Cellular and Molecular Mechanisms and Effects of Berberine on Obesity-Induced Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Study of BBR
2.1.1. Target Genes of BBR
2.1.2. Protein-Protein Interaction (PPI) Network Construction
2.1.3. Functional Pathway Analysis
2.1.4. Molecular Docking Study
2.2. In Vivo Evaluation of Anti-Obesity Effects and Mechanisms of BBR
2.2.1. Experimental Study Design
2.2.2. Experimental Protocol on Glucose and Lipid Metabolism
2.2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.2.4. Fluorescence-Activated Cell Sorting (FACS)
2.2.5. Histological Analysis
2.2.6. Statistical Analysis
3. Results
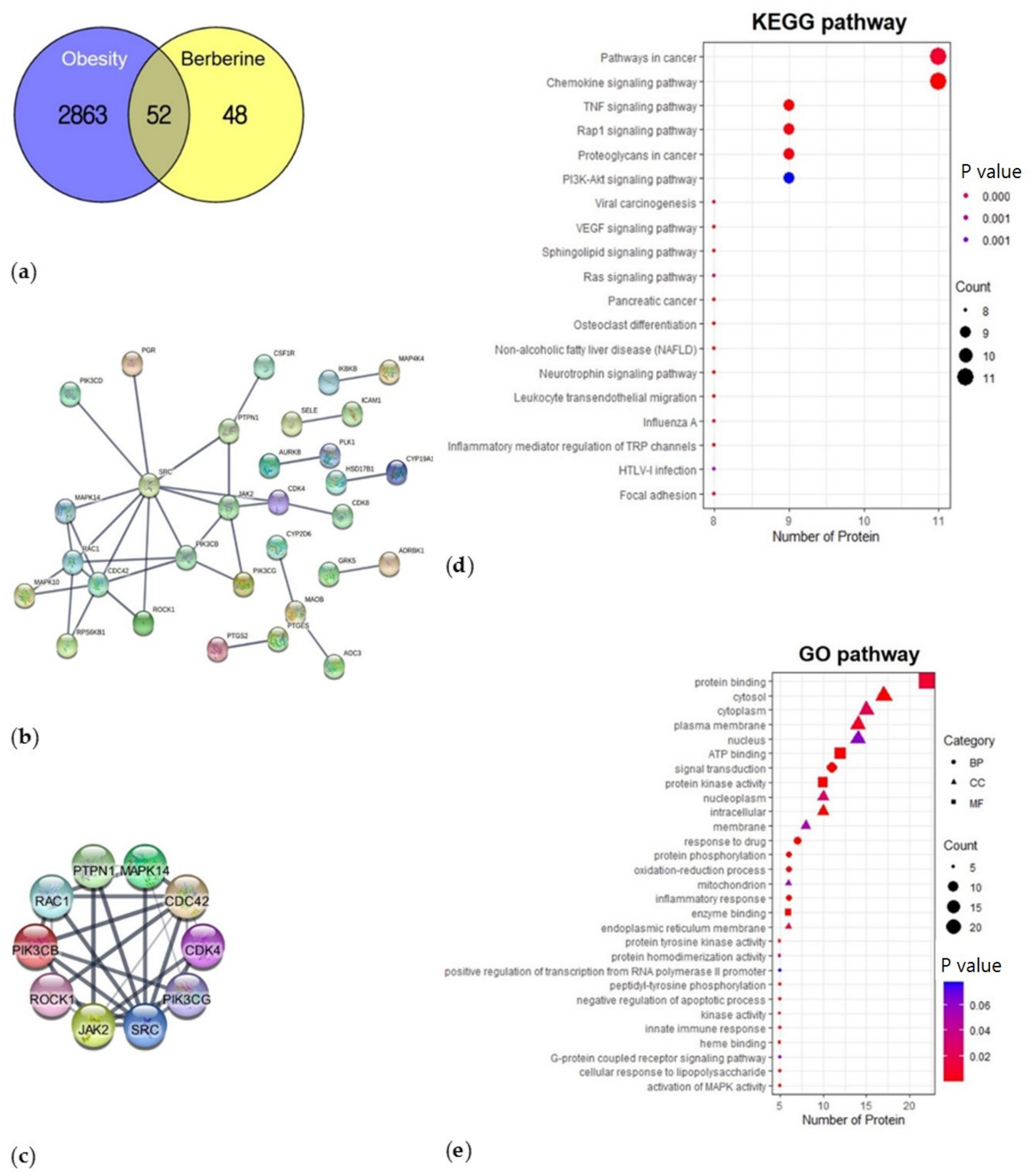
3.1. Network Pharmacological Analysis
3.1.1. Protein-Protein Interaction (PPI)
3.1.2. Functional Enrichment Analysis—KEGG Pathway and GO
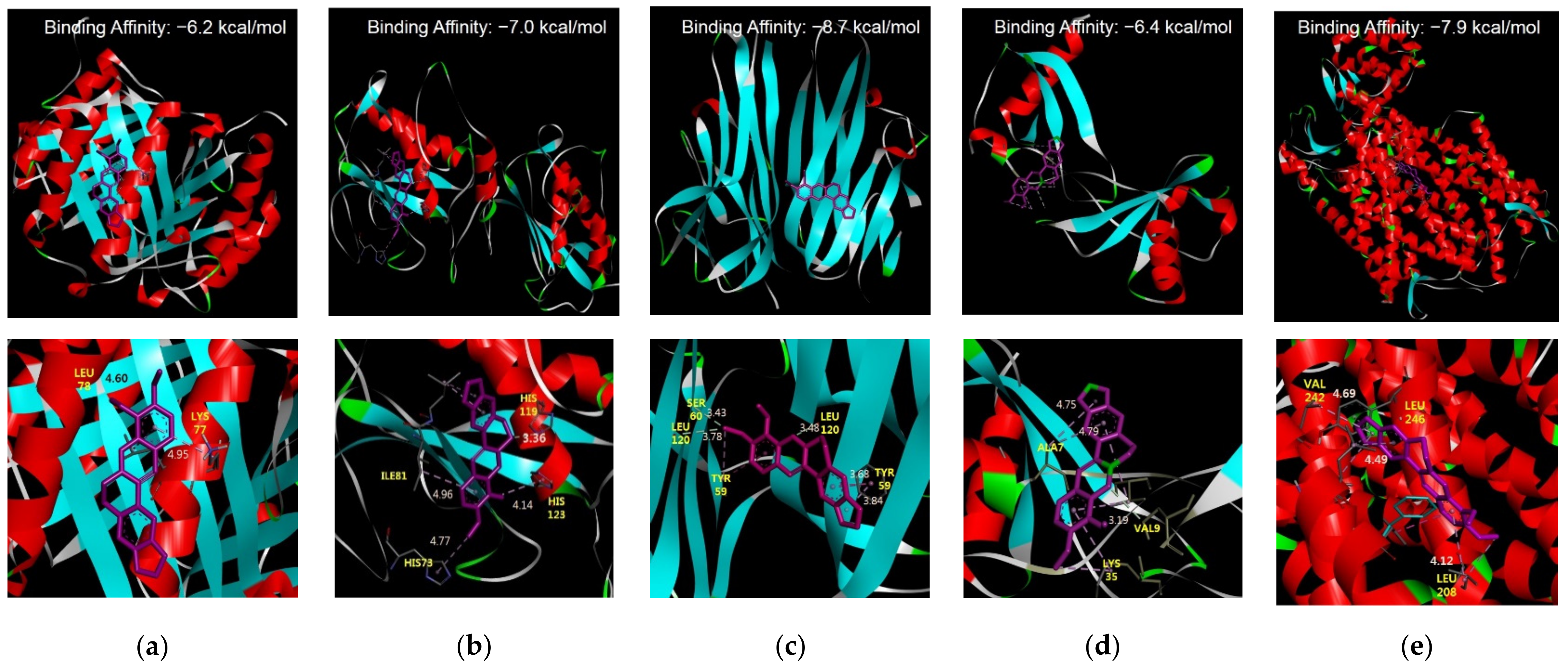
3.1.3. Molecular Docking Studies
3.2. Experimental Evaluation of Anti-Obesity Benefits
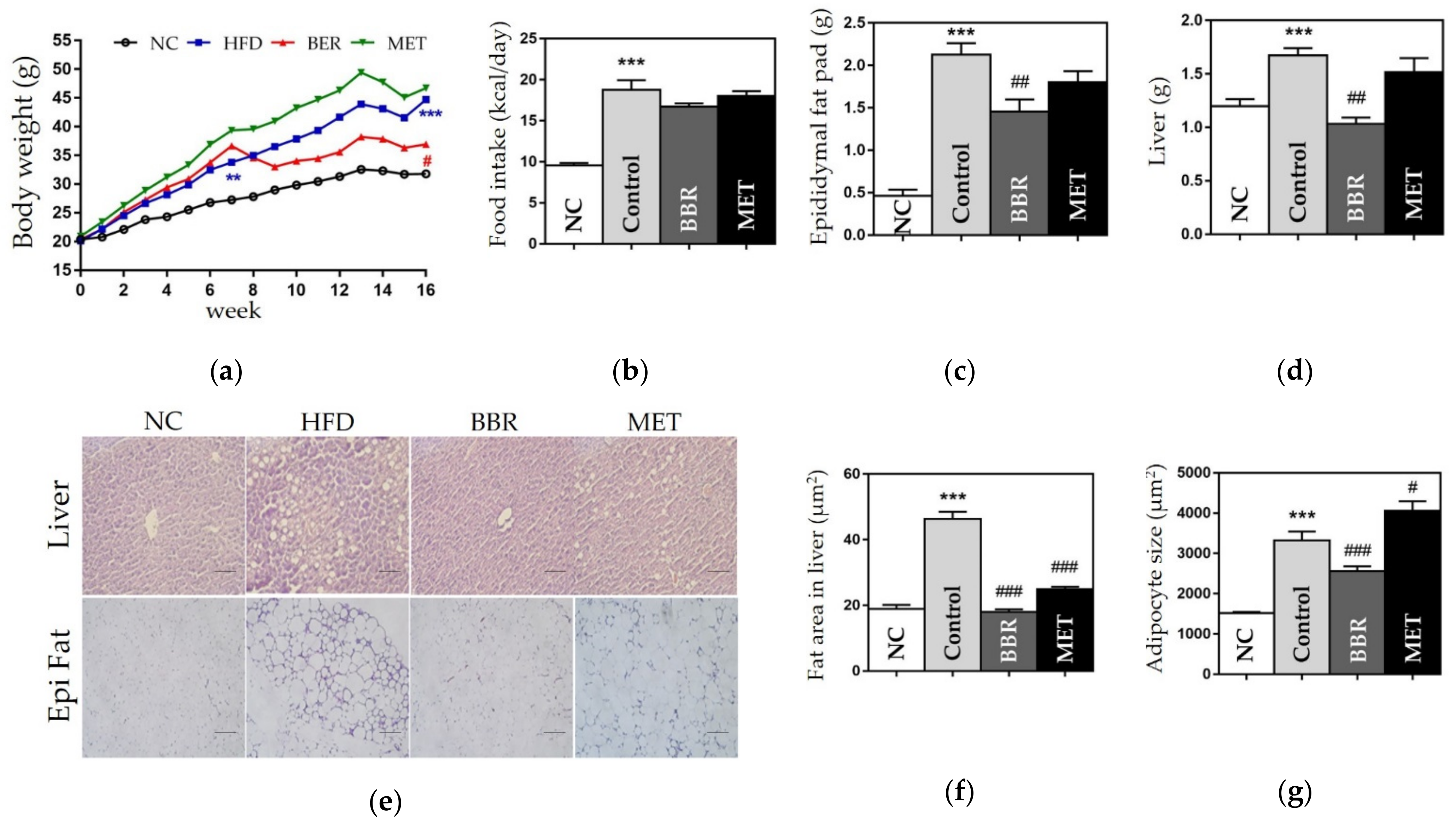
3.2.1. Change in Weight
3.2.2. Effects on the Size of Fat Area
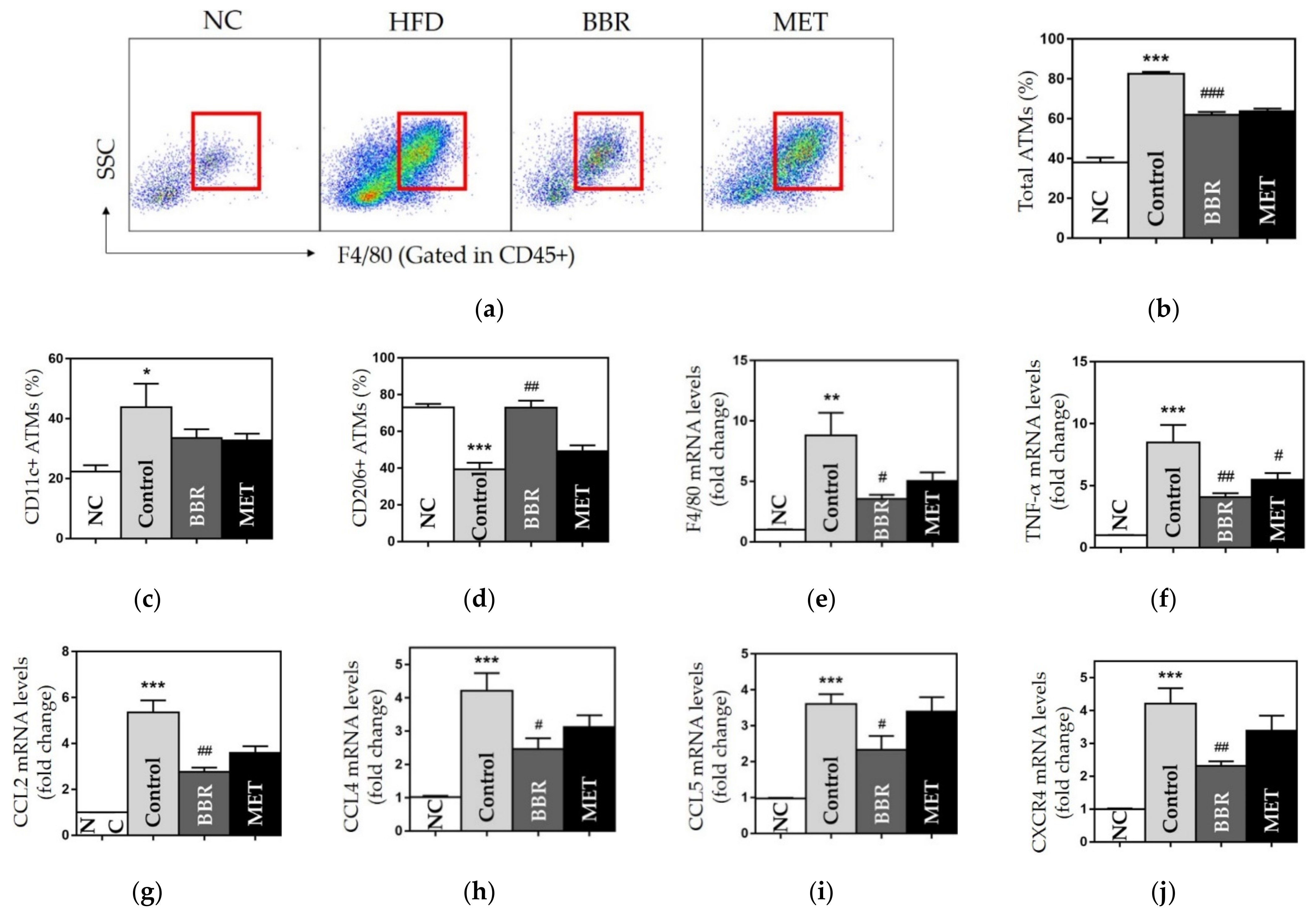
3.2.3. Effects on ATMs
3.2.4. Effects on Chemokines and Inflammatory Cytokines
3.2.5. Effects on Hyperglycemia and Hyperlipidemia
3.2.6. Safety
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Eckel, R.; Grundy, S.; Zimmet, P. The metabolic syndrome. Lancet 2005, 365, 1415–1428. [Google Scholar] [CrossRef]
- Russo, L.; Lumeng, C.N. Properties and functions of adipose tissue macrophages in obesity. Immunology 2018, 155, 407–417. [Google Scholar] [CrossRef]
