C.-B. A complex choreography of cell movements shapes the vertebrate eye. Development 2012, 139, 359–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heermann, S.; Schütz, L.; Lemke, S.; Krieglstein, K.; Wittbrodt, J. Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. eLife 2015, 4, e05216. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.L.; Cepko, C.L. A common progenitor for neurons and glia persists in rat retina late in development. Nature 1987, 328, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Wetts, R.; Fraser, S.E. Multipotent precursors can give rise to all major cell types of the frog retina. Science 1988, 239, 1142–1145. [Google Scholar] [CrossRef]
- Holt, C.E.; Bertsch, T.W.; Ellis, H.M.; Harris, W.A. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1988, 1, 15–26. [Google Scholar] [CrossRef]
- Cepko, C.L.; Austin, C.P.; Yang, X.; Alexiades, M.; Ezzeddine, D. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. USA 1996, 93, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Harris, W.A. Cellular diversification in the vertebrate retina. Curr. Opin. Genet. Dev. 1997, 7, 651–658. [Google Scholar] [CrossRef]
- Belliveau, M.J.; Cepko, C.L. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development 1999, 126, 555–566. [Google Scholar] [CrossRef]
- Livesey, F.J.; Cepko, C.L. Vertebrate neural cell-fate determination: Lessons from the retina. Nat. Rev. Neurosci. 2001, 2, 109–118. [Google Scholar] [CrossRef]
- Aldiri, I.; Xu, B.; Wang, L.; Chen, X.; Hiler, D.; Griffiths, L.; Valentine, M.; Shirinifard, A.; Thiagarajan, S.; Sablauer, A.; et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron 2017, 94, 550–568.e10. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Ratnapriya, R.; Brooks, M.J.; Chaitankar, V.; Wilken, M.S.; Zhang, C.; Starostik, M.R.; Gieser, L.; La Torre, A.; Nishio, M.; et al. Molecular Anatomy of the Developing Human Retina. Dev. Cell. 2017, 43, 763–779.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, M. Intrinsic control of mammalian retinogenesis. Cell Mol. Life Sci. 2013, 70, 2519–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassett, E.A.; Wallace, V.A. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012, 35, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Inoue, T.; Miyoshi, G.; Bessho, Y.; Takahashi, M.; Lee, J.E.; Guillemot, F.; Kageyama, R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 2004, 279, 28492–28498. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-T.; Kim, J.W. Compartmentalization of Vertebrate Optic Neuroepithelium: External Cues and Transcription Factors. Mol. Cells 2012, 33, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Fuhrmann, S.; Zou, C.; Levine, E.M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 2014, 123, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Miesfeld, J.B.; Brown, N.L. Eye organogenesis: A hierarchical view of ocular development. Curr. Top. Dev. Biol. 2019, 132, 351–393. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Simirskii, V.N. Inherited retinal diseases through the eyes of homeobox genes. Int. J. Mol. Sci. 2020, 21, 1602. [Google Scholar] [CrossRef] [Green Version]
- Boije, H.; MacDonald, R.B.; Harris, W.A. Reconciling competence and transcriptional hierarchies with stochasticity in retinal lineages. Curr. Opin. Neurobiol. 2014, 27, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Gomes, F.L.; Zhang, G.; Carbonell, F.; Correa, J.A.; Harris, W.A.; Simons, B.D.; Cayouette, M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 2011, 138, 227–235. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zhang, G.; Almeida, A.D.; Cayouette, M.; Simons, B.D.; Harris, W.A. How variable clones build an invariant retina. Neuron 2012, 75, 786–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, E.S.; Stubbs, J.L.; Levine, E.M. Genetic rescue of cell number in a mouse model of microphthalmia: Interactions between Chx10 and G1-phase cell cycle regulators. Development 2003, 130, 539–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randlett, O.; Macdonald, R.B.; Yoshimatsu, T.; Almeida, A.D.; Suzuki, S.C.; Wong, R.O.; Harris, W.A. Cellular requirements for building a retinal neuropil. Cell Rep. 2013, 3, 282–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldred, M.K.; Charlton-Perkins, M.; Muresan, L.; Harris, W.A. Self-organising aggregates of zebrafish retinal cells for investigating mechanisms of neural lamination. Development 2017, 144, 1097–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trimarchi, J.M.; Stadler, M.B.; Cepko, C.L. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE 2008, 3, e1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakeri, Z.; Penaloza, C.G.; Smith, K.; Ye, Y.; Lockshin, R.A. What cell death does in development. Int. J. Dev. Biol. 2015, 59, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Vecino, E.; Acera, A. Development and programed cell death in the mammalian eye. Int. J. Dev. Biol. 2015, 59, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Valenciano, A.I.; Boya, P.; De La Rosa, E.J. Early neural cell death: Numbers and cues from the developing neuroretina. Int. J. Dev. Biol. 2009, 53, 1515–1528. [Google Scholar] [CrossRef] [Green Version]
- Vecino, E.; Hernandez, M.; Garcia, M. Cell death in the developing vertebrate retina. Int. J. Dev. Biol. 2004, 48, 965–974. [Google Scholar] [CrossRef]
- Braunger, B.M.; Demmer, C.; Tamm, E.R. Programmed cell death during retinal development of the mouse eye. Adv. Exp. Med. Biol. 2014, 801, 9–13. [Google Scholar] [CrossRef]
- Fricker, M.; Tolkovsky, A.M.; Borutaite, V.; Coleman, M.; Brown, G.C. Neuronal Cell Death. Physiol. Rev. 2018, 98, 813–880. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, R.; Niu, S.; Li, Y.; Ma, C.; Zhang, G.; Cong, B. Dynamic changes of proliferation and apoptosis in rat retina development. Int. J. Clin. Exp. Path. 2017, 10, 11679–11684. [Google Scholar]
- Amini, R.; Rocha-Martins, M.; Norden, C. Neuronal Migration and Lamination in the Vertebrate Retina. Front. Neurosci. 2018, 11, 742. [Google Scholar] [CrossRef] [PubMed]
