DSCAM-AS1 Long Non-Coding RNA Exerts Oncogenic Functions in Endometrial Adenocarcinoma via Activation of a Tumor-Promoting Transcriptome Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RNA Isolation and RT-qPCR
2.3. Western Blot Analysis
2.4. Cell Culture, siRNA Transfection and Proliferation Studies
2.5. Apoptosis Assays
2.6. Transcriptome Analyses Using Clariom S Human Microarrays
2.7. In Silico Analyses
2.8. Statistical Analysis
3. Results
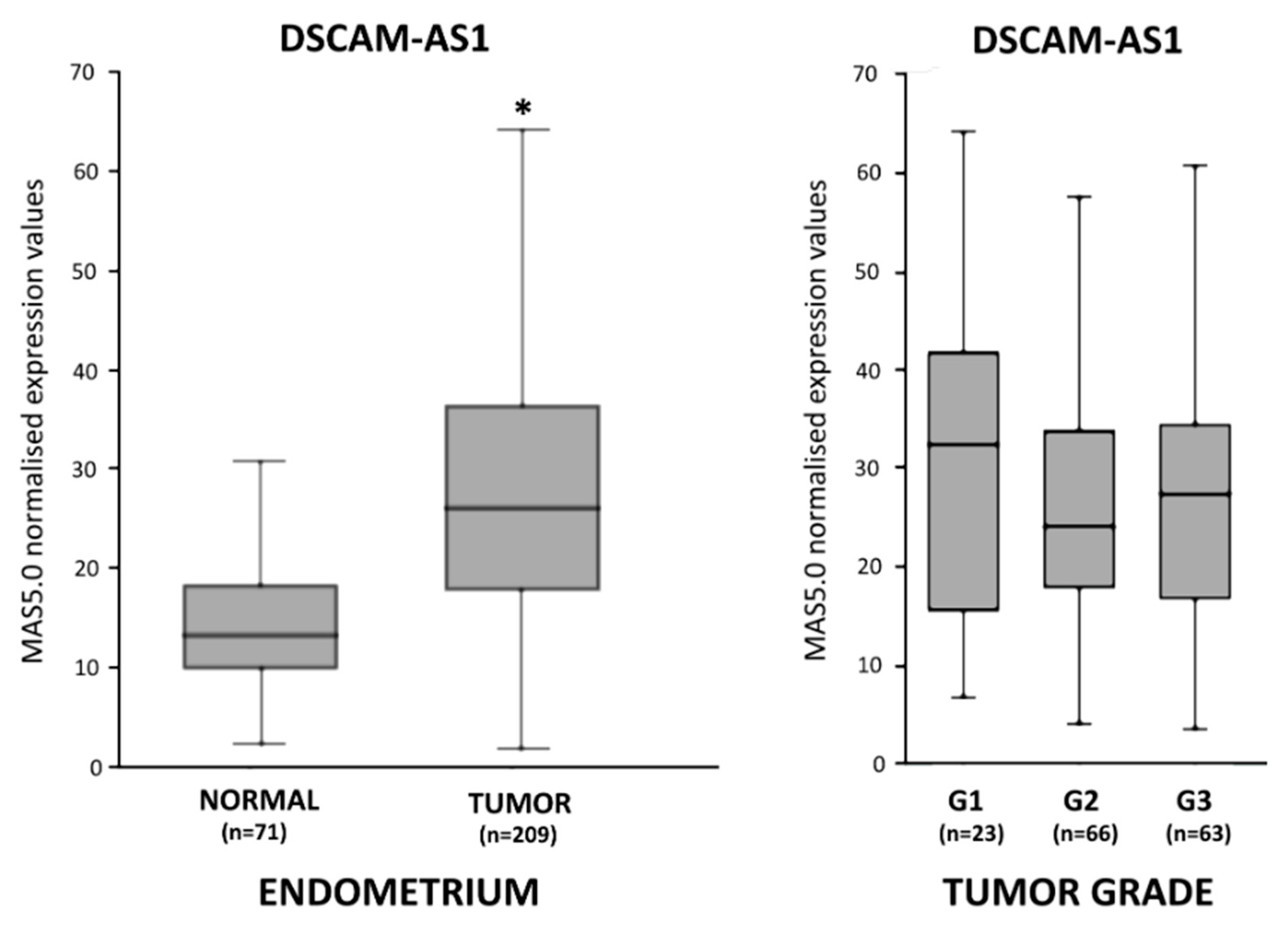
3.1. DSCAM-AS1 Overexpression in Endometrial Adenocarcinoma
3.2. Genes Correlated with DSCAM-AS1 in Endometrial Adenocarcinoma
3.3. Knockdown of DSCAM-AS1 in Endometrial Cancer Cell Lines
3.4. Transcriptome Alterations after DSCAM-AS1 Knockdown in Endometrial Cancer Cells
3.5. Correlation of DSCAM-AS1 with Knockdown DEGs in Endometrial Adenocarcinoma Tissues
3.6. Expression of DSCAM-AS1 and Survival of Endometrial Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer Statistics, 2008. CA A Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.G. Endometrial Carcinoma. N. Engl. J. Med. 1996, 335, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.S.G. Endometrial cancer, types, prognosis, female hormones and antihormones. Climacteric 2011, 14, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Oehler, M.K.; Brand, A.; Wain, G.V. Molecular genetics and endometrial cancer. J. Br. Menopause Soc. 2003, 9, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.K.D.M.; Rajkumar, T.; Mani, S. Perspectives of long non-coding RNAs in cancer. Mol. Biol. Rep. 2017, 44, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Vasilatou, D.; Sioulas, V.D.; Pappa, V.; Papageorgiou, S.G.; Vlahos, N.F. The role of miRNAs in endometrial cancer. Epigenomics 2015, 7, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wan, J.; Chu, J. Long non-coding RNAs and endometrial cancer. Biomed. Pharmacother. 2019, 119, 109396. [Google Scholar] [CrossRef]
- Jiang, Y.; Malouf, G.G.; Zhang, J.; Zheng, X.; Chen, Y.; Thompson, E.J.; Weinstein, J.N.; Yuan, Y.; Spano, J.-P.; Broaddus, R.; et al. Long non-coding RNA profiling links subgroup classification of endometrioid endometrial carcinomas with trithorax and polycomb complex aberrations. Oncotarget 2015, 6, 39865–39876. [Google Scholar] [CrossRef] [Green Version]
- Ravo, M.; Cordella, A.; Saggese, P.; Rinaldi, A.; Castaldi, M.A.; Nassa, G.; Giurato, G.; Zullo, F.; Weisz, A.; Tarallo, R.; et al. Identification of long non-coding RNA expression patterns useful for molecular-based classification of type I endometrial cancers. Oncol. Rep. 2019, 41, 1209–1217. