Applications of Neural Networks in Biomedical Data Analysis
Abstract
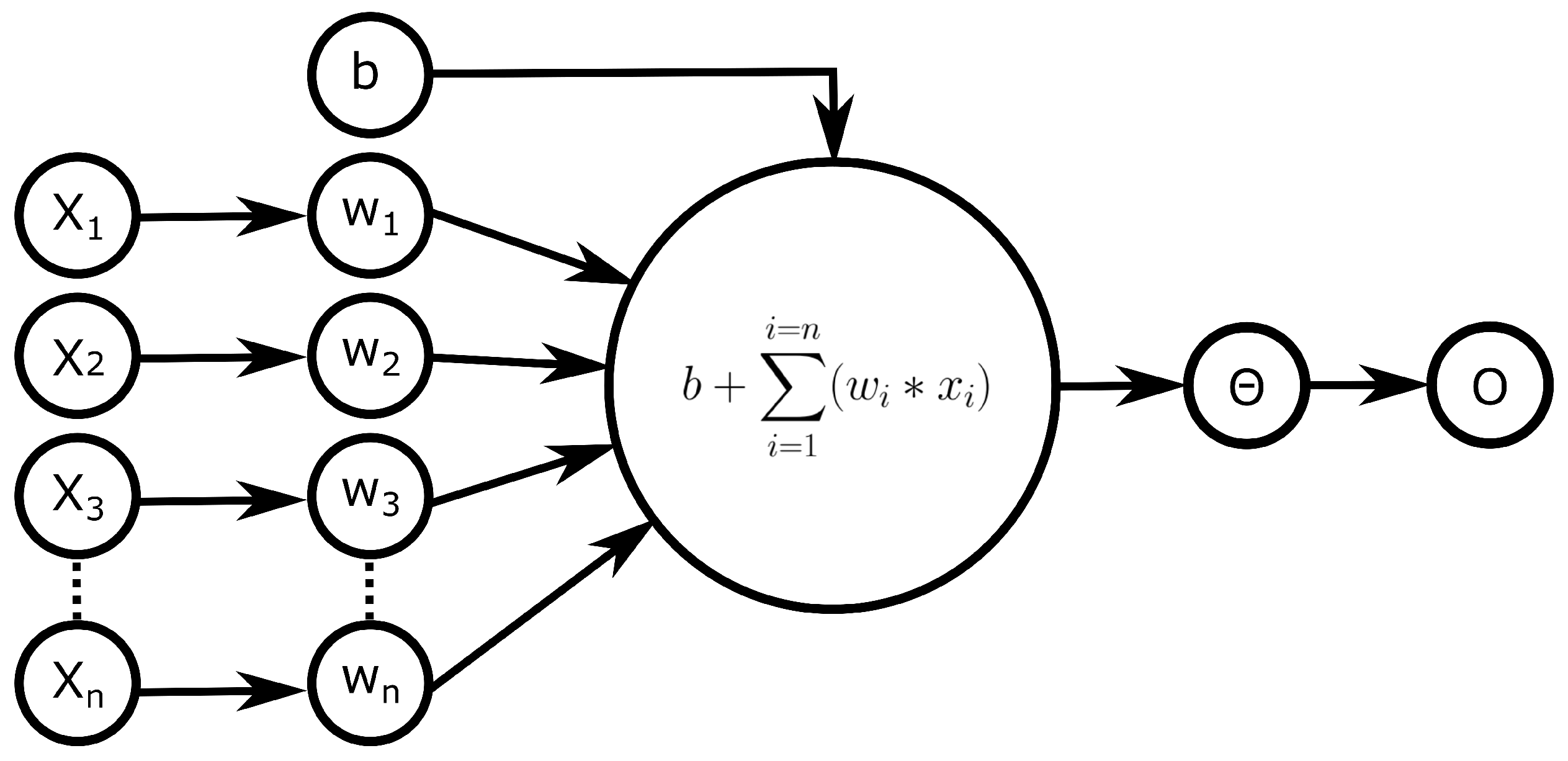
:1. Introduction
| Framework | Programming Languages |
|---|---|
| Tensorflow [37] | Python, R, Java, C++, Go |
| pyTorch [38] | Python |
| sklearn [39] | Python |
| Deeplearning4j [40] | Java |
| caffe [41] | C++, Python, Matlab |
| Keras [42] | Python, R |
| SparkMLlib [43] | Java, Scala, Python, R |
| Deep Java Library [44] | Java |
2. Activation Functions
2.1. Early Activation Functions
2.2. Sigmoid Activation Functions
2.3. Rectified Linear Activation Functions
3. Training
3.1. Gradient Descent
- Initiating a random or all-zero vector value to .
- Modification of in order to decrease .