- Reese, B.E.; Keeley, P.W. Design principles and developmental mechanisms underlying retinal mosaics. Biol. Rev. Camb. Philos. Soc. 2015, 90, 854–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reese, B.E.; Harvey, A.R.; Tan, S.S. Radial and tangential dispersion patterns in the mouse retina are cell-class specific. Proc. Natl. Acad. Sci. USA 1995, 92, 2494–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reese, B.E.; Necessary, B.D.; Tam, P.P.; Faulkner-Jones, B.; Tan, S.S. Clonal expansion and cell dispersion in the developing mouse retina. Eur. J. Neurosci. 1999, 11, 2965–2978. [Google Scholar] [CrossRef]
- Kwan, K.M. Coming into focus: The role of extracellular matrix in vertebrate optic cup morphogenesis. Dev. Dyn. 2014, 243, 1242–1248. [Google Scholar] [CrossRef]
- Fuhrmann, S.; Levine, E.M.; Reh, T.A. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development 2000, 127, 4599–4609. [Google Scholar] [CrossRef]
- Bryan, C.D.; Casey, M.A.; Pfeiffer, R.L.; Jones, B.W.; Kwan, K.M. Optic cup morphogenesis requires neural crest-mediated basement membrane assembly. Development 2020, 147, dev181420. [Google Scholar] [CrossRef] [Green Version]
- Grocott, T.; Johnson, S.; Bailey, A.P.; Streit, A. Neural crest cells organize the eye via TGF-β and canonical Wnt signaling. Nat. Commun. 2011, 2, 266–269. [Google Scholar] [CrossRef] [Green Version]
- Glinka, A.; Wu, W.; Onichtchouk, D.; Blumenstock, C.; Niehrs, C. Head induction by simultaneous repression of Bmp and Wnt signaling in Xenopus. Nature 1997, 389, 517–519. [Google Scholar] [CrossRef] [PubMed]
- Lamba, D.A.; Karl, M.O.; Ware, C.B.; Reh, T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 12769–12774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, T.; Jidigam, V.K.; Patthey, C.; Gunhaga, L. Neural retina identity is specified by lens-derived BMP signals. Development 2015, 142, 1850–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pera, E.M.; Wessely, O.; Li, S.-Y.; De Robertis, E.M. Neural and head induction by insulin-like growth factor signals. Dev. Cell 2001, 1, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Richard-Parpaillon, L.; Heligon, C.; Chesnel, F.; Boujard, D.; Philpott, A. The IGF pathway regulates head formation by inhibiting Wnt signaling in Xenopus. Dev. Biol. 2002, 244, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Cavodeassi, F.; Carreira-Barbosa, F.; Young, R.M.; Concha, M.L.; Allende, M.L.; Houart, C.; Tada, M.; Wilson, S.W. Early Stages of Zebrafish Eye Formation Require the Coordinated Activity of Wnt11, Fz5, and the Wnt/β-Catenin Pathway. Neuron 2005, 47, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, N. WNT/β-Catenin Signaling in Vertebrate Eye Development. Front. Cell Dev. Biol. 2016, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Morales, J.R.; Rodrigo, I.; Bovolenta, P. Eye development: A view from the retina pigmented epithelium. Bioessays 2004, 26, 766–777. [Google Scholar] [CrossRef]
- Wagstaff, P.E.; Berzal, A.H.; Boon, C.J.F.; Quinn, P.M.J.; ten Asbroek, A.L.M.A.; Bergen, A.A. The Role of Small Molecules and Their Effect on the Molecular Mechanisms of Early Retinal Organoid Development. Int. J. Mol. Sci. 2021, 22, 7081. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Min, X.; Quinn, P.M.J.; Giudice, Q.L.; Tao, C.; Polanco, K.; Makrides, N.; Peregrin, J.; Bouaziz, M.; Mao, Y.; et al. Phase transition specified by a binary code patterns the vertebrate eye cup. Sci. Adv. 2021, 7, eabj9846. [Google Scholar] [CrossRef]
- Seritrakul, P.; Gross, J.M. Genetic and epigenetic control of retinal development in zebrafish. Curr. Opin. Neurobiol. 2019, 59, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, K.L.; Zaghloul, N.; Moody, S.A. Transcription factors of the anterior neural plate alter cell movements of epidermal progenitors to specify a retinal fate. Dev. Biol. 2001, 240, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinn, R.; Wittbrodt, J. An eye on eye development. Mech. Dev. 2013, 130, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Zuber, M.E.; Gestri, G.; Viscian, A.S.; Barsacchi, G.; Harris, W.A. Specification of the vertebrate eye by a network of eye field transcription factors. Development 2003, 130, 5155–5167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, A.; Nagano, T.; Takehara, S.; Hibi, M.; Aizawa, S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell 2005, 120, 223–235. [Google Scholar] [CrossRef] [Green Version]
- Luz-Madrigal, A.; Grajales-Esquivel, E.; McCorkle, A.; DiLorenzo, A.M.; Barbosa-Sabanero, K.; Tsonis, P.A.; Del Rio-Tsonis, K. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 2014, 12, 28. [Google Scholar] [CrossRef] [Green Version]
- Steinfeld, J.; Steinfeld, I.; Bausch, A.; Coronato, N.; Hampel, M.-L.; Depner, D.; Layer, P.G.; Vogel-Höpker, A. BMP-induced reprogramming of the neural retina into retinal pigment epithelium requires Wnt signaling. Biol. Open. 2017, 6, 979–992. [Google Scholar] [CrossRef] [Green Version]
- Mitashov, V.I. Mechanisms of retina regeneration in vertebrates. Int. J. Dev. Biol. 1996, 40, 833–844. [Google Scholar]
- Yoshii, C.; Ueda, Y.; Okamoto, M.; Araki, M. Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: Transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev. Biol. 2007, 303, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Rowan, S.; Chen, C.M.; Young, T.L.; Fisher, D.E.; Cepko, C.L. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 2004, 131, 5139–5152. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Raff, M.C. Rod photoreceptor development in vitro: Intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron 1990, 4, 461–467. [Google Scholar] [CrossRef]
- Hatakeyama, J.; Kageyama, R. Retinal cell fate determination and bHLH factors. Semin. Cell Dev. Biol. 2004, 15, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.A.; Cho, J.H.; Wang, J.; Gao, Z.; Pan, P.; Tsai, W.W.; Frishman, L.J.; Klein, W.H. Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development 2013, 140, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquardt, T.; Ashery-Padan, R.; Andrejewski, N.; Scardigli, R.; Guillemot, F.; Gruss, P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 2001, 105, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Oron-Karni, V.; Farhy, C.; Elgart, M.; Marquardt, T.; Remizova, L.; Yaron, O.; Xie, Q.; Cvekl, A.; Ashery- Padan, R. Dual requirement for Pax6 in retinal progenitor cells. Development 2008, 135, 4037–4047. [Google Scholar] [CrossRef] [Green Version]
- Miesfeld, J.B.; Glaser, T.; Brown, N.L. The dynamics of native Atoh7 protein expression during mouse retinal histogenesis, revealed with a new antibody. Gene Expr. Patterns 2018, 27, 114–121. [Google Scholar] [CrossRef] [Green Version]
