High Oxytocin Receptor Expression Linked to Increased Cell Migration and Reduced Survival in Patients with Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression and Survival Analysis
2.2. Chemicals
2.3. Cells and Cell Transfection
2.4. Transwell Cell Migration Assays
2.5. qPCR
2.6. Scratch Wound Healing Assay
2.7. Gelatin Zymography Assay
2.8. Western Blots
2.9. Fluorescence Imaging Plate Reader Functional Calcium Assay
2.10. Functional cAMP Assay
2.11. Statistical Analysis
3. Results
3.1. Oxytocin Receptor Expression Is Highest in Tumour-Adjacent Breast Tissues Followed by Normal and Tumour Breast Tissue
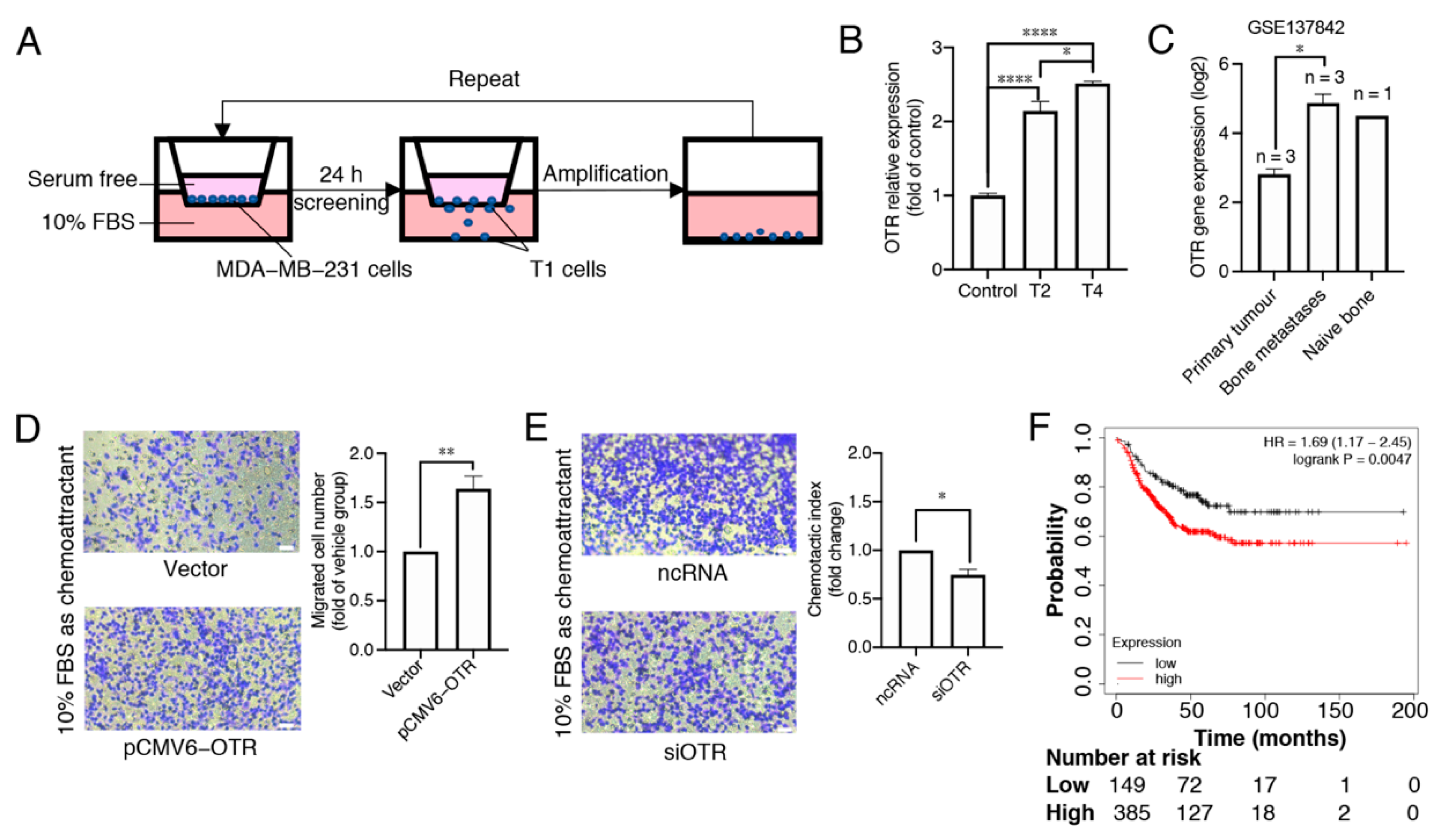
3.2. Oxytocin Receptor Expression Is Higher in Migrated/Metastasised Breast Cancer Cells Than in Primary Cells
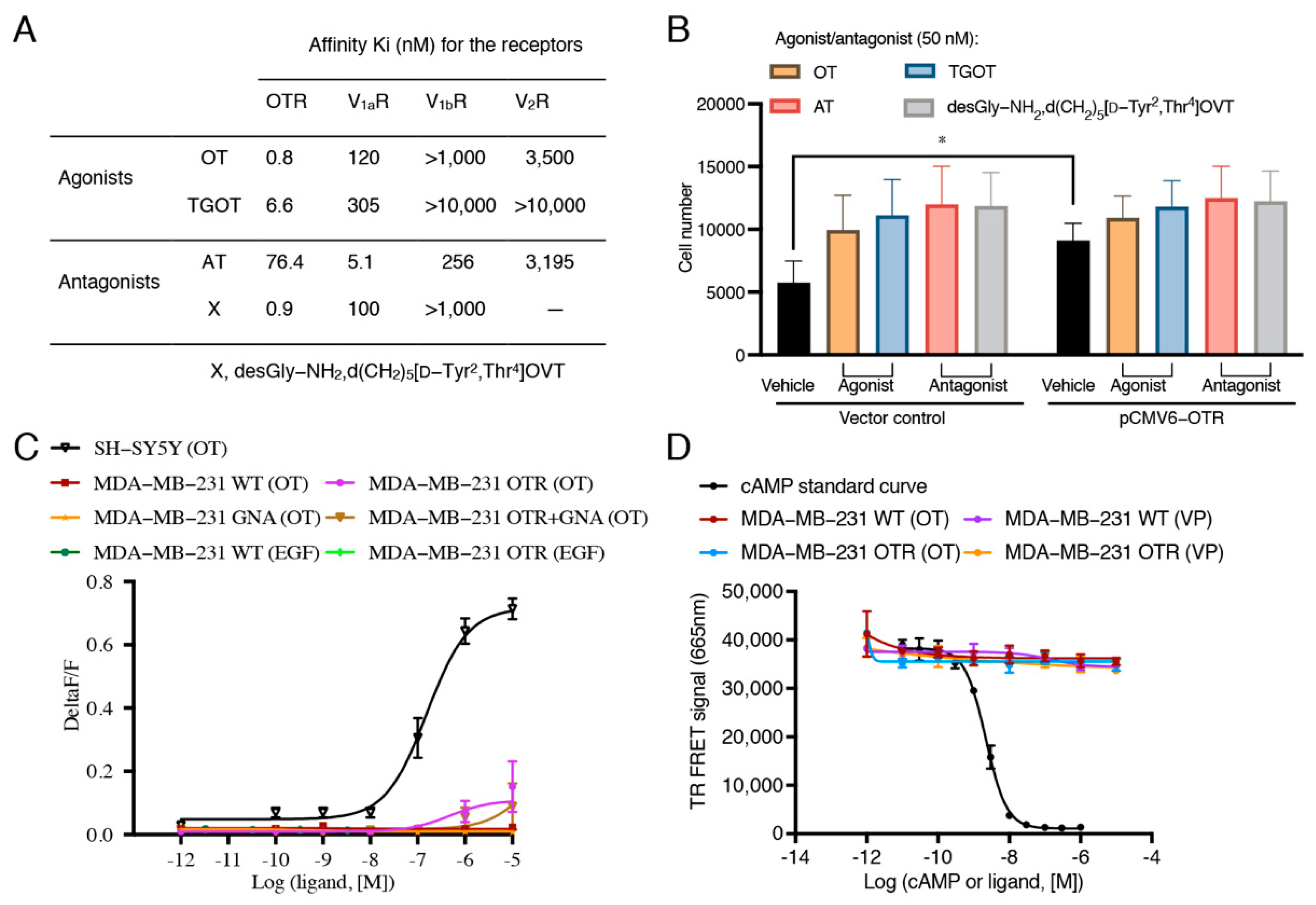
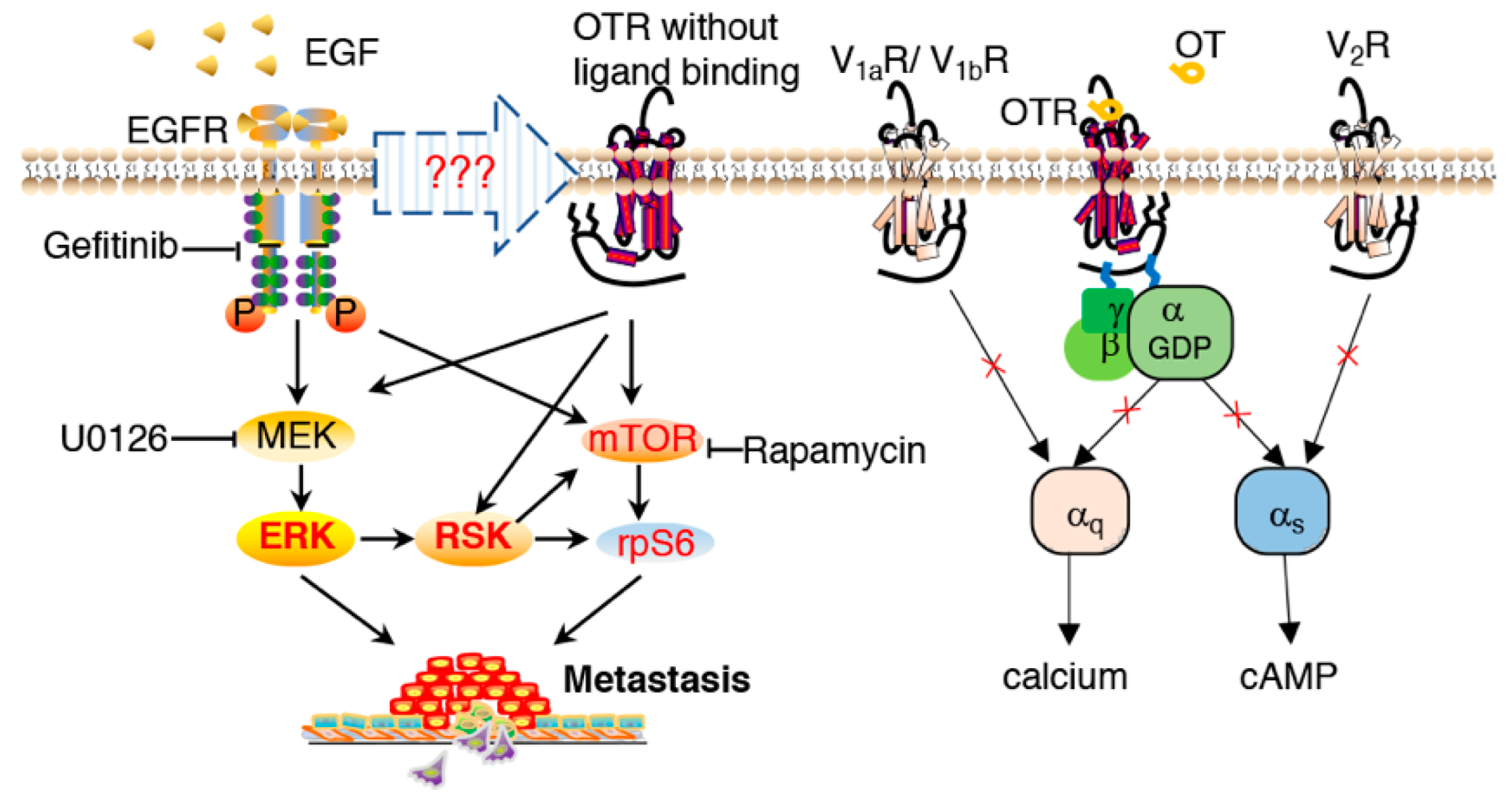
3.3. Oxytocin Receptor Agonists/Antagonists Do Not Affect Cell Migration, and the Oxytocin Receptor Does Not Function via Gq-- and Gs-Pathways in MDA-MB-231 Cells
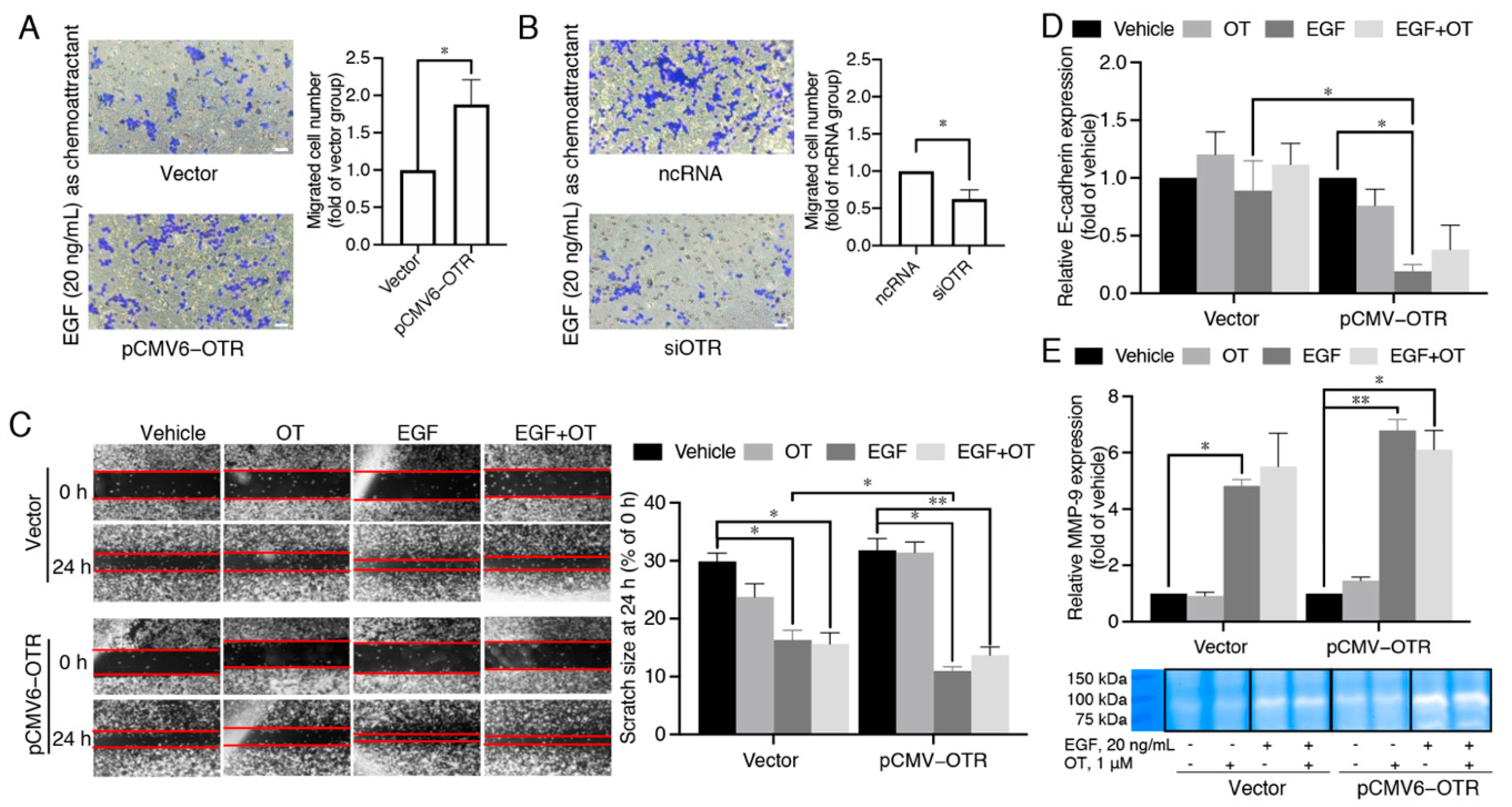
3.4. MDA-MB-231 Cells Overexpressing the Oxytocin Receptor Are More Sensitive to EGF
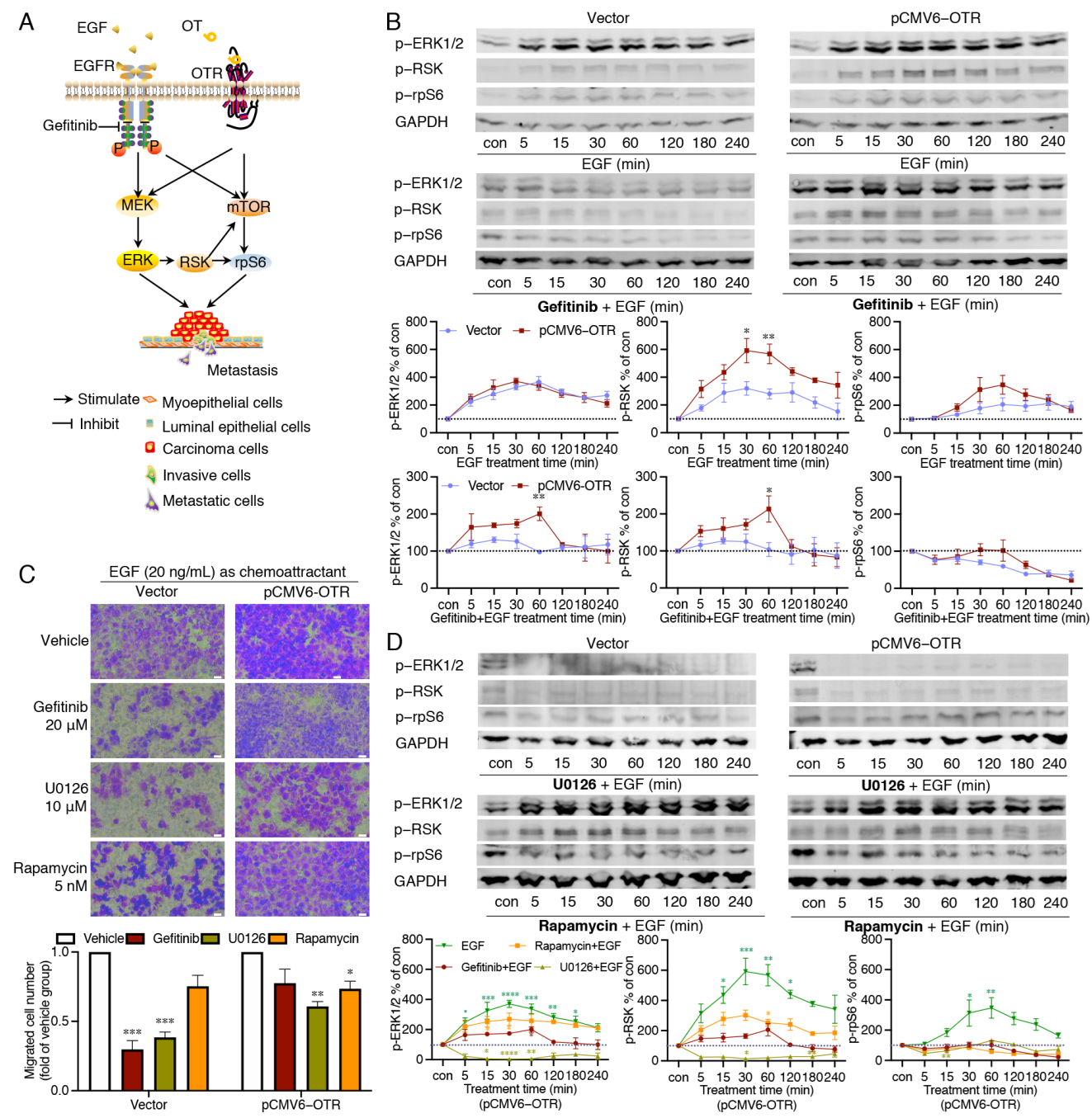
3.5. Oxytocin Receptor Enhances EGF-Stimulated RSK Activation, with the mTOR Pathway Contributing to the Downstream rpS6 Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pasculli, B.; Barbano, R.; Parrella, P. Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin. Cancer Biol. 2018, 51, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Neophytou, C.; Boutsikos, P.; Papageorgis, P. Molecular Mechanisms and Emerging Therapeutic Targets of Triple-Negative Breast Cancer Metastasis. Front. Oncol. 2018, 8, 31. [Google Scholar] [CrossRef]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Massague, J. Genetic determinants of cancer metastasis. Nat. Rev. Genet. 2007, 8, 341–352. [Google Scholar] [CrossRef]
- Yang, R.; Li, Y.; Wang, H.; Qin, T.; Yin, X.; Ma, X. Therapeutic progress and challenges for triple negative breast cancer: Targeted therapy and immunotherapy. Mol. Biomed. 2022, 3, 8. [Google Scholar] [CrossRef]
- Cipriano, E.; Mesquita, A. Emerging Therapeutic Drugs in Metastatic Triple-Negative Breast Cancer. Breast Cancer Basic Clin. Res. 2021, 15, 11782234211002491. [Google Scholar] [CrossRef]