- Saghy, T.; Koroskenyi, K.; Hegedus, K.; Antal, M.; Banko, C.; Bacso, Z.; Papp, A.; Stienstra, R.; Szondy, Z. Loss of transglutaminase 2 sensitizes for diet-induced obesity-related inflammation and insulin resistance due to enhanced macrophage c-Src signaling. Cell Death Dis. 2019, 10, 439. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.K.; Dhir, A. Berberine: A plant alkaloid with therapeutic potential for central nervous system disorders. Phytother. Res. 2010, 24, 317–324. [Google Scholar] [CrossRef]
- Li, M.H.; Zhang, Y.J.; Yu, Y.H.; Yang, S.H.; Iqbal, J.; Mi, Q.Y.; Li, B.; Wang, Z.M.; Mao, W.X.; Xie, H.G.; et al. Berberine improves pressure overload-induced cardiac hypertrophy and dysfunction through enhanced autophagy. Eur. J. Pharmacol. 2014, 728, 67–76. [Google Scholar] [CrossRef]
- Tillhon, M.; Guaman Ortiz, L.M.; Lombardi, P.; Scovassi, A.I. Berberine: New perspectives for old remedies. Biochem. Pharmacol. 2012, 84, 1260–1267. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Yan, J.; Shen, Y.; Tang, K.; Yin, J.; Zhang, Y.; Yang, D.; Liang, H.; Ye, J.; Weng, J. Berberine improves glucose metabolism in diabetic rats by inhibition of hepatic gluconeogenesis. PLoS ONE 2011, 6, e16556. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Kim, W.S.; Kim, K.H.; Yoon, M.J.; Cho, H.J.; Shen, Y.; Ye, J.M.; Lee, C.H.; Oh, W.K.; Kim, C.T.; et al. Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes 2006, 55, 2256–2264. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Ehli, E.A.; Kittelsrud, J.; Ronan, P.J.; Munger, K.; Downey, T.; Bohlen, K.; Callahan, L.; Munson, V.; Jahnke, M.; et al. Lipid-lowering effect of berberine in human subjects and rats. Phytomedicine 2012, 19, 861–867. [Google Scholar] [CrossRef]