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Rudland, P.S.; Sibson, D.R.; Barraclough, R. Identification of mRNAs differentially-expressed between benign and malignant breast tumour cells. Br. J. Cancer 2002, 87, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miano, V.; Ferrero, G.; Reineri, S.; Caizzi, L.; Annaratone, L.; Ricci, L.; Cutrupi, S.; Castellano, I.; Cordero, F.; de Bortoli, M. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget 2016, 7, 3201–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miano, V.; Ferrero, G.; Rosti, V.; Manitta, E.; Elhasnaoui, J.; Basile, G.; de Bortoli, M. Luminal lncRNAs Regulation by ERα-Controlled Enhancers in a Ligand-Independent Manner in Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Kong, D.; Chen, Q.; Ping, Y.; Pang, D. Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer 2017, 16, 129. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Li, A.-Q.; Zhou, P.; Jiang, Y.-Z.; Jin, X.; Liu, Y.-R.; Guo, Y.-J.; Yang, W.-T.; Shao, Z.-M.; Xu, X.-E. DSCAM-AS1 regulates the G1/S cell cycle transition and is an independent prognostic factor of poor survival in luminal breast cancer patients treated with endocrine therapy. Cancer Med. 2018, 7, 6137–6146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Huang, Y.-X.; Wang, D.-L.; Yang, B.; Yan, H.-Y.; Lin, L.-H.; Li, Y.; Chen, J.; Xie, L.-M.; Huang, Y.-S.; et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 2020, 10, 10823–10837. [Google Scholar] [CrossRef]
- Liang, W.-H.; Li, N.; Yuan, Z.-Q.; Qian, X.-L.; Wang, Z.-H. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol. Carcinog. 2019, 58, 461–473. [Google Scholar] [CrossRef]
- Liao, J.; Xie, N. Long noncoding RNA DSCAM-AS1 functions as an oncogene in non-small cell lung cancer by targeting BCL11A. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1087–1092. [Google Scholar] [CrossRef]
- Li, B.; Sun, H.; Zhang, J. LncRNA DSCAM-AS1 promotes colorectal cancer progression by acting as a molecular sponge of miR-384 to modulate AKT3 expression. Aging 2020, 12, 9781–9792. [Google Scholar] [CrossRef]
- Zhang, S.; Ding, L.; Gao, F.; Fan, H. Long non-coding RNA DSCAM-AS1 upregulates USP47 expression through sponging miR-101-3p to accelerate osteosarcoma progression. Biochem. Cell Biol. 2020, 98, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, S.; Zhang, G. LncRNA DSCAM-AS1 Negatively Interacts with miR-124 to Promote Hepatocellular Carcinoma Proliferation. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-L.; Xu, Q.; Wang, X. Long noncoding RNA DSCAM-AS1 is associated with poor clinical prognosis and contributes to melanoma development by sponging miR-136. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2888–2897. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Ebrahimzadeh, K. A Review on the Carcinogenic Roles of DSCAM-AS1. Front. Cell Dev. Biol. 2021, 9, 758513. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, P.; Huang, B.; Cai, P. lncRNA DSCAM-AS1 facilitates the progression of endometrial cancer via miR-136-5p. Oncol. Lett. 2021, 22, 825. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Abdolmaleki, F.; Ghafoui-Fard, S.; Taheri, M.; Mordadi, A.; Afsharpad, M.; Varmazyar, S.; Nazparvar, B.; Oskooei, V.K.; Omrani, M.D. Expression analysis of a panel of long non-coding RNAs (lncRNAs) revealed their potential as diagnostic biomarkers in bladder cancer. Genomics 2020, 112, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-de-Arellano, A.; Villegas-Pineda, J.C.; Hernández-Silva, C.D.; Pereira-Suárez, A.L. The Relevant Participation of Prolactin in the Genesis and Progression of Gynecological Cancers. Front. Endocrinol. 2021, 12, 747810. [Google Scholar] [CrossRef] [PubMed]
- Acs, G.; Xu, X.; Chu, C.; Acs, P.; Verma, A. Prognostic significance of erythropoietin expression in human endometrial carcinoma. Cancer 2004, 100, 2376–2386. [Google Scholar] [CrossRef] [PubMed]
- Pillich, R.T.; Chen, J.; Churas, C.; Liu, S.; Ono, K.; Otasek, D.; Pratt, D. NDEx: Accessing Network Models and Streamlining Network Biology Workflows. Curr. Protoc. 2021, 1, e258. [Google Scholar] [CrossRef] [PubMed]
- Dutta, M.; Das, B.; Mohapatra, D.; Behera, P.; Senapati, S.; Roychowdhury, A. MicroRNA-217 modulates pancreatic cancer progression via targeting ATAD2. Life Sci. 2022, 301, 120592. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Yang, L.; Xu, B.; Wang, M.; Kong, X.; Song, Z. miR-217-5p suppresses epithelial-mesenchymal transition and the NF-κB signaling pathway in breast cancer via targeting of metadherin. Oncol. Lett. 2022, 23, 162. [Google Scholar] [CrossRef]
- Hamidi, A.A.; Zangoue, M.; Kashani, D.; Zangouei, A.S.; Rahimi, H.R.; Abbaszadegan, M.R.; Moghbeli, M. MicroRNA-217: A therapeutic and diagnostic tumor marker. Expert Rev. Mol. Diagn. 2022, 22, 61–76. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Hussen, B.M.; Taheri, M.; Sharifi, G. The Emerging Role of Non-Coding RNAs in Pituitary Gland Tumors and Meningioma. Cancers 2021, 13, 5987. [Google Scholar] [CrossRef]
- Xing, X.; An, M.; Chen, T. LncRNA SNHG20 promotes cell proliferation and invasion by suppressing miR-217 in ovarian cancer. Genes Genom. 2021, 43, 1095–1104. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, X.; Jiang, Y.; Jia, X.; Guo, Y. miR-217-5p Inhibits Invasion and Metastasis of Prostate Cancer by Targeting Clusterin. Mamm. Genome 2021, 32, 371–380. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, X.; Wang, Y.; Li, L.; Wang, Q.; Zheng, J. Expression and Prognostic Significance of Wnt7a in Human Endometrial Carcinoma. Obstet. Gynecol. Int. 2012, 2012, 134962. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Qian, Y.; Ye, M.; Fu, Z.; Jia, X.; Li, W.; Xu, P.; Lv, M.; Huang, L.; Wang, L.; et al. Distinct expression profile of lncRNA in endometrial carcinoma. Oncol. Rep. 2016, 36, 3405–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ZHAI, W.E.; LI, X.U.; WU, S.; ZHANG, Y.A.; PANG, H.; CHEN, W.E. Microarray expression profile of lncRNAs and the upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int. J. Oncol. 2015, 46, 2125–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Zhang, Z.; Zhao, H.; Bao, S.; Sun, J. A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer 2018, 18, 1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolle, M.; Bullock, M.; Ling, H.; Pichler, M.; Haybaeck, J. Long Non-Coding RNAs in Endometrial Carcinoma. Int. J. Mol. Sci. 2015, 16, 26463–26472. [Google Scholar] [CrossRef] [Green Version]
- Takenaka, K.; Chen, B.J.; Modesitt, S.C.; Byrne, F.L.; Hoehn, K.L.; Janitz, M. The emerging role of long non-coding RNAs in endometrial cancer. Cancer Genet. 2016, 209, 445–455. [Google Scholar] [CrossRef]
- Khorshidi, H.; Azari, I.; Oskooei, V.K.; Taheri, M.; Ghafouri-Fard, S. DSCAM-AS1 up-regulation in invasive ductal carcinoma of breast and assessment of its potential as a diagnostic biomarker. Breast Dis. 2019, 38, 25–30. [Google Scholar] [CrossRef]
- Qiu, Z.; Pan, X.X.; You, D.Y. LncRNA DSCAM-AS1 promotes non-small cell lung cancer progression via regulating miR-577/HMGB1 axis. Neoplasma 2020, 67, 871–879. [Google Scholar] [CrossRef]
- Yu, C.-L.; Xu, N.-W.; Jiang, W.; Zhang, H.; Ma, Y. LncRNA DSCAM-AS1 promoted cell proliferation and invasion in osteosarcoma by sponging miR-101. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7709–7717. [Google Scholar] [CrossRef]
- Ji, D.; Hu, G.; Zhang, X.; Yu, T.; Yang, J. Long non-coding RNA DSCAM-AS1 accelerates the progression of hepatocellular carcinoma via sponging miR-338-3p. Am. J. Transl. Res. 2019, 11, 4290–4302. [Google Scholar]
- Liang, J.; Zhang, S.; Wang, W.; Xu, Y.; Kawuli, A.; Lu, J.; Xiu, X. Long non-coding RNA DSCAM-AS1 contributes to the tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3 axis. Biosci. Rep. 2020, 40, BSR20192061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharaibeh, L.; Elmadany, N.; Alwosaibai, K.; Alshaer, W. Notch1 in Cancer Therapy: Possible Clinical Implications and Challenges. Mol. Pharmacol. 2020, 98, 559–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, W.; Huang, K.; Wang, Y.; Li, J.; Yang, X. MicroRNA-34a inhibits cells proliferation and invasion by downregulating Notch1 in endometrial cancer. Oncotarget 2017, 8, 111258–111270. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Ditzel, H.J.; Duijf, P.H.G.; Khaze, V.; Gjerstorff, M.F.; Baradaran, B. HMGA2 as a Critical Regulator in Cancer Development. Genes 2021, 12, 269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yi, Q.; Tang, L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 2019, 38, 250. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, B.; Hasenburg, A.; Waizenegger, M.; Orlowska-Volk, M.; Stickeler, E.; zur Hausen, A. Expression of focal adhesion kinase in patients with endometrial cancer: A clinicopathologic study. Int. J. Gynecol. Cancer 2009, 19, 1221–1225. [Google Scholar] [CrossRef]