- Aysola, K.; Desai, A.; Welch, C.; Xu, J.; Qin, Y.; Reddy, V.; Matthews, R.; Owens, C.; Okoli, J.; Beech, D.J.; et al. Triple Negative Breast Cancer—An Overview. Hered. Genet. 2013, 2013, 1. [Google Scholar] [CrossRef]
- Twelves, C.; Jove, M.; Gombos, A.; Awada, A. Cytotoxic chemotherapy: Still the mainstay of clinical practice for all subtypes metastatic breast cancer. Crit. Rev. Oncol. 2016, 100, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Jurek, B.; Neumann, I.D. The Oxytocin Receptor: From Intracellular Signaling to Behavior. Physiol. Rev. 2018, 98, 1805–1908. [Google Scholar] [CrossRef] [PubMed]
- Lerman, B.; Harricharran, T.; Ogunwobi, O.O. Oxytocin and cancer: An emerging link. World J. Clin. Oncol. 2018, 9, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Cassoni, P.; Sapino, A.; Marrocco, T.; Chini, B.; Bussolati, G. Oxytocin and oxytocin receptors in cancer cells and proliferation. J. Neuroendocr. 2004, 16, 362–364. [Google Scholar] [CrossRef]
- Cassoni, P.; Marrocco, T.; Deaglio, S.; Sapino, A.; Bussolati, G. Biological relevance of oxytocin and oxytocin receptors in cancer cells and primary tumors. Ann. Oncol. 2001, 12 (Suppl. 2), S37–S39. [Google Scholar] [CrossRef]
- Viero, C.; Shibuya, I.; Kitamura, N.; Verkhratsky, A.; Fujihara, H.; Katoh, A.; Ueta, Y.; Zingg, H.H.; Chvatal, A.; Sykova, E.; et al. REVIEW: Oxytocin: Crossing the bridge between basic science and pharmacotherapy. CNS Neurosci. Ther. 2010, 16, e138–e156. [Google Scholar] [CrossRef]
- Ito, Y.; Kobayashi, T.; Kimura, T.; Matsuura, N.; Wakasugi, E.; Takeda, T.; Shimano, T.; Kubota, Y.; Nobunaga, T.; Makino, Y.; et al. Investigation of the oxytocin receptor expression in human breast cancer tissue using newly established monoclonal antibodies. Endocrinology 1996, 137, 773–779. [Google Scholar] [CrossRef]
- Bussolati, G.; Cassoni, P.; Ghisolfi, G.; Negro, F.; Sapino, A. Immunolocalization and gene expression of oxytocin receptors in carcinomas and non-neoplastic tissues of the breast. Am. J. Pathol. 1996, 148, 1895–1903. [Google Scholar]
- Copland, J.A.; Jeng, Y.J.; Strakova, Z.; Ives, K.L.; Hellmich, M.R.; Soloff, M.S. Demonstration of functional oxytocin receptors in human breast Hs578T cells and their up-regulation through a protein kinase C-dependent pathway. Endocrinology 1999, 140, 2258–2267. [Google Scholar] [CrossRef][Green Version]