- Jasoni, C.L.; Reh, T.A. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J. Comp. Neurol. 1996, 369, 319–327. [Google Scholar] [CrossRef]
- Nakamura, K.; Harada, C.; Namekata, K.; Harada, T. Expression of olig2 in retinal progenitor cells. NeuroReport 2006, 17, 345–349. [Google Scholar] [CrossRef]
- Shibasaki, K.; Takebayashi, H.; Ikenaka, K.; Feng, L.; Gan, L. Expression of the basic helix-loop-factor Olig2 in the developing retina: Olig2 as a new marker for retinal progenitors and late-born cells. Gene Expr. Patterns 2007, 7, 57–65. [Google Scholar] [CrossRef]
- Brzezinski, J.A., 4th; Kim, E.J.; Johnson, J.E.; Reh, T.A. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development 2011, 138, 3519–3531. [Google Scholar] [CrossRef] [Green Version]
- Elliott, J.; Jolicoeur, C.; Ramamurthy, V.; Cayouette, M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 2008, 60, 26–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsawa, R.; Kageyama, R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008, 1192, 90–98. [Google Scholar] [CrossRef]
- Clark, A.M.; Yun, S.; Veien, E.S.; Wu, Y.Y.; Chow, R.L.; Dorsky, R.I.; Levine, E.M. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 2008, 1192, 99–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usui, A.; Mochizuki, Y.; Iida, A.; Miyauchi, E.; Satoh, S.; Sock, E.; Nakauchi, H.; Aburatani, H.; Murakami, A.; Wegner, M.; et al. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development 2013, 140, 740–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, P.; Hoang, T.; Santiago, C.P.; Thomas, E.D.; Timms, A.E.; Appel, H.; Gimmen, M.; Le, N.; Jiang, L.; Kim, D.W.; et al. Gene regulatory networks controlling temporal patterning, neurogenesis, and cell-fate specification in mammalian retina. Cell Rep. 2021, 37, 109994. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.C.; Harris, W.A. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev. Biol. 2005, 285, 101–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Serb, J.M.; Greenlee, M.H. Mouse retinal development: A dark horse model for systems biology research. Bioinform. Biol. Insights 2011, 5, 99–113. [Google Scholar] [CrossRef] [Green Version]
- Buono, L.; Martinez-Morales, J.R. Retina development in vertebrates: Systems biology approaches to understanding genetic programs: On the contribution of next-generation sequencing methods to the characterization of the regulatory networks controlling vertebrate eye development. Bioessays 2020, 42, e1900187. [Google Scholar] [CrossRef]
- Norrie, J.L.; Lupo, M.S.; Xu, B.; Al Diri, I.; Valentine, M.; Putnam, D.; Griffiths, L.; Zhang, J.; Johnson, D.; Easton, J.; et al. Nucleome Dynamics during Retinal Development. Neuron 2019, 104, 512–528.e11. [Google Scholar] [CrossRef]
- Raeisossadati, R.; Ferrari, M.F.R.; Kihara, A.H.; AlDiri, I.; Gross, J.M. Epigenetic regulation of retinal development. Epigenetics Chromatin 2021, 14, 11. [Google Scholar] [CrossRef]
- Daghsni, M.; Aldiri, I. Building a Mammalian Retina: An Eye on Chromatin Structure. Front. Genet. 2021, 12, 775205. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007, 282, 25053–25066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, D.G.; Oliveira-Fernandes, M.; Lavker, R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006, 12, 1175–1184. [Google Scholar] [PubMed]
- Hackler, L., Jr.; Wan, J.; Swaroop, A.; Qian, J.; Zack, D.J. MicroRNA profile of the developing mouse retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1823–1831. [Google Scholar] [CrossRef]
- Cremisi, F. MicroRNAs and cell fate in cortical and retinal development. Front. Cell Neurosci. 2013, 7, 141. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Taylor, R.J.; La Torre, A.; Wilken, M.S.; Cox, K.E.; Reh, T.A.; Vetter, M.L. Ezh2 Maintains Retinal Progenitor Proliferation, Transcriptional Integrity, and the Timing of Late Differentiation. Dev. Biol. 2015, 403, 128–138. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, N.; Kuzelova, A.; Ebert, A.; Strnad, H.; Lachova, J.; Machon, O.; Busslinger, M.; Kozmik, Z. Polycomb Repression Complex 2 Is Required for the Maintenance of Retinal Progenitor Cells and Balanced Retinal Differentiation. Dev. Biol. 2018, 433, 47–60. [Google Scholar] [CrossRef]
- Chow, R.L.; Altmann, C.R.; Lang, R.A.; Hemmati-Brivanlou, A. Pax6 induces ectopic eyes in a vertebrate. Development 1999, 126, 4213–4222. [Google Scholar] [CrossRef]
- Gehring, W.J. Chance and Necessity in Eye Evolution. Genome Biol. Evol. 2011, 3, 1053–1066. [Google Scholar] [CrossRef] [Green Version]
- Grocott, T.; Lozano-Velasco, E.; Mok, J.F.; Münsterberg, A.E. The Pax6 master control gene initiates spontaneous retinal development via a self-organising Turing network. Development 2020, 147, dev185827. [Google Scholar] [CrossRef] [PubMed]
- Turing, A. The Chemical Basis of Morphogenesis (PDF). Philos. Trans. R. Soc. Lond. B 1952, 237, 37–72. [Google Scholar]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Souza, S.; Lang, L.A. Retinal ganglion cell interactions shape the developing mammalian visual system. Development 2020, 147, dev196535. [Google Scholar] [CrossRef]
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Polacheck, W.J.; German, A.E.; Mammoto, A.; Ingber, D.E.; Kamm, R.D. Mechanotransduction of fluid stresses governs 3D cell migration. Proc. Natl. Acad. Sci. USA 2014, 111, 2447–2452. [Google Scholar] [CrossRef] [Green Version]
- Okuda, S.; Takata, N.; Hasegawa, Y.; Kawada, M.; Inoue, Y.; Adachi, T.; Sasai, Y.; Eiraku, M. Strain-triggered mechanical feedback in self-organizing optic-cup morphogenesis. Sci. Adv. 2018, 4, eaau1354. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Adachi, N.; Sasai, Y. Relaxation-expansion model for self-driven retinal morphogenesis: A hypothesis from the perspective of biosystems dynamics at the multi-cellular level. Bioessays 2012, 34, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Ching Chan, B.H.; Moosajee, M.; Rainger, J. Closing the Gap: Mechanisms of Epithelial Fusion during Optic Fissure Closure. Front. Cell Dev. Biol. 2020, 8, 620774. [Google Scholar] [CrossRef]