3.1.1. Batch Gradient Descent
3.1.2. Stochastic Gradient Descent
3.1.3. Mini-Batch Gradient Descent
3.2. Optimisation Algorithms
3.2.1. Momentum
3.2.2. Nesterov Accelerated Gradient
3.2.3. Adagrad
3.2.4. Adadelta
3.2.5. Root Mean Square Propagation
3.2.6. Adam
3.2.7. AdaMax
3.3. Choosing the Right Optimiser
3.4. Back Propagation
4. Potential Training Problems
5. Network Types
5.1. Convolutional and Generative Adversarial Neural Networks
5.2. Recurrent Neural Networks
5.3. Graph Neural Networks
5.4. Transformers
5.5. Challenges of Neural Networks
6. Usage of Neural Networks for Medical Data Analysis
6.1. Scalar Data
- Heart rate.
- Blood pressure.
- Blood type.
- Glucose level.
- Spectroscopic measurements.
- Biomarker concentration.
6.2. Images
- MRI/CT images.
- Tissue section images.
- Immunofluorescence images.
- Retinal images.
6.3. Series Data
- Biomarker concentration over time.
- ECG.
- Live cell imaging.
6.4. Graph Data
- Protein/molecule structures.
- Patient data.
7. Publication Development between 2000 and 2021
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAM | Adaptive Moment Estimation Method |
| AdaDelta | Adaptive Delta |
| Adagrad | Adaptive Subgradient Method |
| ANN | Artificial Neural Network |
| AI | Artificial Intelligence |
| API | Application Programming Interface |
| Backpropagation | Backward Propagation of Error |
| BGD | Batch Gradient Descent |
| BPTT | Backpropagation Through Time |
| CHP | Clever Hans Predictors |
| CNN | Convolutional Neural Networks |
| CT | Computed Tomography |
| COVID-19 | Coronavirus Disease 2019 |
| ECG | Electrocardiography |
| FCN | Fully Convolutional Network |
| GCN | Graph Convolutional Network |
| GNN | Graph Neural Network |
| GRU | Gated Recurrent Unit |
| GPU | Graphics Processing Unit |
| HCA | High-Content Analysis |
| ID | Identifier |
| LSTM | Long Short-Term Memory Network |
| Mini BGD | Mini Batch Gradient Descent |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| MLP | Multilayer Perceptron |
| NAG | Nesterov Accelerated Gradient |
| NLP | Natural Language Processing |
| PPG | Photoplethysmography |
| R-CNN | Regional Convolutional Neural Network |
| ReLU | Rectified Linear Unit function |
| RMS | Root Mean Square |
| RMSprop | Root Mean Square Propagation |
| RNN | Recurrent Neural Network |
| RU | Recurrent Unit |
| SGD | Stochastic Gradient Descent |
| tanh | Tangens Hyperbolicus |
| U-Net | Architecture for Semantic Segmentation |
References
- Zafeiris, D.; Rutella, S.; Ball, G.R. An Artificial Neural Network Integrated Pipeline for Biomarker Discovery Using Alzheimer’s Disease as a Case Study. Comput. Struct. Biotechnol. J. 2018, 16, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.A.; Kohane, I.S.; Manrai, A.K. Biomedical informatics and machine learning for clinical genomics. Hum. Mol. Genet. 2018, 27, R29–R34. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, L.P.; Glory-Afshar, E.; Kangas, J.; Quinn, S.; Shariff, A.; Murphy, R.F. Principles of Bioimage Informatics: Focus on Machine Learning of Cell Patterns. In Linking Literature, Information, and Knowledge for Biology; Hutchison, D., Kanade, T., Kittler, J., Kleinberg, J.M., Mattern, F., Mitchell, J.C., Naor, M., Nierstrasz, O., Pandu Rangan, C., Steffen, B., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6004, pp. 8–18. [Google Scholar] [CrossRef] [Green Version]
- Peng, H. Bioimage informatics: A new area of engineering biology. Bioinformatics 2008, 24, 1827–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Baracchi, D.; Ni, R.; Zhao, Y.; Argenti, F.; Piva, A. A Survey of Deep Learning-Based Source Image Forensics. J. Imaging 2020, 6, 9. [Google Scholar] [CrossRef] [Green Version]
- Thurzo, A.; Kosnáčová, H.S.; Kurilová, V.; Kosmeľ, S.; Beňuš, R.; Moravanský, N.; Kováč, P.; Kuracinová, K.M.; Palkovič, M.; Varga, I. Use of Advanced Artificial Intelligence in Forensic Medicine, Forensic Anthropology and Clinical Anatomy. Healthcare 2021, 9, 1545. [Google Scholar] [CrossRef]
- Schneider, J.; Weiss, R.; Ruhe, M.; Jung, T.; Roggenbuck, D.; Stohwasser, R.; Schierack, P.; Rödiger, S. Open source bioimage informatics tools for the analysis of DNA damage and associated biomarkers. J. Lab. Precis. Med. 2019, 4, 21. [Google Scholar] [CrossRef]