- Sapino, A.; Cassoni, P.; Stella, A.; Bussolati, G. Oxytocin receptor within the breast: Biological function and distribution. Anticancer Res. 1998, 18, 2181–2186. [Google Scholar]
- Liu, H.; Gruber, C.W.; Alewood, P.F.; Moller, A.; Muttenthaler, M. The oxytocin receptor signalling system and breast cancer: A critical review. Oncogene 2020, 39, 5917–5932. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- Saunus, J.M.; Smart, C.E.; Kutasovic, J.R.; Johnston, R.L.; Kalita-de Croft, P.; Miranda, M.; Rozali, E.N.; Vargas, A.C.; Reid, L.E.; Lorsy, E.; et al. Multidimensional phenotyping of breast cancer cell lines to guide preclinical research. Breast Cancer Res. Treat. 2018, 167, 289–301. [Google Scholar] [CrossRef]
- Lefley, D.; Howard, F.; Arshad, F.; Bradbury, S.; Brown, H.; Tulotta, C.; Eyre, R.; Alferez, D.; Wilkinson, J.M.; Holen, I.; et al. Development of clinically relevant in vivo metastasis models using human bone discs and breast cancer patient-derived xenografts. Breast Cancer Res. 2019, 21, 130. [Google Scholar] [CrossRef]
- Manning, M.; Stoev, S.; Chini, B.; Durroux, T.; Mouillac, B.; Guillon, G. Peptide and non-peptide agonists and antagonists for the vasopressin and oxytocin V1a, V1b, V2 and OT receptors: Research tools and potential therapeutic agents. Prog. Brain Res. 2008, 170, 473–512. [Google Scholar] [CrossRef]
- Manning, M.; Misicka, A.; Olma, A.; Bankowski, K.; Stoev, S.; Chini, B.; Durroux, T.; Mouillac, B.; Corbani, M.; Guillon, G. Oxytocin and vasopressin agonists and antagonists as research tools and potential therapeutics. J. Neuroendocr. 2012, 24, 609–628. [Google Scholar] [CrossRef]
- Busnelli, M.; Sauliere, A.; Manning, M.; Bouvier, M.; Gales, C.; Chini, B. Functional selective oxytocin-derived agonists discriminate between individual G protein family subtypes. J. Biol. Chem. 2012, 287, 3617–3629. [Google Scholar] [CrossRef]
- Taylor, A.H.; Ang, V.T.; Jenkins, J.S.; Silverlight, J.J.; Coombes, R.C.; Luqmani, Y.A. Interaction of vasopressin and oxytocin with human breast carcinoma cells. Cancer Res. 1990, 50, 7882–7886. [Google Scholar] [PubMed]