- Chakraborty, S.; Njah, K.; Pobbati, A.V.; Lim, Y.B.; Raju, A.; Lakshmanan, M.; Tergaonkar, V.; Lim, C.T.; Hong, W. Agrin as a mechanotransduction signal regulating YAP through the Hippo pathway. Cell Rep. 2017, 18, 2464–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miesfeld, J.B.; Gestri, G.; Clark, B.S.; Flinn, M.A.; Poole, R.J.; Bader, J.R.; Besharse, J.C.; Wilson, S.W.; Link, B.A. Yap and Taz regulate retinal pigment epithelial cell fate. Development 2015, 142, 3021–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiore, L.; Takata, N.; Acosta, A.; Ma, W.; Pandit, T.; Oxendine, M.; Oliver, G. Optic vesicle morphogenesis requires primary cilia. Dev. Biol. 2020, 462, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mármol, T.; Ledesma-Terrón, M.; Tabanera, N.; Martin-Bermejo, M.J.; Cardozo, J.M.J.; Cavodeassi, F.; Bovolenta, P. Stretching of the retinal pigment epithelium contributes to zebrafish optic cup morphogenesis. eLife 2021, 10, e63396. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.S.; Taber, L.A. How mechanical forces shape the developing eye. Prog. Biophys. Mol. Biol. 2018, 137, 25–36. [Google Scholar] [CrossRef]
- Cavodeassi, F. Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models. J. Dev. Biol. 2018, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Fortune, B. Pulling and Tugging on the Retina: Mechanical Impact of Glaucoma Beyond the Optic Nerve Head. Investig. Ophthalmol. Vis. Sci. 2019, 60, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Nicolás-Pérez, M.; Kuchling, F.; Letelier, J.; Polvillo, R.; Wittbrodt, J.; Martínez-Morales, J.R. Analysis of cellular behavior and cytoskeletal dynamics reveal a constriction mechanism driving optic cup morphogenesis. eLife 2016, 5, e15797. [Google Scholar] [CrossRef]
- Eiraku, M.; Sasai, Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr. Opin. Neurobiol. 2012, 22, 768–777. [Google Scholar] [CrossRef]
- Roberts, P.A.; Gaffney, E.A.; Luthert, P.J.; Foss, A.J.E.; Byrne, H.M. Mathematical and computational models of the retina in health, development and disease. Prog. Retin. Eye Res. 2016, 53, 48–69. [Google Scholar] [CrossRef]
- Oltean, A.; Huang, J.; Beebe, D.C.; Taber, L.A. Tissue growth constrained by extracellular matrix drives invagination during optic cup morphogenesis. Biomech. Model. Mechanobiol. 2016, 15, 1405–1421. [Google Scholar] [CrossRef] [PubMed]
- Clements, R.; Turk, R.; Campbell, K.P.; Wright, K.M. Dystroglycan Maintains Inner Limiting Membrane Integrity to Coordinate Retinal Development. J. Neurosci. 2017, 37, 8559–8574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulombre, A. The role of intraocular pressure in the development of the chick eye. J. Exp. Zool. 1956, 133, 211–225. [Google Scholar] [CrossRef]
- Butler, M.T.; Wallingford, J.B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 375–388. [Google Scholar] [CrossRef]
- Álvarez-Hernán, G.; Garrido-Jiménez, S.; Román, A.C.; Carvajal-González, H.M.; Francisco-Morcillo, J. Distribution of planar cell polarity proteins in the developing avian retina. Exp. Eye Res. 2021, 209, 108681. [Google Scholar] [CrossRef]
- Hasegawa, M. Restitution of the eye after removal of the retina and lens in the newt Triturus pyrrhogaster. Embryologia 1975, 8, 362–386. [Google Scholar] [CrossRef]
- Keefe, J.R. An analysis of urodelean retinal regeneration. I-IV. J. Exp. Zool. 1973, 184, 185–257. [Google Scholar] [CrossRef]
- Mitashov, V.I. Retinal regeneration in amphibians. Int. J. Dev. Biol. 1997, 41, 893–905. [Google Scholar]
- Grigoryan, E.N. Endogenous cell sources for eye retina regeneration in vertebrate animals and human. Russ. J. Dev. Biol. 2018, 49, 314–326. [Google Scholar] [CrossRef]
- Chiba, C.; Mitashov, V.I. Cellular and molecular events in the adult newt retinal regeneration. In Strategies for Retinal Tissue Repair and Regeneration in Vertebrates: From Fish to Human; Chiba, C., Ed.; Research Signpost: Kerala, India, 2007; pp. 15–33. [Google Scholar]
- Yasumuro, H.; Sakurai, K.; Toyama, F.; Maruo, F.; Chiba, C. Implications of a Multi-Step Trigger of Retinal Regeneration in the Adult Newt. Biomedicines 2017, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Sarthy, P.V.; Lam, D.M.K. Retinal regeneration in the adult newt, Notophthalmus viridescens: Appearance of neurotransmitter synthesis and the electroretinogram. Dev. Brain Res. 1983, 6, 99–105. [Google Scholar] [CrossRef]
- Cheon, E.W.; Kaneko, Y.; Saito, T. Regeneration of the newt retina: Order of appearance of photoreceptors and ganglion cells. Comp. Neurol. 1998, 396, 267–274. [Google Scholar] [CrossRef]
- Grigoryan, E.N.; Bazhin, A.V.; Krasnov, M.S.; Philippov, P.P. Study of calcium-binding protein recoverin expression in normal, surviving and regenerating retina of the newt Pleurodeles waltl. Klet. Tekhnol. Biol. Med. 2009, N3, 169–173. [Google Scholar]
- Markitantova, Y.V.; Makar’ev, E.O.; Smirnova, Y.A.; Zinov’eva, R.D.; Mitashov, V.I. Analysis of the expression pattern of regulatory genes pax6, prox1, and six3 during regeneration of eye structures in the newt. Biol. Bull. 2004, 31, 428–436. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N.; Zinovieva, R.D. Identification of the pitx1 embryogenesis regulatory gene in a regenerating newt retina. Dokl. Biol. Sci. 2010, 435, 421–424. [Google Scholar] [CrossRef]
- Sakami, S.; Hisatomi, O.; Sakakibara, S.; Liu, J. Downregulation of Otx2 in the dedifferentiated RPE cells of regenerating newt retina. Dev. Brain Res. 2005, 155, 49–59. [Google Scholar] [CrossRef]
- Avdonin, P.P.; Markitantova, Y.V.; Zinovieva, R.D.; Mitashov, V.I. Expression of regulatory genes Pax6, Otx2, Six3, and FGF2 during newt retina regeneration. Izv. Akad. Nauk Ser. Biol. 2008, 4, 414–421. [Google Scholar]
- Maki, N.; Suetsugu-Maki, R.; Tarui, H.; Agata, K.; Del Rio-Tsonis, K.; Tsonis, P.A. Expression of stem cell pluripotency factors during regeneration in newts. Dev. Dynam. 2009, 238, 1613–1616. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.R.; Nakamura, K.; Casco-Robles, M.M.; Kunahong, A.; Inami, W.; Toyama, F.; Maruo, F.; Chiba, C. The newt reprograms mature RPE cells into a unique multipotent state for retinal regeneration. Sci. Rep. 2014, 4, 6043. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, Y.; Hirota, K.; Matsumoto, G.; Hanyu, Y. Expression pattern of a newt Notch homologue in regenerating newt retina. Brain Res. Dev. Brain Res. 2001, 31, 53–62. [Google Scholar] [CrossRef]