- Meijering, E.; Carpenter, A.E.; Peng, H.; Hamprecht, F.A.; Olivo-Marin, J.C. Imagining the future of bioimage analysis. Nat. Biotechnol. 2016, 34, 1250–1255. [Google Scholar] [CrossRef]
- Chessel, A. An Overview of data science uses in bioimage informatics. Methods 2017, 115, 110–118. [Google Scholar] [CrossRef]
- Cardona, A.; Tomancak, P. Current challenges in open-source bioimage informatics. Nat. Methods 2012, 9, 661–665. [Google Scholar] [CrossRef]
- Rödiger, S.; Schierack, P.; Böhm, A.; Nitschke, J.; Berger, I.; Frömmel, U.; Schmidt, C.; Ruhland, M.; Schimke, I.; Roggenbuck, D.; et al. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. Adv. Biochem. Eng. 2013, 133, 35–74. [Google Scholar] [CrossRef]
- Willitzki, A.; Hiemann, R.; Peters, V.; Sack, U.; Schierack, P.; Rödiger, S.; Anderer, U.; Conrad, K.; Bogdanos, D.P.; Reinhold, D.; et al. New platform technology for comprehensive serological diagnostics of autoimmune diseases. Clin. Dev. Immunol. 2012, 2012, 284740. [Google Scholar] [CrossRef] [Green Version]
- Sowa, M.; Großmann, K.; Scholz, J.; Röber, N.; Rödiger, S.; Schierack, P.; Conrad, K.; Roggenbuck, D.; Hiemann, R. The CytoBead assay—A novel approach of multiparametric autoantibody analysis in the diagnostics of systemic autoimmune diseases. J. Lab. Med. 2015, 38, 000010151520150036. [Google Scholar] [CrossRef]
- Reddig, A.; Rübe, C.E.; Rödiger, S.; Schierack, P.; Reinhold, D.; Roggenbuck, D. DNA damage assessment and potential applications in laboratory diagnostics and precision medicine. J. Lab. Precis. Med. 2018, 3, 31. [Google Scholar] [CrossRef]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef]
- Feng, J.; Lin, J.; Zhang, P.; Yang, S.; Sa, Y.; Feng, Y. A novel automatic quantification method for high-content screening analysis of DNA double strand-break response. Sci. Rep. 2017, 7, 9581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millard, B.L.; Niepel, M.; Menden, M.P.; Muhlich, J.L.; Sorger, P.K. Adaptive informatics for multifactorial and high-content biological data. Nat. Methods 2011, 8, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Shariff, A.; Kangas, J.; Coelho, L.P.; Quinn, S.; Murphy, R.F. Automated Image Analysis for High-Content Screening and Analysis. J. Biomol. Screen. 2010, 15, 726–734. [Google Scholar] [CrossRef] [Green Version]
- Caie, P.D.; Walls, R.E.; Ingleston-Orme, A.; Daya, S.; Houslay, T.; Eagle, R.; Roberts, M.E.; Carragher, N.O. High-Content Phenotypic Profiling of Drug Response Signatures across Distinct Cancer Cells. Mol. Cancer Ther. 2010, 9, 1913–1926. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Tsourkas, A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009, 37, e100. [Google Scholar] [CrossRef] [Green Version]
- Carragher, N.O.; Brunton, V.G.; Frame, M.C. Combining imaging and pathway profiling: An alternative approach to cancer drug discovery. Drug Discov. Today 2012, 17, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Ruhe, M.; Rabe, D.; Jurischka, C.; Schröder, J.; Schierack, P.; Deckert, P.M.; Rödiger, S. Molecular biomarkers of DNA damage in diffuse large-cell lymphoma—A review. J. Lab. Precis. Med. 2019, 4, 5. [Google Scholar] [CrossRef]
- Rabbi, F.; Dabbagh, S.R.; Angin, P.; Yetisen, A.K.; Tasoglu, S. Deep Learning-Enabled Technologies for Bioimage Analysis. Micromachines 2022, 13, 260. [Google Scholar] [CrossRef]
- Everingham, M.; Eslami, S.M.A.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes Challenge: A Retrospective. Int. J. Comput. Vis. 2015, 111, 98–136. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Ciresan, D.C.; Meier, U.; Masci, J.; Gambardella, L.M.; Schmidhuber, J. Flexible, High Performance Convolutional Neural Networks for Image Classification. In Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence, Barcelona, Spain, 16–22 July 2011. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.Z.; Wu, Y.; Gao, Q.Z.; Zhao, L.; Xu, Y.Y. Automated classification of protein subcellular localization in immunohistochemistry images to reveal biomarkers in colon cancer. BMC Bioinform. 2020, 21, 398. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Gerst, R.; Cseresnyés, Z.; Foo, W.; Sommerfeld, O.; Press, A.T.; Bauer, M.; Figge, M.T. Spatial quantification of clinical biomarker pharmacokinetics through deep learning-based segmentation and signal-oriented analysis of MSOT data. Photoacoustics 2022, 26, 100361. [Google Scholar] [CrossRef]