- Cassoni, P.; Marrocco, T.; Bussolati, B.; Allia, E.; Munaron, L.; Sapino, A.; Bussolati, G. Oxytocin induces proliferation and migration in immortalized human dermal microvascular endothelial cells and human breast tumor-derived endothelial cells. Mol. Cancer Res. 2006, 4, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Cassoni, P.; Sapino, A.; Fortunati, N.; Munaron, L.; Chini, B.; Bussolati, G. Oxytocin inhibits the proliferation of MDA-MB231 human breast-cancer cells via cyclic adenosine monophosphate and protein kinase A. Int. J. Cancer 1997, 72, 340–344. [Google Scholar] [CrossRef]
- Garona, J.; Pifano, M.; Orlando, U.D.; Pastrian, M.B.; Iannucci, N.B.; Ortega, H.H.; Podesta, E.J.; Gomez, D.E.; Ripoll, G.V.; Alonso, D.F. The novel desmopressin analogue [V4Q5]dDAVP inhibits angiogenesis, tumour growth and metastases in vasopressin type 2 receptor-expressing breast cancer models. Int. J. Oncol. 2015, 46, 2335–2345. [Google Scholar] [CrossRef]
- Muttenthaler, M.; Andersson, A.; Vetter, I.; Menon, R.; Busnelli, M.; Ragnarsson, L.; Bergmayr, C.; Arrowsmith, S.; Deuis, J.R.; Chiu, H.S.; et al. Subtle modifications to oxytocin produce ligands that retain potency and improved selectivity across species. Sci. Signal. 2017, 10, eaan3398. [Google Scholar] [CrossRef]
- Giannone, F.; Malpeli, G.; Lisi, V.; Grasso, S.; Shukla, P.; Ramarli, D.; Sartoris, S.; Monsurro, V.; Krampera, M.; Amato, E.; et al. The puzzling uniqueness of the heterotrimeric G15 protein and its potential beyond hematopoiesis. J. Mol. Endocrinol. 2010, 44, 259–269. [Google Scholar] [CrossRef]
- Kankanamge, D.; Tennakoon, M.; Weerasinghe, A.; Cedeno-Rosario, L.; Chadee, D.N.; Karunarathne, A. G protein alphaq exerts expression level-dependent distinct signaling paradigms. Cell. Signal. 2019, 58, 34–43. [Google Scholar] [CrossRef]
- Gruber, C.W.; Koehbach, J.; Muttenthaler, M. Exploring bioactive peptides from natural sources for oxytocin and vasopressin drug discovery. Futur. Med. Chem. 2012, 4, 1791–1798. [Google Scholar] [CrossRef]
- Ariana, M.; Pornour, M.; Mehr, S.S.; Vaseghi, H.; Ganji, S.M.; Alivand, M.R.; Salari, M.; Akbari, M.E. Preventive effects of oxytocin and oxytocin receptor in breast cancer pathogenesis. Pers. Med. 2019, 16, 25–34. [Google Scholar] [CrossRef]
- Bastian, S.E.; Dunbar, A.J.; Priebe, I.K.; Owens, P.C.; Goddard, C. Measurement of betacellulin levels in bovine serum, colostrum and milk. J. Endocrinol. 2001, 168, 203–212. [Google Scholar] [CrossRef]
- Klein, B.Y.; Tamir, H.; Hirschberg, D.L.; Glickstein, S.B.; Welch, M.G. Oxytocin modulates mTORC1 pathway in the gut. Biochem. Biophys. Res. Commun. 2013, 432, 466–471. [Google Scholar] [CrossRef]