- Nakamura, K.; Chiba, C. Evidence for Notch signaling involvement in retinal regeneration of adult newt. Brain Res. 2007, 1136, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Mercer, S.E.; Cheng, C.H.; Atkinson, D.L.; Krcmery, J.; Guzman, C.E.; Kent, D.T.; Zukor, K.; Marx, K.A.; Odelberg, S.J.; Simon, H.G. Multi-tissue microarray analysis identifies a molecular signature of regeneration. PLoS ONE 2012, 7, e52375. [Google Scholar] [CrossRef] [Green Version]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N. FGF2 signaling pathway components in tissues of the posterior eye sector in the adult newt Pleurodeles waltl. Izv. Akad Nauk Ser. Biol. 2014, 4, 325–333. [Google Scholar] [CrossRef]
- Mitusda, S.; Yoshii, C.; Ikegami, Y.; Araki, M. Tissue interaction between the retinal pigment epithelium and the choroid triggers retinal regeneration of the newt Cynops pyrrhogaster. Dev. Biol. 2005, 280, 122–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, Y.; Matsumoto, G.; Hanyu, Y. The occurrence of apoptosis during retinal regeneration in adult newts. Brain Res. Dev. Brain Res. 1999, 117, 225–228. [Google Scholar] [CrossRef]
- Behrndt, M.; Salbreux, G.; Campinho, P.; Hauschild, R.; Oswald, F.; Roensch, J.; Grill, S.W.; Heisenberg, C.P. Forces driving epithelial spreading in zebrafish gastrulation. Science 2012, 338, 257–260. [Google Scholar] [CrossRef]
- Hasegawa, A.; Hisatomi, O.; Yamamoto, S.; Ono, E.; Tokunaga, F. Stathmin expression during newt retina regeneration. Exp Eye Res. 2007, 85, 518–527. [Google Scholar] [CrossRef]
- Grigoryan, E.N.; Mitashov, V.I. Cultivation of retinal pigment epithelium in the cavity of lensectomized newt eye. Ontog. 1985, 16, 34–43. [Google Scholar]
- Sologub, A.A. Mechanisms of repression and derepression of artificial transformation of pigmented epithelium into retina in Xenopus laevis. Rouxs Arch. Dev. Biol. 1977, 182, 277–291. [Google Scholar] [CrossRef]
- Sakaguchi, D.S.; Janick, L.M.; Reh, T.A. Basic Fibroblast Growth Factor (FGF-2) induced transdifferentiation of retinal pigment epithelium: Generation of retinal neurons and glia. Dev. Dyn. 1997, 209, 387–398. [Google Scholar] [CrossRef]
- Mitashov, V.I.; Grigoryan, E.N.; Chernova, N.V. Radioautographic study of nonhistone protein synthesis in the process of lens and retina regeneration in adult tritons. Ontogenez 1983, 14, 390–397. [Google Scholar] [PubMed]
- Grigoryan, E.N.; Markitantova, Y.V. Cellular and molecular preconditions for retinal pigment epithelium (RPE) natural reprogramming during retinal regeneration in Urodela. Biomedicines 2016, 4, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigoryan, E.N. Potential endogenous cell sources for retinal regeneration in vertebrates and humans: Progenitor traits and specialization. Biomedicines 2020, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, E.N. High regenerative ability of tailed amphibians (Urodela) as a result of the expression of juvenile traits by mature animals. Russ. J. Dev. Biol. 2016, 47, 83–92. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Molecular factors of the maintenance and activation of the juvenile phenotype of cellular sources for eye tissue regeneration. Biochemistry 2018, 83, 1627–1642. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Study of Natural Long-life Juvenility and Tissue Regeneration in Caudate Amphibians and Potential Application of Resulting Data in Biomedicine. J. Dev. Biol. 2021, 9, 2. [Google Scholar] [CrossRef]
- Araki, M. Regeneration of the amphibian retina: Role of tissue interaction and related signaling molecules on RPE transdifferentiation. Dev. Growth Differ. 2007, 49, 109–120. [Google Scholar] [CrossRef]
- Vergara, M.N.; Del Rio-Tsonis, K. Retinal regeneration in the Xenopus laevis tadpole: A new model system. Mol. Vis. 2009, 15, 1000–1013. [Google Scholar]
- Herbst, C. Uber das Auseinandergehen von Furchungs- und Gewebezellen in kalkfreiem Medium. Arch. Entwickl. Organ. 1900, 9, 424–463. [Google Scholar] [CrossRef]
- Wilson, H.V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 1907, 5, 245–258. [Google Scholar] [CrossRef]
- Moscona, A.; Moscona, H. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J. Anat. 1952, 86, 287–301. [Google Scholar] [PubMed]
- Moscona, A. Rotation-mediated histogenetic aggregation of dissociated cells. A quantifiable approach to cell interactions in vitro. Exp. Cell Res. 1961, 22, 455–475. [Google Scholar] [CrossRef]
- Sheffield, J.B.; Moscona, A.A. Early stages in the reaggregation of embryonic chick neural retina cells. Exp. Cell Res. 1969, 57, 462–466. [Google Scholar] [CrossRef]
- Layer, P.G.; Willbold, E. Embryonic chicken retinal cells can regenerate all cell layers in vitro, but ciliary pigmented cells induce their correct polarity. Cell Tissue Res. 1989, 258, 233–242. [Google Scholar] [CrossRef]
- Layer, P.G.; Willbold, E. Histogenesis of the avian retina in reaggregation culture: From dissociated cells to laminar neuronal networks. Int. Rev. Cytol. 1993, 146, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Layer, P.G.; Willbold, E. Regeneration of the avian retina by retinospheroid technology. Prog. Retin. Eye Res. 1994, 13, 197–230. [Google Scholar] [CrossRef]
- Wolburg, H.; Willbold, E.; Layer, P.G. Müller glia endfeet, a basal lamina and the polarity of retinal layers form properly in vitro only in the presence of marginal pigmented epithelium. Cell Tissue Res. 1991, 264, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Willbold, E.; Layer, P.G. Müller glia cells and their possible roles during retina differentiation in vivo and in vitro. Histol. Histopathol. 1998, 13, 531–552. [Google Scholar] [CrossRef]
- Rothermel, A.; Willbold, E.; Degrip, W.J.; Layer, P.G. Pigmented epithelium induces complete retinal reconstitution from dispersed embryonic chick retinae in reaggregation culture. Proc. Biol. Sci. 1997, 264, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Layer, P.G.; Robitzki, A.; Rothermel, A.; Willbold, E. Of layers and spheres: The reaggregate approach in tissue engineering. Trends Neurosci. 2002, 25, 131–134. [Google Scholar] [CrossRef]
- Willbold, E.; Layer, P.G. A hidden retinal regenerative capacity from the chick ciliary margin is reactivated in vitro, that is accompanied by down-regulation of butyrylcholinesterase. Eur. J. Neurosci. 1992, 4, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Layer, P.G.; Rothermel, A.; Willbold, E. From stem cells towards neural layers: A lesson from reaggregated embryonic retinal cells. Neuroreport 2001, 12, A39–A46. [Google Scholar] [CrossRef] [PubMed]