- Talotta, F.; Casalino, L.; Verde, P. The nuclear oncoprotein Fra-1: A transcription factor knocking on therapeutic applications’ door. Oncogene 2020, 39, 4491–4506. [Google Scholar] [CrossRef]
- Sung, N.J.; Kim, N.H.; Surh, Y.-J.; Park, S.-A. Gremlin-1 Promotes Metastasis of Breast Cancer Cells by Activating STAT3-MMP13 Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 9227. [Google Scholar] [CrossRef]
- Li, T.-T.; Liu, M.-R.; Pei, D.-S. Friend or foe, the role of EGR-1 in cancer. Med. Oncol. 2019, 37, 7. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, K.; Cheng, B.; Gao, Q.; Wang, L.; Yang, X. TRAIL inhibits proliferation and promotes apoptosis of 3AO ovarian cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2014, 30, 453–457. [Google Scholar]
- Miller, C.; Sassoon, D.A. Wnt-7a maintains appropriate uterine patterning during the development of the mouse female reproductive tract. Development 1998, 125, 3201–3211. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhu, H.; Gao, Z.; Li, J.; Zhuang, J.; Dong, Y.; Shen, B.; Li, M.; Zhou, H.; Guo, H.; et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J. Biol. Chem. 2018, 293, 6693–6706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Jiang, Y.; He, R.; Tao, L.; Yang, M.; Fu, X.; Yang, J.; Zhu, K. High expression of WNT7A predicts poor prognosis and promote tumor metastasis in pancreatic ductal adenocarcinoma. Sci. Rep. 2018, 8, 607. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Avasarala, S.; van Scoyk, M.; Arcaroli, J.; Brzezinski, C.; Zhang, W.; Edwards, M.G.; Rathinam, M.K.K.; Zhou, T.; Tauler, J.; et al. Wnt7a is a novel inducer of β-catenin-independent tumor-suppressive cellular senescence in lung cancer. Oncogene 2015, 34, 5406. [Google Scholar] [CrossRef] [Green Version]
- Yi, K.; Min, K.-W.; Wi, Y.C.; Kim, Y.; Shin, S.-J.; Chung, M.S.; Jang, K.; Paik, S.S. Wnt7a Deficiency Could Predict Worse Disease-Free and Overall Survival in Estrogen Receptor-Positive Breast Cancer. J. Breast Cancer 2017, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Solano, M.; Meza-Canales, I.D.; Torres-Reyes, L.A.; Alvarez-Zavala, M.; Alvarado-Ruíz, L.; Rincon-Orozco, B.; Garcia-Chagollan, M.; Ochoa-Hernández, A.B.; Ortiz-Lazareno, P.C.; Rösl, F.; et al. Expression of WNT genes in cervical cancer-derived cells: Implication of WNT7A in cell proliferation and migration. Exp. Cell Res. 2015, 335, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Xie, S.; Zhou, J.; Zhu, Y. Nomegestrol Acetate Suppresses Human Endometrial Cancer RL95-2 Cells Proliferation In Vitro and In Vivo Possibly Related to Upregulating Expression of SUFU and Wnt7a. Int. J. Mol. Sci. 2017, 18, 1337. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.-K.; Fan, C.-C.; Lin, P.-S.; Liao, P.-Y.; Tung, J.-C.; Hsieh, C.-H.; Hung, M.-C.; Chen, C.-H.; Chang, W.-C. Sciellin mediates mesenchymal-to-epithelial transition in colorectal cancer hepatic metastasis. Oncotarget 2016, 7, 25742–25754. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A.; Banyai, D.; Semjen, D.; Beothe, T.; Kovacs, G. Sciellin is a marker for papillary renal cell tumours. Virchows Arch. 2015, 467, 695–700. [Google Scholar] [CrossRef]
- Li Santi, A.; Napolitano, F.; Montuori, N.; Ragno, P. The Urokinase Receptor: A Multifunctional Receptor in Cancer Cell Biology. Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 4111. [Google Scholar] [CrossRef]
- Xue, A.; Xue, M.; Jackson, C.; Smith, R.C. Suppression of urokinase plasminogen activator receptor inhibits proliferation and migration of pancreatic adenocarcinoma cells via regulation of ERK/p38 signaling. Int. J. Biochem. Cell Biol. 2009, 41, 1731–1738. [Google Scholar] [CrossRef] [PubMed]
- Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef] [PubMed]





| (a) GSEA OF GENES POSITIVELY CORRELATED WITH DSCAM-AS1 IN ENDOMETRIAL CANCER | ||||
| Gene Set Name (# Genes) | Description | Genes in Overlap | p-value | FDR |
| LIU COMMON CANCER GENES (63) | Low abundance transcripts common to nasopharyngeal carcinoma (NPC), breast and liver tumors | 12 | 2.96 × 10−12 | 9.43 × 1−9 |
| miR-373-5p (956) | Genes predicted to be high confidence targets of miRBase v22 hsa-miR-373-5p in miRDB v6.0 | 31 | 1.81 × 10−6 | 1.73 × 10−3 |
| miR-371-5p (959) | Genes predicted to be high confidence targets of miRBase v22 hsa-miR-371-5p in miRDB v6.0 | 31 | 1.92 × 10−6 | 1.73 × 10−3 |
| miR-616-5p (961) | Genes predicted to be high confidence targets of miRBase v22 hsa-miR-616-5p in miRDB v6.0 | 31 | 1.99 × 10−6 | 1.73 × 10−3 |
| REACTOME TRAF6 MEDIATED IRF7 ACTIVATION (30) | TRAF6 mediated IRF7 activation | 5 | 1.53 × 10−5 | 2.35 × 10−3 |
| (b) GSEA OF GENES NEGATIVELY CORRELATED WITH DSCAM-AS1 IN ENDOMETRIAL CANCER | ||||
| SCGGAAGY ELK1 02 (1242) | Genes with occurrence of the motif M3 SCGGAAGY, the ELK1 transcription factor binding site V$ELK1_02 (v7.4 TRANSFAC) | 36 | 9.7 × 10−23 | 1.09 × 10-−9 |
| GOBP PROTEOLYSIS (1790) | The hydrolysis of proteins into smaller polypeptides and/or amino acids | 35 | 9.81 × 10−17 | 2.5 × 10−13 |
| GOBP MACROMOLECULE CATABOLIC PROCESS (1331) | The chemical reactions and pathways resulting in the breakdown of a macromolecule | 34 | 9.16 × 10−20 | 7.01 × 10−16 |