- Oura, P.; Junno, A.; Junno, J.A. Deep learning in forensic gunshot wound interpretation—A proof-of-concept study. Int. J. Leg. Med. 2021, 135, 2101–2106. [Google Scholar] [CrossRef]
- Zeng, J.; Zeng, J.; Qiu, X. Deep learning based forensic face verification in videos. In Proceedings of the 2017 International Conference on Progress in Informatics and Computing (PIC), Nanjing, China, 15–17 December 2017; pp. 77–80. [Google Scholar] [CrossRef]
- Homma, N.; Zhang, X.; Qureshi, A.; Konno, T.; Kawasumi, Y.; Usui, A.; Funayama, M.; Bukovsky, I.; Ichiji, K.; Sugita, N.; et al. A Deep Learning Aided Drowning Diagnosis for Forensic Investigations using Post-Mortem Lung CT Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1262–1265. [Google Scholar] [CrossRef]
- Bayar, B.; Stamm, M.C. A Deep Learning Approach to Universal Image Manipulation Detection Using a New Convolutional Layer. In Proceedings of the 4th ACM Workshop on Information Hiding and Multimedia Security, Vigo, Spain, 20–22 June 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 5–10. [Google Scholar] [CrossRef]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. In Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation (OSDI’16), Savannah, GA, USA, 2–4 November 2016; p. 283. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., Alché-Buc, F.D., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Eclipse Deeplearning4J. Available online: https://github.com/eclipse/deeplearning4j (accessed on 28 May 2022).
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolutional Architecture for Fast Feature Embedding. In Proceedings of the 22nd ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014; ACM: Orlando, FL, USA, 2014; pp. 675–678. [Google Scholar] [CrossRef]
- Chollet, F. Keras. Available online: https://github.com/fchollet/keras (accessed on 30 May 2022).
- Meng, X.; Bradley, J.; Yavuz, B.; Sparks, E.; Venkataraman, S.; Liu, D.; Freeman, J.; Tsai, D.; Amde, M.; Owen, S.; et al. MLlib: Machine Learning in Apache Spark. J. Mach. Learn. Res. 2016, 17, 1235–1241. [Google Scholar]
- Deep Java Library (DJL). Available online: https://github.com/deepjavalibrary/djl (accessed on 28 May 2022).
- Nwankpa, C.; Ijomah, W.; Gachagan, A.; Marshall, S. Activation Functions: Comparison of trends in Practice and Research for Deep Learning. arXiv 2018, arXiv:1811.03378. [Google Scholar]
- Legua, M.P.; Morales, I.; Sánchez Ruiz, L.M. The Heaviside Step Function and MATLAB. In Computational Science and Its Applications—ICCSA 2008; Gervasi, O., Murgante, B., Laganà, A., Taniar, D., Mun, Y., Gavrilova, M.L., Eds.; Series Title: Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2008; Volume 5072, pp. 1212–1221. [Google Scholar] [CrossRef]
- Lederer, J. Activation Functions in Artificial Neural Networks: A Systematic Overview. arXiv 2021, arXiv:2101.09957. [Google Scholar] [CrossRef]
- Roodschild, M.; Gotay Sardiñas, J.; Will, A. A new approach for the vanishing gradient problem on sigmoid activation. Prog. Artif. Intell. 2020, 9, 351–360. [Google Scholar] [CrossRef]
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical Evaluation of Rectified Activations in Convolutional Network. arXiv 2015, arXiv:1505.00853. [Google Scholar] [CrossRef]
- Lu, L. Dying ReLU and Initialization: Theory and Numerical Examples. Commun. Comput. Phys. 2020, 28, 1671–1706. [Google Scholar] [CrossRef]
- Maas, A.L. Rectifier Nonlinearities Improve Neural Network Acoustic Models. Proc. ICML 2013, 30, 3. [Google Scholar]
- Clevert, D.A.; Unterthiner, T.; Hochreiter, S. Fast and Accurate Deep Network Learning by Exponential Linear Units (ELUs). arXiv 2016, arXiv:1511.07289. [Google Scholar]
- Agarap, A.F. Deep Learning using Rectified Linear Units (ReLU). arXiv 2019, arXiv:1803.08375. [Google Scholar]
- Jain, A.; Jain, V. Effect of Activation Functions on Deep Learning Algorithms Performance for IMDB Movie Review Analysis. In Proceedings of the International Conference on Artificial Intelligence and Applications, Virtual, 9–10 December 2021; Bansal, P., Tushir, M., Balas, V.E., Srivastava, R., Eds.; Springer: Singapore, 2021; pp. 489–497. [Google Scholar] [CrossRef]
- Lau, M.M.; Lim, K.H. Review of Adaptive Activation Function in Deep Neural Network. In Proceedings of the 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Sarawak, Malaysia, 3–6 December 2018; pp. 686–690. [Google Scholar] [CrossRef]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2017, arXiv:1609.04747. [Google Scholar] [CrossRef]