- Conde, E.; Angulo, B.; Tang, M.; Morente, M.; Torres-Lanzas, J.; Lopez-Encuentra, A.; Lopez-Rios, F.; Sanchez-Cespedes, M. Molecular context of the EGFR mutations: Evidence for the activation of mTOR/S6K signaling. Clin. Cancer Res. 2006, 12, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Doehn, U.; Hauge, C.; Frank, S.R.; Jensen, C.J.; Duda, K.; Nielsen, J.V.; Cohen, M.S.; Johansen, J.V.; Winther, B.R.; Lund, L.R.; et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol. Cell 2009, 35, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Carriere, A.; Cargnello, M.; Julien, L.A.; Gao, H.; Bonneil, E.; Thibault, P.; Roux, P.P. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 2008, 18, 1269–1277. [Google Scholar] [CrossRef]
- Roux, P.P.; Shahbazian, D.; Vu, H.; Holz, M.K.; Cohen, M.S.; Taunton, J.; Sonenberg, N.; Blenis, J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 2007, 282, 14056–14064. [Google Scholar] [CrossRef]
- Leslie, M. EGF is internalized and degraded. J. Cell Biol. 2005, 170, 339. [Google Scholar] [CrossRef]
- Cheng, Y.; Tian, H. Current Development Status of MEK Inhibitors. Molecules 2017, 22, 1551. [Google Scholar] [CrossRef]
- Sriram, K.; Wiley, S.Z.; Moyung, K.; Gorr, M.W.; Salmeron, C.; Marucut, J.; French, R.P.; Lowy, A.M.; Insel, P.A. Detection and Quantification of GPCR mRNA: An Assessment and Implications of Data from High-Content Methods. ACS Omega 2019, 4, 17048–17059. [Google Scholar] [CrossRef]
- Petryszak, R.; Keays, M.; Tang, Y.A.; Fonseca, N.A.; Barrera, E.; Burdett, T.; Fullgrabe, A.; Fuentes, A.M.; Jupp, S.; Koskinen, S.; et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016, 44, D746–D752. [Google Scholar] [CrossRef]
- Amico, J.A.; Rauk, P.N.; Cai, H.M. Estradiol and progesterone regulate oxytocin receptor binding and expression in human breast cancer cell lines. Endocrine 2002, 18, 79–84. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, K.A.; Heaphy, C.M.; Mai, M.; Vargas, K.M.; Jones, A.C.; Vo, P.; Butler, K.S.; Joste, N.E.; Bisoffi, M.; Griffith, J.K. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int. J. Cancer 2011, 129, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shen, M.; Qi, Q.; Zhang, H.; Guo, S.W. Corroborating evidence for platelet-induced epithelial-mesenchymal transition and fibroblast-to-myofibroblast transdifferentiation in the development of adenomyosis. Hum. Reprod. 2016, 31, 734–749. [Google Scholar] [CrossRef]