- Bytyqi, A.H.; Bachmann, G.; Rieke, M.; Paraoanu, L.E.; Layer, P.G. Cell-by-cell Reconstruction in Reaggregates from Neonatal Gerbil Retina Begins from the Inner Retina and Is Promoted by Retinal Pigmented Epithelium. Eur. J. Neurosci. 2007, 26, 1560–1574. [Google Scholar] [CrossRef] [PubMed]
- Caffe, A.R.; Visser, H.; Jansen, H.G.; Sanyal, S. Histotypic differentiation of neonatal mouse retina in organ culture. Curr. Eye Res. 1989, 8, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Raymond, S.M.; Jackson, I.J. The retinal pigmented epithelium is required for development and maintenance of the mouse neural retina. Curr. Biol. 1995, 5, 1286–1295. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, G.; Frohns, F.; Thangaraj, G.; Bausch, A.; Layer, P.G. IPL Sublamination in Chicken Retinal Spheroids Is Initiated via Müller Cells and Cholinergic Differentiation, and Is Disrupted by NMDA Signaling. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4759–4773. [Google Scholar] [CrossRef] [Green Version]
- Nelson, B.R.; Ueki, Y.; Reardon, S.; Karl, M.O.; Georgi, S.; Hartman, B.H.; Lamba, D.A.; Reh, T.A. Genome-wide analysis of Müller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS ONE 2011, 6, e22817. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, R.B.; Randlett, O.; Oswald, J.; Yoshimatsu, T.; Franze, K.; Harris, W.A. Müller glia provide essential tensile strength to the developing retina. J. Cell Biol. 2015, 210, 1075–1083. [Google Scholar] [CrossRef] [Green Version]
- Campbell, L.J.; Levendusky, J.L.; Steines, S.A.; Hyde, D.R. Retinal regeneration requires dynamic Notch signaling. Neural Regen. Res. 2022, 17, 1199–1209. [Google Scholar] [CrossRef]
- Ueki, Y.; Wilken, M.S.; Cox, K.E.; Chipman, L.B.; Bermingham-Mcdonogh, O.; Reh, T.A. A transient wave of BMP signaling in the retina is necessary for Müller glial differentiation. Development 2015, 142, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Almeida, A.D.; Boije, H.; Chow, R.W.; He, J.; Tham, J.; Suzuki, S.C.; Harris, W.A. Spectrum of Fates: A new approach to the study of the developing zebrafish retina. Development 2014, 141, 1971–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldred, M.K.; Muresan, L.; Harris, W.A. Disaggregation and Reaggregation of Zebrafish Retinal Cells for the Analysis of Neuronal Layering. Methods Mol. Biol. 2019, 1576, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Willbold, E.; Rothermel, A.; Tomlinson, S.; Layer, P.G. Müller glia cells reorganize reaggregating chicken retinal cells into correctly laminated in vitro retinae. Glia 2000, 29, 45–57. [Google Scholar] [CrossRef]
- Lindqvist, N.; Liu, Q.; Zajadacz, J.; Franze, K.; Reichenbach, A. Retinal glial (Müller) cells: Sensing and responding to tissue stretch. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1683–1690. [Google Scholar] [CrossRef] [Green Version]
- Reichenbach, A.; Bringmann, A. New functions of Müller cells. Glia 2013, 61, 651–678. [Google Scholar] [CrossRef]
- Puech, P.-H.; Poole, K.; Knebel, D.; Muller, D.J. A new technical approach to quantify cell–cell adhesion forces by AFM. Ultramicroscopy 2006, 106, 637–644. [Google Scholar] [CrossRef]
- Maître, J.-L.; Berthoumieux, H.; Krens, S.F.G.; Salbreux, G.; Julicher, F.; Paluch, E.; Heisenberg, C.-P. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 2012, 338, 253–256. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takada, S.; Takada, R.; Takeichi, M. Identification of the laminar-inducing factor: Wnt-signal from the anterior rim induces correct laminar formation of the neural retina in vitro. Dev. Biol. 2003, 260, 414–425. [Google Scholar] [CrossRef] [Green Version]
- Volpert, K.N.; Tombran-Tink, J.; Barnstable, C.; Layer, P.G. PEDF and GDNF are key regulators of photoreceptor development and retinal neurogenesis in reaggregates from chick embryonic retina. J. Ocul. Biol. Dis. Inform. 2009, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, N.; Zinyk, D.; Ringuette, R.; Wallace, V.; Schuurmans, C. Heterochronic Pellet Assay to Test Cell-cell Communication in the Mouse Retina. Bio Protoc. 2017, 7, e2117. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.L.; Knoblich, J.A. 2014. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y.; Eiraku, M.; Suga, H. In vitro organogenesis in three dimensions: Self-organising stem cells. Development 2012, 139, 4111–4121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, J.S.; Shearer, R.L.; Capowski, E.E.; Wright, L.S.; Wallace, K.A.; McMillan, E.L.; Zhang, S.C.; Gamm, D.M. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 16698–16703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, J.S.; Howden, S.E.; Wallace, K.A.; Verhoeven, A.D.; Wright, L.S.; Capowski, E.E.; Pinilla, I.; Martin, J.M.; Tian, S.; Stewart, R.; et al. Optic Vesicle-like Structures Derived from Human Pluripotent Stem Cells Facilitate a Customized Approach to Retinal Disease Treatment. Stem Cells 2011, 29, 1206–1218. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef] [Green Version]
- Eldred, K.C.; Hadyniak, S.E.; Hussey, K.A.; Brenerman, B.; Zhang, P.W.; Chamling, X.; Sluch, V.M.; Welsbie, D.S.; Hattar, S.; Taylor, J.; et al. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 2018, 362, eaau6348. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Mallela, R.K.; Cornuet, P.K.; Reifler, A.N.; Chervenak, A.P.; West, M.D.; Wong, K.Y.; Nasonkin, I.O. Characterization of Three-Dimensional Retinal Tissue Derived from Human Embryonic Stem Cells in Adherent Monolayer Cultures. Stem Cells Dev. 2015, 24, 2778–2795. [Google Scholar] [CrossRef]
- Parfitt, D.A.; Lane, A.; Ramsden, C.M.; Carr, A.J.F.; Munro, P.M.; Jovanovic, K.; Schwarz, N.; Kanuga, N.; Muthiah, M.N.; Hull, S.; et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016, 18, 769–781. [Google Scholar] [CrossRef] [Green Version]
- Volkner, M.; Zschätzsch, M.; Rostovskaya, M.; Overall, R.W.; Busskamp, V.; Anastassiadis, K.; Karl, M.O. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Rep. 2016, 6, 525–538. [Google Scholar] [CrossRef] [Green Version]
- Wahlin, K.J.; Maruotti, J.A.; Sripathi, S.R.; Ball, J.; Angueyra, J.M.; Kim, C.; Grebe, R.; Li, W.; Jones, B.W.; Zack, D.J. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 766. [Google Scholar] [CrossRef]
- Martin, I.; Wendt, D.; Heberer, M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004, 22, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, J. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef] [PubMed]