- Zhou, X. Understanding the Convolutional Neural Networks with Gradient Descent and Backpropagation. J. Phys. Conf. Ser. 2018, 1004, 012028. [Google Scholar] [CrossRef] [Green Version]
- Kochenderfer, M.J.; Wheeler, T.A. Algorithms for Optimization; The MIT Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Ketkar, N. Deep Learning with Python: A Hands-On Introduction, 1st ed.; Imprint Apress: Berkeley, CA, USA, 2017. [Google Scholar] [CrossRef]
- Masters, D.; Luschi, C. Revisiting Small Batch Training for Deep Neural Networks. arXiv 2018, arXiv:1804.07612. [Google Scholar] [CrossRef]
- Bengio, Y. Practical Recommendations for Gradient-Based Training of Deep Architectures. In Neural Networks: Tricks of the Trade; Montavon, G., Orr, G.B., Müller, K.R., Eds.; Series Title: Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7700, pp. 437–478. [Google Scholar] [CrossRef] [Green Version]
- Yaqub, M.; Feng, J.; Zia, M.; Arshid, K.; Jia, K.; Rehman, Z.; Mehmood, A. State-of-the-Art CNN Optimizer for Brain Tumor Segmentation in Magnetic Resonance Images. Brain Sci. 2020, 10, 427. [Google Scholar] [CrossRef]
- Wang, H.; Dalkilic, B.; Gemmeke, H.; Hopp, T.; Hesser, J. Ultrasound Image Reconstruction Using Nesterov’s Accelerated Gradient. In Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018; pp. 1–3. [Google Scholar] [CrossRef]
- Dean, J.; Corrado, G.; Monga, R.; Chen, K.; Devin, M.; Mao, M.; Ranzato, M.a.; Senior, A.; Tucker, P.; Yang, K.; et al. Large Scale Distributed Deep Networks. In Proceedings of the Advances in Neural Information Processing Systems, Harrah’s and Harveys, Lake Tahoe, NV, USA, 3–6 December 2012; Volume 25. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C. Glove: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; Association for Computational Linguistics: Doha, Qatar, 2014; pp. 1532–1543. [Google Scholar] [CrossRef]
- Fang, J.K.; Fong, C.M.; Yang, P.; Hung, C.K.; Lu, W.L.; Chang, C.W. AdaGrad Gradient Descent Method for AI Image Management. In Proceedings of the 2020 IEEE International Conference on Consumer Electronics-Taiwan (ICCE-Taiwan), Taoyuan, Taiwan, 28–30 September 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Alfian, G.; Syafrudin, M.; Ijaz, M.; Syaekhoni, M.; Fitriyani, N.; Rhee, J. A Personalized Healthcare Monitoring System for Diabetic Patients by Utilizing BLE-Based Sensors and Real-Time Data Processing. Sensors 2018, 18, 2183. [Google Scholar] [CrossRef] [Green Version]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the Gradient by a Running Average of Its Recent Magnitude; Technical Report; University of Toronto: Toronto, ON, Canada, 2012. [Google Scholar]
- Fei, Z.; Wu, Z.; Xiao, Y.; Ma, J.; He, W. A new short-arc fitting method with high precision using Adam optimization algorithm. Optik 2020, 212, 164788. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Paola, J.D.; Schowengerdt, R.A. A review and analysis of backpropagation neural networks for classification of remotely-sensed multi-spectral imagery. Int. J. Remote Sens. 1995, 16, 3033–3058. [Google Scholar] [CrossRef]
- Li, J.; Cheng, J.h.; Shi, J.y.; Huang, F. Brief Introduction of Back Propagation (BP) Neural Network Algorithm and Its Improvement. In Advances in Computer Science and Information Engineering; Jin, D., Lin, S., Eds.; Series Title: Advances in Intelligent and Soft Computing; Springer: Berlin/Heidelberg, Germany, 2012; Volume 169, pp. 553–558. [Google Scholar] [CrossRef]
- Kim, P. MATLAB Deep Learning: With Machine Learning, Neural Networks and Artificial Intelligence; Springer: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Czum, J.M. Dive Into Deep Learning. J. Am. Coll. Radiol. 2020, 17, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Thimm, G.; Fiesler, E. High-order and multilayer perceptron initialization. IEEE Trans. Neural Netw. 1997, 8, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Masood, S.; Chandra, P. Training neural network with zero weight initialization. In Proceedings of the CUBE International Information Technology Conference on CUBE ’12, Pune, India, 3–5 September 2012; ACM Press: Pune, India, 2012; p. 235. [Google Scholar] [CrossRef]