- Zingg, H.H.; Laporte, S.A. The oxytocin receptor. Trends Endocrinol. Metab. 2003, 14, 222–227. [Google Scholar] [CrossRef]
- Soloff, M.S.; Alexandrova, M.; Fernstrom, M.J. Oxytocin receptors: Triggers for parturition and lactation? Science 1979, 204, 1313–1315. [Google Scholar] [CrossRef]
- Li, D.; San, M.; Zhang, J.; Yang, A.; Xie, W.; Chen, Y.; Lu, X.; Zhang, Y.; Zhao, M.; Feng, X.; et al. Oxytocin receptor induces mammary tumorigenesis through prolactin/p-STAT5 pathway. Cell Death Dis 2021, 12, 588. [Google Scholar] [CrossRef]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef]
- Kassam, F.; Enright, K.; Dent, R.; Dranitsaris, G.; Myers, J.; Flynn, C.; Fralick, M.; Kumar, R.; Clemons, M. Survival outcomes for patients with metastatic triple-negative breast cancer: Implications for clinical practice and trial design. Clin. Breast Cancer 2009, 9, 29–33. [Google Scholar] [CrossRef]
- Kose, M. GPCRs and EGFR-Cross-talk of membrane receptors in cancer. Bioorg Med. Chem. Lett. 2017, 27, 3611–3620. [Google Scholar] [CrossRef]
- Guzzi, F.; Zanchetta, D.; Cassoni, P.; Guzzi, V.; Francolini, M.; Parenti, M.; Chini, B. Localization of the human oxytocin receptor in caveolin-1 enriched domains turns the receptor-mediated inhibition of cell growth into a proliferative response. Oncogene 2002, 21, 1658–1667. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rimoldi, V.; Reversi, A.; Taverna, E.; Rosa, P.; Francolini, M.; Cassoni, P.; Parenti, M.; Chini, B. Oxytocin receptor elicits different EGFR/MAPK activation patterns depending on its localization in caveolin-1 enriched domains. Oncogene 2003, 22, 6054–6060. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Huang, C.C.; Hsu, K.S. Oxytocin promotes long-term potentiation by enhancing epidermal growth factor receptor-mediated local translation of protein kinase Mzeta. J. Neurosci. 2012, 32, 15476–15488. [Google Scholar] [CrossRef]