- Gong, J.; Li, J.; Dong, H.; Chen, G.; Qin, X.; Hu, M.; Yuan, F.; Fang, K.; Wang, D.; Jiang, S.; et al. Inhibitory effects of berberine on proinflammatory M1 macrophage polarization through interfering with the interaction between TLR4 and MyD88. BMC Complement. Altern. Med. 2019, 19, 314. [Google Scholar] [CrossRef]
- Mo, C.; Wang, L.; Zhang, J.; Numazawa, S.; Tang, H.; Tang, X.; Han, X.; Li, J.; Yang, M.; Wang, Z.; et al. The crosstalk between Nrf2 and AMPK signal pathways is important for the anti-inflammatory effect of berberine in LPS-stimulated macrophages and endotoxin-shocked mice. Antioxid. Redox. Signal. 2014, 20, 574–588. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, Y.; Gong, Z.; Sheng, X.; Li, Z.; Zhang, W.; Qin, Y. Berberine inhibits 3T3-L1 adipocyte differentiation through the PPARgamma pathway. Biochem. Biophys. Res. Commun. 2006, 348, 571–578. [Google Scholar] [CrossRef]
- Lin, J.; Cai, Q.; Liang, B.; Wu, L.; Zhuang, Y.; He, Y.; Lin, W. Berberine, a Traditional Chinese Medicine, Reduces Inflammation in Adipose Tissue, Polarizes M2 Macrophages, and Increases Energy Expenditure in Mice Fed a High-Fat Diet. Med. Sci. Monit. 2019, 25, 87–97. [Google Scholar] [CrossRef]
- Cui, G.; Qin, X.; Zhang, Y.; Gong, Z.; Ge, B.; Zang, Y.Q. Berberine differentially modulates the activities of ERK, p38 MAPK, and JNK to suppress Th17 and Th1 T cell differentiation in type 1 diabetic mice. J. Biol. Chem. 2009, 284, 28420–28429. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Liang, S.; Guo, C.; Yu, X.; Zhao, J.; Zhang, H.; Shang, W. Inhibition of M1 macrophage activation in adipose tissue by berberine improves insulin resistance. Life Sci. 2016, 166, 82–91. [Google Scholar] [CrossRef]