- Lapuschkin, S.; Wäldchen, S.; Binder, A.; Montavon, G.; Samek, W.; Müller, K.R. Unmasking Clever Hans predictors and assessing what machines really learn. Nat. Commun. 2019, 10, 1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.; Debray, T.P.A.; et al. Prediction models for diagnosis and prognosis of COVID-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AIX-COVNET; Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Waibel, A.; Hanazawa, T.; Hinton, G.; Shikano, K.; Lang, K. Phoneme recognition using time-delay neural networks. IEEE Trans. Acoust. Speech Signal Process. 1989, 37, 328–339. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Santiago, Chile, 7–13 December 2015; pp. 3431–3440. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Wu, J.; Han, F.; Yuan, Y.; Zhao, W.; Ibragimov, B.; Gu, J.; Xing, L. Superpixel-based and boundary-sensitive convolutional neural network for automated liver segmentation. Phys. Med. Biol. 2018, 63, 095017. [Google Scholar] [CrossRef]
- Gheshlaghi, S.H.; Ranjbar, A.; Suratgar, A.A.; Menhaj, M.B.; Faraji, F. A superpixel segmentation based technique for multiple sclerosis lesion detection. arXiv 2019, arXiv:1907.03109. [Google Scholar]
- Fang, L.; Wang, X.; Wang, M. Superpixel/voxel medical image segmentation algorithm based on the regional interlinked value. Pattern Anal. Appl. 2021, 24, 1685–1698. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2016, arXiv:1511.06434. [Google Scholar]
- Jenni, S.; Favaro, P. On Stabilizing Generative Adversarial Training With Noise. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 12145–12153. [Google Scholar] [CrossRef] [Green Version]
- Mo, S.; Cho, M.; Shin, J. Freeze the discriminator: A simple baseline for fine-tuning gans. arXiv 2020, arXiv:2002.10964. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Gers, F. Learning to forget: Continual prediction with LSTM. In Proceedings of the 9th International Conference on Artificial Neural Networks: ICANN ’99, Edinburgh, UK, 7–10 September 1999; Volume 1999, pp. 850–855. [Google Scholar] [CrossRef]
- Gers, F.; Schmidhuber, J. Recurrent nets that time and count. In Proceedings of the IEEE-INNS-ENNS International Joint Conference on Neural Networks. IJCNN 2000. Neural Computing: New Challenges and Perspectives for the New Millennium, Como, Italy, 24–27 July 2000; Volume 3, pp. 189–194. [Google Scholar] [CrossRef]
- Cho, K.; van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations using RNN Encoder–Decoder for Statistical Machine Translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1724–1734. [Google Scholar] [CrossRef]
- Karimi Jafarbigloo, S.; Danyali, H. Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI Trans. Intell. Technol. 2021, 6, 426–439. [Google Scholar] [CrossRef]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Bi-directional ConvLSTM U-Net with densley connected convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, Seoul, Korea, 27–28 October 2019. [Google Scholar] [CrossRef] [Green Version]
- Scarselli, F.; Gori, M.; Tsoi, A.C.; Hagenbuchner, M.; Monfardini, G. The Graph Neural Network Model. IEEE Trans. Neural Netw. 2009, 20, 61–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017, arXiv:1609.02907. [Google Scholar] [CrossRef]
- Wu, F.; Souza, A.; Zhang, T.; Fifty, C.; Yu, T.; Weinberger, K. Simplifying Graph Convolutional Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; Volume 97, pp. 6861–6871. [Google Scholar]
- Zhou, Y.; Graham, S.; Alemi Koohbanani, N.; Shaban, M.; Heng, P.A.; Rajpoot, N. CGC-Net: Cell Graph Convolutional Network for Grading of Colorectal Cancer Histology Images. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), Seoul, Korea, 27–28 October 2019; pp. 388–398. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Xie, W.; Wang, L.; Zhang, S.; Li, W. Determination of biomarkers from microarray data using graph neural network and spectral clustering. Sci. Rep. 2021, 11, 23828. [Google Scholar] [CrossRef]
- Li, W.; Xie, W.; Zhang, S.; Wang, L.; Yang, J.; Zhao, D. A Novel Biomarker Selection Method Combining Graph Neural Network and Gene Relationships Applied to Microarray Data. Preprint 2022. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar] [CrossRef]