| GOMF RNA BINDING (1972) | Binding to an RNA molecule | 31 | 2.34 × 10−12 | 4.06 × 10−9 |
| GOBP PROTEIN CATABOLIC PROCESS (977) | The chemical reactions and pathways resulting in the breakdown of a protein | 27 | 1.33 × 10−16 | 2.54 × 10−13 |
| GOBP CELLULAR PROTEIN CATABOLIC PROCESS (819) | The chemical reactions and pathways resulting in the breakdown of a protein by individual cells. | 26 | 1.9 × 10−17 | 7.26 × 10−14 |
| HALLMARK MYC TARGETS V1 (200) | A subgroup of genes regulated by MYC - version 1 (v1). | 17 | 9.3 × 10−19 | 4.65 × 10−17 |
| REACTOME SWITCHING OF ORIGINS TO A POSTREPLICATIVE STATE (91) | Switching of origins to a post-replicative state | 14 | 2.29 × 10−19 | 1.85 × 10−16 |
| WP PROTEASOME DEGRADATION (64) | Proteasome degradation | 12 | 7.18 × 10−18 | 4.77 × 10−15 |
| REACTOME REGULATION OF PTEN STABILITY AND ACTIVITY (69) | Regulation of PTEN stability and activity | 11 | 1.19 × 10−15 | 8.37 × 10−14 |
| Genes Regulated upon DSCAM-AS1 Silencing (Top 10) | |||||
|---|---|---|---|---|---|
| HEC-1B | RL95-2 | ||||
| Gene Symbol | Gene Name | FC | Gene Symbol | Gene Name | FC |
| SCEL | sciellin | −5.99 | EHF | ets homologous factor | −3.14 |
| TMC7 | transmembrane channel like 7 | −4.93 | MAP2 | microtubule associated protein 2 | −2.84 |
| ELL2 | elongation factor, RNA polymerase II, 2 | −4.70 | LNPEP | UTR3 best transcript NM_175920 | −2.79 |
| UNC13D | unc-13 homolog D (C. elegans) | −3.46 | KRT23 | keratin 23, type I | −2.72 |
| PLAU | plasminogen activator, urokinase | −3.39 | FADS1 | fatty acid desaturase 1 | −2.69 |
| STEAP2 | STEAP family member 2, metalloreductase | 5.59 | HOXB9 | homeobox B9 | 2.78 |
| SLC3A1 | solute carrier family 3, member 1 | 6.12 | SPIN3 | spindlin family, member 3 | 2.89 |
| THG1L | tRNA-histidine guanylyltransferase 1-like | 6.20 | IGF2 | insulin-like growth factor 2 | 2.94 |
| WNT7A | wingless-type MMTV integration site family, member 7A | 7.15 | FOLR1 | folate receptor 1 (adult) | 2.98 |
| TNSF10 | tumor necrosis factor (ligand) superfamily, member 10 | 7.92 | PAQR8 | progestin and adipoQ receptor family member VIII | 3.04 |
| Gene Ontology Annotation of Genes Downregulated after DSCAM-AS1 Knockdown in EC cells | ||
| Gene Ontology Category “Biological Processes” (Activated by DSCAM-AS1) | Corrected p-Value | Downregulated Genes after DSCAM-AS1 KD (Cut-Off: 2.5-Fold), Annotated to the GO Terms |
| positive regulation of cell migration GO: 0030335 | 3.61 × 10−16 | CLDN4, MAP2K3, PLP1, DOCK1, PLAU, BDKRB1, NOTCH1, CYR61, C10orf54, RIN2, ANXA3, PTK2, EDN2, TJP1, IFNG, PTN, SERPINE1 |
| negative regulation of cell death GO:0060548 | 1.62 × 10−6 | NOTCH1, UNC5B, HMGA2, PTK2, TJP1, ZFPM2, MECP2, GREM1, SERPINE1, PROK2, CYR61, CD34 |