- Noh, J.W.; Yang, H.K.; Jun, M.S.; Lee, B.C. Puerarin Attenuates Obesity-Induced Inflammation and Dyslipidemia by Regulating Macrophages and TNF-Alpha in Obese Mice. Biomedicines 2022, 10, 175. [Google Scholar] [CrossRef]
- Chen, J. The Src/PI3K/Akt pathway may play a key role in the production of IL-17 in obesity. J. Leukoc. Biol. 2010, 87, 355, author reply 357. [Google Scholar] [CrossRef]
- Kong, L.; Ge, B.X. MyD88-independent activation of a novel actin-Cdc42/Rac pathway is required for Toll-like receptor-stimulated phagocytosis. Cell Res. 2008, 18, 745–755. [Google Scholar] [CrossRef]
- Huang, Q.Y.; Lai, X.N.; Qian, X.L.; Lv, L.C.; Li, J.; Duan, J.; Xiao, X.H.; Xiong, L.X. Cdc42: A Novel Regulator of Insulin Secretion and Diabetes-Associated Diseases. Int. J. Mol. Sci. 2019, 20, 179. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Yu, D.; Ning, S.; Zhang, H.; Jiang, L.; He, L.; Li, M.; Sun, M. Augmented Rac1 Expression and Activity are Associated with Oxidative Stress and Decline of beta Cell Function in Obesity. Cell Physiol. Biochem. 2015, 35, 2135–2148. [Google Scholar] [CrossRef]
- Breasson, L.; Becattini, B.; Sardi, C.; Molinaro, A.; Zani, F.; Marone, R.; Botindari, F.; Bousquenaud, M.; Ruegg, C.; Wymann, M.P.; et al. PI3Kgamma activity in leukocytes promotes adipose tissue inflammation and early-onset insulin resistance during obesity. Sci. Signal. 2017, 10, eaaf2969. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, N.; Ueki, K.; Okazaki, Y.; Iwane, A.; Kubota, N.; Ohsugi, M.; Awazawa, M.; Kobayashi, M.; Sasako, T.; Kaneko, K.; et al. Blockade of class IB phosphoinositide-3 kinase ameliorates obesity-induced inflammation and insulin resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 5753–5758. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.J.; Kim, D.H.; Kim, D.G.; Lee, H.J.; Min, W.; Rhee, M.H.; Cho, J.Y.; Watarai, M.; Kim, S. Toll-like receptor 4-linked Janus kinase 2 signaling contributes to internalization of Brucella abortus by macrophages. Infect. Immun. 2013, 81, 2448–2458. [Google Scholar] [CrossRef] [Green Version]
- Lo, U.; Selvaraj, V.; Plane, J.M.; Chechneva, O.V.; Otsu, K.K.; Deng, W. p38alpha (MAPK14) critically regulates the immunological response and the production of specific cytokines and chemokines in astrocytes. Sci. Rep. 2014, 4, 7405. [Google Scholar] [CrossRef] [Green Version]
- Lagarrigue, S.; Lopez-Mejia, I.C.; Denechaud, P.D.; Escote, X.; Castillo-Armengol, J.; Jimenez, V.; Chavey, C.; Giralt, A.; Lai, Q.; Zhang, L.; et al. CDK4 is an essential insulin effector in adipocytes. J. Clin. Investig. 2016, 126, 335–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaman, L.D.; Boss, O.; Peroni, O.D.; Kim, J.K.; Martino, J.L.; Zabolotny, J.M.; Moghal, N.; Lubkin, M.; Kim, Y.B.; Sharpe, A.H.; et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell Biol. 2000, 20, 5479–5489. [Google Scholar] [CrossRef] [Green Version]
- Vemula, S.; Shi, J.; Hanneman, P.; Wei, L.; Kapur, R. ROCK1 functions as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Blood 2010, 115, 1785–1796. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Vazquez, I.; Fernandez-Veledo, S.; Kramer, D.K.; Vila-Bedmar, R.; Garcia-Guerra, L.; Lorenzo, M. Insulin resistance associated to obesity: The link TNF-alpha. Arch. Physiol. Biochem. 2008, 114, 183–194. [Google Scholar] [CrossRef]