- Parvaiz, A.; Khalid, M.A.; Zafar, R.; Ameer, H.; Ali, M.; Fraz, M.M. Vision Transformers in Medical Computer Vision—A Contemplative Retrospection. arXiv 2022, arXiv:2203.15269. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Lum, T.; Mahdavi, M.; Frenkel, O.; Lee, C.; Jafari, M.H.; Dezaki, F.T.; Woudenberg, N.V.; Gu, A.N.; Abolmaesumi, P.; Tsang, T. Imaging Biomarker Knowledge Transfer for Attention-Based Diagnosis of COVID-19 in Lung Ultrasound Videos. In Proceedings of the International Workshop on Advances in Simplifying Medical Ultrasound, Strasbourg, France, 27 September–1 October 2021; pp. 159–168. [Google Scholar] [CrossRef]
- Lan, E. Performer: A Novel PPG to ECG Reconstruction Transformer For a Digital Biomarker of Cardiovascular Disease Detection. arXiv 2022, arXiv:2204.11795. [Google Scholar]
- Oh, S.J.; Schiele, B.; Fritz, M. Towards Reverse-Engineering Black-Box Neural Networks. In Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer International Publishing: Cham, Switzerland, 2019; pp. 121–144. [Google Scholar] [CrossRef] [Green Version]
- Buhrmester, V.; Münch, D.; Arens, M. Analysis of Explainers of Black Box Deep Neural Networks for Computer Vision: A Survey. Mach. Learn. Knowl. Extr. 2021, 3, 966–989. [Google Scholar] [CrossRef]
- Starke, G.; De Clercq, E.; Elger, B. Towards a pragmatist dealing with algorithmic bias in medical machine learning. Med. Health Care Philos. 2021, 24, 341–349. [Google Scholar] [CrossRef]
- Tommasi, T.; Patricia, N.; Caputo, B.; Tuytelaars, T. A Deeper Look at Dataset Bias. In Domain Adaptation in Computer Vision Applications; Csurka, G., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 37–55. [Google Scholar] [CrossRef] [Green Version]
- Al-shayea, Q.K. Artificial Neural Networks in Medical Diagnosis. Int. J. Comput. Sci. Issues (IJCSI) 2011, 8, 150–154. [Google Scholar]
- Amato, F.; López, A.; Peña-Méndez, E.M.; Vaňhara, P.; Hampl, A.; Havel, J. Artificial neural networks in medical diagnosis. J. Appl. Biomed. 2013, 11, 47–58. [Google Scholar] [CrossRef]
- Zherebtsov, E.; Dremin, V.; Popov, A.; Doronin, A.; Kurakina, D.; Kirillin, M.; Meglinski, I.; Bykov, A. Hyperspectral imaging of human skin aided by artificial neural networks. Biomed. Opt. Express 2019, 10, 3545–3559. [Google Scholar] [CrossRef] [Green Version]
- Fredriksson, I.; Larsson, M.; Strömberg, T. Machine learning for direct oxygen saturation and hemoglobin concentration assessment using diffuse reflectance spectroscopy. J. Biomed. Opt. 2020, 25, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Moncayo, S.; Manzoor, S.; Ugidos, T.; Navarro-Villoslada, F.; Caceres, J. Discrimination of human bodies from bones and teeth remains by Laser Induced Breakdown Spectroscopy and Neural Networks. Spectrochim. Acta Part At. Spectrosc. 2014, 101, 21–25. [Google Scholar] [CrossRef]
- Al-Hetlani, E.; Halámková, L.; Amin, M.O.; Lednev, I.K. Differentiating smokers and nonsmokers based on Raman spectroscopy of oral fluid and advanced statistics for forensic applications. J. Biophotonics 2020, 13, e201960123. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.W.; Hui, R.; Tian, Z. Classification of CT brain images based on deep learning networks. Comput. Methods Programs Biomed. 2017, 138, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Yu-Jen Chen, Y.J.; Hua, K.L.; Hsu, C.H.; Cheng, W.H.; Hidayati, S.C. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015, 8, 2015–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, M.; Chakraborty, C.; Racoceanu, D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput. Med. Imaging Graph. 2018, 64, 29–40. [Google Scholar] [CrossRef]
- Jafarbiglo, S.K.; Danyali, H.; Helfroush, M.S. Nuclear atypia grading in histopathological images of breast cancer using convolutional neural networks. In Proceedings of the 2018 4th Iranian Conference on Signal Processing and Intelligent Systems (ICSPIS), Tehran, Iran, 25–27 December 2018; pp. 89–93. [Google Scholar]
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis. Oncol. 2020, 4, 14. [Google Scholar] [CrossRef]
- Alex, V.; Safwan, K.P.M.; Chennamsetty, S.S.; Krishnamurthi, G. Generative Adversarial Networks for Brain Lesion Detection. Med. Imaging 2017, 10133, 101330G. [Google Scholar] [CrossRef]
- Son, J.; Park, S.J.; Jung, K.H. Retinal vessel segmentation in fundoscopic images with generative adversarial networks. arXiv 2017, arXiv:1706.09318. [Google Scholar]