- Ovando-Roche, P.; West, E.L.; Branch, M.J.; Sampson, R.D.; Fernando, M.; Munro, P.; Georgiadis, A.; Rizzi, M.; Kloc, M.; Naeem, A.; et al. Use of bioreactors for culturing human retinal organoids improves photoreceptor yields. Stem Cell Res. Ther. 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Reichman, S.; Slembrouck, A.; Gagliardi, G.; Chaffiol, A.; Terray, A.; Nanteau, C.; Potey, A.; Belle, M.; Rabesandratana, O.; Duebel, J.; et al. Generation of storable retinal organoids and retinal pigmented epithelium from adherent human iPS cells in xeno-free and feeder-free conditions. Stem Cells 2017, 35, 1176–1188. [Google Scholar] [CrossRef] [Green Version]
- DiStefano, T.; Chen, H.Y.; Panebianco, C.; Kaya, K.D.; Brooks, M.J.; Gieser, L.; Morgan, N.Y.; Pohida, T.; Swaroop, A. Accelerated and improved differentiation of retinal organoids from pluripotent stem cells in rotating-wall vessel bioreactors. Stem Cell Rep. 2018, 10, 300–313. [Google Scholar] [CrossRef] [Green Version]
- Capowski, E.E.; Samimi, K.; Mayerl, S.J.; Phillips, M.J.; Pinilla, I.; Howden, S.E.; Saha, J.; Jansen, A.D.; Edwards, K.L.; Jager, L.D.; et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2019, 146, dev171686. [Google Scholar] [CrossRef] [Green Version]
- Marcos, L.F.; Wilson, S.L.; Roach, P. Tissue engineering of the retina: From organoids to microfluidic chips. J. Tissue Eng. 2021, 12, 20417314211059876. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [Green Version]
- Muncie, J.M.; Weaver, V.M. The physical and biochemical properties of the extracellular matrix regulate cell fate. Curr. Top. Dev. Biol. 2018, 130, 1–37. [Google Scholar] [CrossRef]
- Capowski, E.E.; Simonett, J.M.; Clark, E.M.; Wright, L.S.; Howden, S.E.; Wallace, K.A.; Petelinsek, A.M.; Pinilla, I.; Phillips, M.J.; Meyer, J.S.; et al. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum. Mol. Genet. 2014, 23, 6332–6344. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.J.; Perez, E.T.; Martin, J.M.; Reshel, S.T.; Wallace, K.A.; Capowski, E.E.; Singh, R.; Wright, L.S.; Clark, E.M.; Barney, P.M.; et al. Modeling human retinal development with patient-specific iPS cells reveals multiple roles for VSX2. Stem Cells 2014, 32, 1480–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capowski, E.E.; Wright, L.S.; Liang, K.; Phillips, M.J.; Wallace, K.; Petelinsek, A.; Hagstrom, A.; Pinilla, I.; Borys, K.; Lien, J.; et al. Regulation of WNT signaling by VSX2 during optic vesicle patterning in human induced pluripotent stem cells. Stem Cells 2016, 34, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, A.; Ozone, C.; Nakano, T.; Saito, K.; Eiraku, M.; Sasai, Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015, 6, 6286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellough, C.B.; Collin, J.; Khazim, M.; White, K.; Sernagor, E.; Steel, D.H.; Lako, M. IGF-1 signaling plays an important role in the formation of three-dimensional laminated neural retina and other ocular structures from human embryonic stem cells. Stem Cells 2015, 33, 2416–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamm, D.M.; Clark, E.; Capowski, E.E.; Singh, R. The role of FGF9 in the production of neural retina and RPE in a pluripotent stem cell model of early human retinal development. Am. J. Ophthalmol. 2019, 206, 113–131. [Google Scholar] [CrossRef] [PubMed]
- O’Hara-Wright, M.; Gonzalez-Cordero, A. Retinal organoids: A window into human retinal development. Development 2020, 47, dev189746. [Google Scholar] [CrossRef] [PubMed]
- Collin, J.; Queen, R.; Zerti, D.; Dorgau, B.; Hussain, R.; Coxhead, J.; Cockell, S.; Lako, M. Deconstructing retinal organoids: Single cell RNA-Seq reveals the cellular components of human pluripotent stem cell-derived retina. Stem Cells 2019, 37, 593–598. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Guo, Y.; Zhou, Y.; Mao, S.; Yan, X.; Zeng, Y.; Ding, C.; Chan, H.F.; Tang, S.; Tang, L.; et al. Transcriptomic analysis of the developmental similarities and differences between the native retina and retinal organoids. Investig. Ophthalmol. Vis. Sci. 2020, 61, 6. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659.e4. [Google Scholar] [CrossRef]
- Welby, E.; Lakowski, J.; Di Foggia, V.; Budinger, D.; Gonzalez-Cordero, A.; Lun, A.T.; Epstein, M.; Patel, A.; Cuevas, E.; Kruczek, K.; et al. Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Report. 2017, 9, 1898–1915. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lowe, A.; Dharmat, R.; Lee, S.; Owen, L.A.; Wang, J.; Shakoor, A.; Li, Y.; Morgan, D.J.; Hejazi, A.A.; et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. USA 2019, 116, 10824–10833. [Google Scholar] [CrossRef] [Green Version]
- Langer, K.B.; Ohlemacher, S.K.; Phillips, M.J.; Fligor, C.M.; Jiang, P.; Gamm, D.M.; Meyer, J.S. Retinal Ganglion Cell Diversity and Subtype Specification from Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1282–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; An, Q.; Xi, H.; Yang, X.-J.; Zhang, X.; Yuan, S.; Wang, J.; Hu, Y.; Liu, Q.; Fan, G. Single-cell RNA sequencing of hESC-derived 3D retinal organoids reveals novel genes regulating RPC commitment in early human retinogenesis. Stem Cell Rep. 2019, 13, 747–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llonch, S.; Carido, M.; Ader, M. Organoid technology for retinal repair. Dev. Biol. 2018, 433, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Nasonkin, I.O. Limitations and Promise of Retinal Tissue from Human Pluripotent Stem Cells for Developing Therapies of Blindness. Front. Cell Neurosci. 2020, 14, 179. [Google Scholar] [CrossRef] [PubMed]
- Afanasyeva, T.A.V.; Corral-Serrano, J.C.; Garanto, A.; Roepman, R.; Cheetham, M.E.; Collin, R.W.J. A look into retinal organoids: Methods, analytical techniques, and applications. Cell Mol. Life Sci. 2021, 78, 6505–6532. [Google Scholar] [CrossRef] [PubMed]
- Dakubo, G.D.; Mazerolle, C.; Furimsky, M.; Yu, C.; St-Jacques, B.; McMahon, A.P.; Wallace, V.A. Indian hedgehog signaling from endothelial cells is required for sclera and retinal pigment epithelium development in the mouse eye. Dev. Biol. 2008, 320, 242–255. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.N.; Walker, H.M.; Thompson, H.; Collinson, J.M.; Vargesson, N.; Erskine, L. Lens-regulated retinoic acid signaling controls expansion of the developing eye. Development 2018, 145, dev167171. [Google Scholar] [CrossRef] [Green Version]