| positive regulation of angiogenesis GO:0045766 | 1.30 × 10−6 | BMPER, GREM1, SERPINE1, ANXA3, HMGA2, TJP1, CD34 |
| negative regulation of apoptotic process GO:0043066 | 9.46 × 10−5 | NOTCH1, UNC5B, HMGA2, PTK2, TJP1, MECP2, GREM1, SERPINE1, PROK2, CYR61 |
| positive regulation of cell proliferation GO:0008284 | 0.00014 | NOTCH1, HMGA2, PTK2, FOSL1, GREM1, EGR1 |
| Gene Ontology Annotation of Genes Upregulated after DSCAM-AS1 Knockdown in EC cells | ||
| Gene Ontology Category “Biological Processes” (Inhibited by DSCAM-AS1) | Corrected p-Value | Upregulated Genes after DSCAM-AS1 KD (Cut-Off: 2.5-Fold), Annotated to the GO Terms |
| positive regulation of developmental process GO:0051094 | 8.59 × 10−14 | SEMA4A, IRX3, DKK1, SMAD7, SEMA4D, LINGO2, C5AR1, IGF1R, PLXNB2, CAMK2B, WNT7A, RELN, PLXNB1, FN1, TGFBR1, INSR, SLITRK5 |
| positive regulation of apoptotic process GO:0043065 | 3.87 × 10−8 | TNFSF10, HTRA1, SKIL, CYP1B1, BMP4, BMP2, TGFBR1, DKKL1, FOXO3, ZC3H12A, IGFBP3 |
| positive regulation of cell junction assembly GO:1901890 | 1.24 × 10−8 | SEMA4A, IRX3, ACE2, WNT7A, SEMA4D, EPB41L5, LINGO2, SLITRK5 |
| positive regulation of cell development GO:0010720 | 2.04 × 10−6 | PLXNB2, CAMK2B, SMAD7, RELN, SEMA4D, PLXNB1, EPHA4, FN1, CX3CL1 |
| positive regulation of cell differentiation GO:0045597 | 1.34 × 10−5 | IRX3, DKK1, PLXNB2, CAMK2B, SMAD7, RELN, SEMA4D, PLXBN1, EPHA4, FN1, CX3CL1, TGFBR1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Treeck, O.; Weber, F.; Fritsch, J.; Skrzypczak, M.; Schüler-Toprak, S.; Buechler, C.; Ortmann, O. DSCAM-AS1 Long Non-Coding RNA Exerts Oncogenic Functions in Endometrial Adenocarcinoma via Activation of a Tumor-Promoting Transcriptome Profile. Biomedicines 2022, 10, 1727. https://doi.org/10.3390/biomedicines10071727
Treeck O, Weber F, Fritsch J, Skrzypczak M, Schüler-Toprak S, Buechler C, Ortmann O. DSCAM-AS1 Long Non-Coding RNA Exerts Oncogenic Functions in Endometrial Adenocarcinoma via Activation of a Tumor-Promoting Transcriptome Profile. Biomedicines. 2022; 10(7):1727. https://doi.org/10.3390/biomedicines10071727
Chicago/Turabian StyleTreeck, Oliver, Florian Weber, Juergen Fritsch, Maciej Skrzypczak, Susanne Schüler-Toprak, Christa Buechler, and Olaf Ortmann. 2022. "DSCAM-AS1 Long Non-Coding RNA Exerts Oncogenic Functions in Endometrial Adenocarcinoma via Activation of a Tumor-Promoting Transcriptome Profile" Biomedicines 10, no. 7: 1727. https://doi.org/10.3390/biomedicines10071727
APA StyleTreeck, O., Weber, F., Fritsch, J., Skrzypczak, M., Schüler-Toprak, S., Buechler, C., & Ortmann, O. (2022). DSCAM-AS1 Long Non-Coding RNA Exerts Oncogenic Functions in Endometrial Adenocarcinoma via Activation of a Tumor-Promoting Transcriptome Profile. Biomedicines, 10(7), 1727. https://doi.org/10.3390/biomedicines10071727