- Krishnaswamy, N.; Lacroix-Pepin, N.; Chapdelaine, P.; Taniguchi, H.; Kauffenstein, G.; Chakravarti, A.; Danyod, G.; Fortier, M.A. Epidermal growth factor receptor is an obligatory intermediate for oxytocin-induced cyclooxygenase 2 expression and prostaglandin F2 alpha production in bovine endometrial epithelial cells. Endocrinology 2010, 151, 1367–1374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tani, T.; Yasuda, H.; Hamamoto, J.; Kuroda, A.; Arai, D.; Ishioka, K.; Ohgino, K.; Miyawaki, M.; Kawada, I.; Naoki, K.; et al. Activation of EGFR Bypass Signaling by TGFalpha Overexpression Induces Acquired Resistance to Alectinib in ALK-Translocated Lung Cancer Cells. Mol. Cancer Ther. 2016, 15, 162–171. [Google Scholar] [CrossRef]
- Barr, S.; Thomson, S.; Buck, E.; Russo, S.; Petti, F.; Sujka-Kwok, I.; Eyzaguirre, A.; Rosenfeld-Franklin, M.; Gibson, N.W.; Miglarese, M.; et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin. Exp. Metastasis 2008, 25, 685–693. [Google Scholar] [CrossRef]
- Cassoni, P.; Marrocco, T.; Sapino, A.; Allia, E.; Bussolati, G. Oxytocin synthesis within the normal and neoplastic breast: First evidence of a local peptide source. Int. J. Oncol. 2006, 28, 1263–1268. [Google Scholar] [CrossRef][Green Version]
- Murrell, T.G. The potential for oxytocin (OT) to prevent breast cancer: A hypothesis. Breast Cancer Res. Treat. 1995, 35, 225–229. [Google Scholar] [CrossRef]
- Imanieh, M.H.; Bagheri, F.; Alizadeh, A.M.; Ashkani-Esfahani, S. Oxytocin has therapeutic effects on cancer, a hypothesis. Eur. J. Pharmacol. 2014, 741, 112–123. [Google Scholar] [CrossRef]

| Datasets Containing Breast Cancer and Normal Breast Tissues Identified Using Genevestigator in Figure 1B | |||
|---|---|---|---|
| GEO Accession | Overall Design and Sample Information(Affymetrix Human Genome U133 Plus 2.0 Array Platform, OTR Gene ID, 206825_at) | ||
| GSE22544 | 16 invasive ductal carcinomas samples (including 2 node metastasis samples) analysed with the U133 Plus 2.0 array compared to 4 normal control samples | ||
| GSE21422 | Dataset including 5 healthy tissue samples, 9 ductal carcinomas in situ, and 5 invasive ductal carcinomas | ||
| GSE7904 | 62 samples including 43 tumours, 7 normal breast, and 12 normal organelles | ||
| GSE25407 | 3 examples of Stage-I breast tumour and 3 samples of breast reduction mammoplasty tissue were expanded as explant cultures for RNA extraction and hybridisation to Affymetrix microarrays | ||
| GSE7307 | Affymetrix human U133 plus 2.0 array was used to transcriptionally profile both normal and diseased human tissues representing over 90 distinct tissue types (herein, breast tissues analysed only) | ||