- Lowe, A.; Harris, R.; Bhansali, P.; Cvekl, A.; Liu, W. Intercellular Adhesion-Dependent Cell Survival and ROCK-Regulated Actomyosin-Driven Forces Mediate Self-Formation of a Retinal Organoid. Stem Cell Rep. 2016, 6, 743–756. [Google Scholar] [CrossRef] [Green Version]
- McMurtrey, R.J. Analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in three-dimensional tissue constructs with applications and insights in cerebral organoids. Tissue Eng. Part C Methods 2016, 22, 221–249. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Cordero, A.; Kruczek, K.; Naeem, A.; Fernando, M.; Kloc, M.; Ribeiro, J.; Goh, D.; Duran, Y.; Blackford, S.J.; Abelleira-Hervas, L.; et al. Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Rep. 2017, 9, 820–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya, K.D.; Chen, H.Y.; Brooks, M.J.; Kelley, R.A.; Shimada, H.; Nagashima, K.; de Val, N.; Drinnan, C.T.; Gieser, L.; Kruczek, K.; et al. Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. Mol. Vis. 2019, 25, 663–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, S.; Anderson, C.; Zhao, F.; Qin, Y.; Wu, D.; Wu, X.; Liu, J.; He, X.; Zhao, J.; et al. Requirement of Smad4 from ocular surface ectoderm for retinal development. PLoS ONE 2016, 11, e0159639. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, T.; Gruss, P. Generating neuronal diversity in the retina: One for nearly all. Trends Neurosci. 2002, 25, 32–38. [Google Scholar] [CrossRef]
- Montague, P.R.; Friedlander, M.J. Expression of an intrinsic growth strategy by mammalian retinal neurons. Proc. Natl. Acad. Sci. USA 1989, 86, 7223–7227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montague, P.R.; Friedlander, M.J. Morphogenesis and territorial coverage by isolated mammalian retinal ganglion cells. J. Neurosci. 1991, 11, 1440–1457. [Google Scholar] [CrossRef] [Green Version]
- Fligor, C.M.; Langer, K.B.; Sridhar, A.; Ren, Y.; Shields, P.K.; Edler, M.C.; Ohlemacher, S.K.; Sluch, V.M.; Zack, D.J.; Zhang, C.; et al. Three-dimensional retinal organoids facilitate the investigation of retinal ganglion cell development, organization and neurite outgrowth from human pluripotent stem cells. Sci. Rep. 2018, 8, 14520. [Google Scholar] [CrossRef]
- Cohen-Cory, S.; Lom, B. Neurotrophic regulation of retinal ganglion cell synaptic connectivity: From axons and dendrites to synapses. Int. J. Dev. Biol. 2004, 48, 947–956. [Google Scholar] [CrossRef]
- Hamon, A.; Roger, J.E.; Yang, X.J.; Perron, M. Müller glial cell dependent regeneration of the neural retina: An over-view across vertebrate model systems. Dev. Dyn. 2016, 245, 727–738. [Google Scholar] [CrossRef]
- Gao, M.-L.; Zhang, X.; Han, F.; Xu, J.; Yu, S.-J.; Jin, K.; Jin, Z.-B. Functional microglia derived from human pluripotent stem cells empower retinal organs. Sci. China Life Sci. 2022, 65, 1057–1071. [Google Scholar] [CrossRef]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell types of the human retina and its organoids at single-cell resolution. Cell 2020, 182, 1623–1640.e34. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Han, J.; Woo, D.H.; Kim, S.E.; Kim, S.K.; Kang, H.G.; Kim, J.H. Biochemical and morphological effects of hypoxic environment on human embryonic stem cells in long-term culture and differentiating embryoid bodies. Mol. Cells 2011, 31, 123–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Hou, Z.; Gulbranson, D.R.; Thomson, J.A. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 2010, 7, 240–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakada, F.; Jin, Z.B.; Hirami, Y.; Ikeda, H.; Danjyo, T.; Watanabe, K.; Sasai, Y.; Takahashi, M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J. Cell Sci. 2009, 122, 3169–3179. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhang, Y.; Zhang, Y.-Y.; Li, Y.-P.; Hua, Z.-Q.; Zhang, C.-J.; Wu, K.-C.; Yu, F.; Zhang, Y.; Su, J.; et al. Human embryonic stem cell-derived organoid retinoblastoma reveals a cancerous origin. Proc. Natl. Acad. Sci. USA 2020, 117, 33628–33638. [Google Scholar] [CrossRef]
- Gao, M.-L.; Lei, X.-L.; Han, F.; He, K.-W.; Jin, S.-Q.; Zhang, Y.-Y.; Jin, Z.-B. Patient-Specific Retinal Organoids Recapitulate Disease Features of Late-Onset Retinitis Pigmentosa. Front. Cell Dev. Biol. 2020, 8, 128. [Google Scholar] [CrossRef]
- Foltz, L.P.; Howden, S.E.; Thomson, J.A.; Clegg, D.O. Functional Assessment of Patient-Derived Retinal Pigment Epithelial Cells Edited by CRISPR/Cas9. Int. J. Mol. Sci. 2018, 19, 4127. [Google Scholar] [CrossRef] [Green Version]
- Foltz, L.P.; Clegg, D.O. Patient-derived induced pluripotent stem cells for modeling genetic retinal dystrophies. Prog. Retin. Eye Res. 2019, 68, 54–66. [Google Scholar] [CrossRef]
- Jin, Z.-B.; Gao, M.-L.; Deng, W.-L.; Wu, K.-C.; Sugita, S.; Mandai, M.; Takahashi, M. Stemming retinal regeneration with pluripotent stem cells. Prog. Retin. Eye Res. 2019, 69, 38–56. [Google Scholar] [CrossRef]
- Pan, D.; Xia, X.-X.; Zhou, H.; Jin, S.-Q.; Lu, Y.-Y.; Liu, H.; Gao, M.-L.; Jin, Z.-B. COCO enhances the efficiency of photoreceptor precursor differentiation in early human embryonic stem cell-derived retinal organoids. Stem Cell Res. Ther. 2020, 11, 366. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Pigment epithelia of the eye: Cell-type conversion in regeneration and disease. Life 2022, 12, 382. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigoryan, E.N. Self-Organization of the Retina during Eye Development, Retinal Regeneration In Vivo, and in Retinal 3D Organoids In Vitro. Biomedicines 2022, 10, 1458. https://doi.org/10.3390/biomedicines10061458
Grigoryan EN. Self-Organization of the Retina during Eye Development, Retinal Regeneration In Vivo, and in Retinal 3D Organoids In Vitro. Biomedicines. 2022; 10(6):1458. https://doi.org/10.3390/biomedicines10061458
Chicago/Turabian StyleGrigoryan, Eleonora N. 2022. "Self-Organization of the Retina during Eye Development, Retinal Regeneration In Vivo, and in Retinal 3D Organoids In Vitro" Biomedicines 10, no. 6: 1458. https://doi.org/10.3390/biomedicines10061458
APA StyleGrigoryan, E. N. (2022). Self-Organization of the Retina during Eye Development, Retinal Regeneration In Vivo, and in Retinal 3D Organoids In Vitro. Biomedicines, 10(6), 1458. https://doi.org/10.3390/biomedicines10061458