- Zhang, Q.; Wang, H.; Lu, H.; Won, D.; Yoon, S.W. Medical Image Synthesis with Generative Adversarial Networks for Tissue Recognition. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; pp. 199–207. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 417–425. [Google Scholar] [CrossRef]
- Lipton, Z.C.; Berkowitz, J.; Elkan, C. A Critical Review of Recurrent Neural Networks for Sequence Learning. arXiv 2015, arXiv:1506.00019. [Google Scholar] [CrossRef]
- Futoma, J.; Hariharan, S.; Heller, K.; Sendak, M.; Brajer, N.; Clement, M.; Bedoya, A.; O’Brien, C. An Improved Multi-Output Gaussian Process RNN with Real-Time Validation for Early Sepsis Detection. In Proceedings of the 2nd Machine Learning for Healthcare Conference, Boston, MA, USA, 18–19 August 2017; Doshi-Velez, F., Fackler, J., Kale, D., Ranganath, R., Wallace, B., Wiens, J., Eds.; PMLR: Boston, MA, USA, 2017; Volume 68, pp. 243–254. [Google Scholar]
- Giunchiglia, E.; Nemchenko, A.; van der Schaar, M. RNN-SURV: A Deep Recurrent Model for Survival Analysis. In Proceedings of the Artificial Neural Networks and Machine Learning—ICANN 2018, Rhodes, Greece, 4–7 October 2018; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 23–32. [Google Scholar] [CrossRef]
- Reddy, B.K.; Delen, D. Predicting hospital readmission for lupus patients: An RNN-LSTM-based deep-learning methodology. Comput. Biol. Med. 2018, 101, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, E.Z.; Chen, T.; Patel, V.M.; Sun, S. Pyramid Convolutional RNN for MRI Reconstruction. IEEE Trans. Med. Imaging 2020. [Google Scholar] [CrossRef]
- Li, Y.; Qian, B.; Zhang, X.; Liu, H. Graph Neural Network-Based Diagnosis Prediction. Big Data 2020, 8, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, T.; Ding, H.; Tang, B.; Wang, X.; Chen, Q.; Yan, J.; Zhou, Y. A hybrid method of recurrent neural network and graph neural network for next-period prescription prediction. Int. J. Mach. Learn. Cybern. 2020, 11, 2849–2856. [Google Scholar] [CrossRef]
- Li, X.; Dvornek, N.C.; Zhou, Y.; Zhuang, J.; Ventola, P.; Duncan, J.S. Graph Neural Network for Interpreting Task-fMRI Biomarkers. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Series Title: Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11768, pp. 485–493. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Wang, R.; Zheng, Y.; Jiang, Z.; Zhang, H.; Yu, L. Cervical cell classification with graph convolutional network. Comput. Methods Programs Biomed. 2021, 198, 105807. [Google Scholar] [CrossRef]
- Steinkraus, D.; Buck, I.; Simard, P. Using GPUs for machine learning algorithms. In Proceedings of the Eighth International Conference on Document Analysis and Recognition (ICDAR’05), Seoul, Korea, 31 August–1 September 2005; Volume 2, pp. 1115–1120. [Google Scholar] [CrossRef]






| Name | Function [Range] | Function (Red) and Derivative (Blue) |
|---|---|---|
| linear |  | |
| Heaviside |  | |
| Rectified Linear Unit |  | |
| Logistic/sigmoid |  | |
| tanh |  | |
| elu |  |
| Algorithm | Advantages | Disadvantages |
|---|---|---|
| Batch Gradient Descent |
|
|
| Stochastic Gradient Descent |
|
|
| Mini-Batch Gradient Descent |
|
|
| Architecture | Search Terms |
|---|---|
| General | Artificial Neural Network; Deep Learning |
| Convolutional Neural Networks | Convolutional Neural Network; Fully Convolutional Neural Network |
| Generative Adversarial Neural Networks | Generative Adversarial Neural Network |
| Recurrent Neural Networks | Recurrent Neural Network |
| Graph Neural Networks | Graph Neural Networks |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, R.; Karimijafarbigloo, S.; Roggenbuck, D.; Rödiger, S. Applications of Neural Networks in Biomedical Data Analysis. Biomedicines 2022, 10, 1469. https://doi.org/10.3390/biomedicines10071469
Weiss R, Karimijafarbigloo S, Roggenbuck D, Rödiger S. Applications of Neural Networks in Biomedical Data Analysis. Biomedicines. 2022; 10(7):1469. https://doi.org/10.3390/biomedicines10071469
Chicago/Turabian StyleWeiss, Romano, Sanaz Karimijafarbigloo, Dirk Roggenbuck, and Stefan Rödiger. 2022. "Applications of Neural Networks in Biomedical Data Analysis" Biomedicines 10, no. 7: 1469. https://doi.org/10.3390/biomedicines10071469
APA StyleWeiss, R., Karimijafarbigloo, S., Roggenbuck, D., & Rödiger, S. (2022). Applications of Neural Networks in Biomedical Data Analysis. Biomedicines, 10(7), 1469. https://doi.org/10.3390/biomedicines10071469