| GSE10810 | 58 samples including 31 tumours and 27 controls; some of the samples are paired | ||
| GSE31448 | Tumour tissues from 353 patients with invasive adenocarcinoma who underwent initial surgery. Dataset also includes 4 normal breast samples | ||
| GSE10780 | 143 histologically normal breast tissues and 42 invasive ductal carcinoma tissues | ||
| GSE20711 | Dataset includes 2 normal breast tissues and 88 breast tumour tissues | ||
| GSE3744 | Dataset includes 7 normal breast tissues and 40 breast tumour tissues | ||
| Datasets from the GEO database containing paired adjacent and breast tumour tissues in Figure 1C # | |||
| GEO accession | Samples | Platform | OTR gene ID a |
| GSE109169 | 25 sets of paired adjacent/tumour breast tissue specimens | GPL5175 [HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [transcript (gene) version] | 2661992 |
| GSE139038 | 65 samples including 41 breast tumours, 18 adjacent tissues [paired normal], and 6 apparently normal tissues from breasts operated on for non-malignant conditions | GPL27630 Print_1437 (Block_Column_Row IDs) | 7_14_20 |
| GSE76250 | 198 samples including 165 TNBC tissues and 33 paired adjacent breast tissues | GPL17586 [HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] | TC03001145.hg.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Muttenthaler, M. High Oxytocin Receptor Expression Linked to Increased Cell Migration and Reduced Survival in Patients with Triple-Negative Breast Cancer. Biomedicines 2022, 10, 1595. https://doi.org/10.3390/biomedicines10071595
Liu H, Muttenthaler M. High Oxytocin Receptor Expression Linked to Increased Cell Migration and Reduced Survival in Patients with Triple-Negative Breast Cancer. Biomedicines. 2022; 10(7):1595. https://doi.org/10.3390/biomedicines10071595
Chicago/Turabian StyleLiu, Huiping, and Markus Muttenthaler. 2022. "High Oxytocin Receptor Expression Linked to Increased Cell Migration and Reduced Survival in Patients with Triple-Negative Breast Cancer" Biomedicines 10, no. 7: 1595. https://doi.org/10.3390/biomedicines10071595
APA StyleLiu, H., & Muttenthaler, M. (2022). High Oxytocin Receptor Expression Linked to Increased Cell Migration and Reduced Survival in Patients with Triple-Negative Breast Cancer. Biomedicines, 10(7), 1595. https://doi.org/10.3390/biomedicines10071595