- Engin, A.B.; Engin, A.; Gonul, I. The effect of adipocyte-macrophage crosstalk in obesity-related breast cancer. J. Mol. Endocrinol. 2019, 62, R201–R222. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef] [Green Version]
- Geeraerts, X.; Bolli, E.; Fendt, S.M.; van Ginderachter, J.A. Macrophage Metabolism As Therapeutic Target for Cancer, Atherosclerosis, and Obesity. Front. Immunol. 2017, 8, 289. [Google Scholar] [CrossRef] [Green Version]
- Lumeng, C.N.; Deyoung, S.M.; Bodzin, J.L.; Saltiel, A.R. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 2007, 56, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Chylikova, J.; Dvorackova, J.; Tauber, Z.; Kamarad, V. M1/M2 macrophage polarization in human obese adipose tissue. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2018, 162, 79–82. [Google Scholar] [CrossRef] [Green Version]
- Ruytinx, P.; Proost, P.; van Damme, J.; Struyf, S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front. Immunol. 2018, 9, 1930. [Google Scholar] [CrossRef] [Green Version]
- Castoldi, A.; Naffah de Souza, C.; Camara, N.O.; Moraes-Vieira, P.M. The Macrophage Switch in Obesity Development. Front. Immunol. 2015, 6, 637. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Liu, J.; Li, H. Metformin ameliorates olanzapine-induced insulin resistance via suppressing macrophage infiltration and inflammatory responses in rats. Biomed. Pharmacother. 2021, 133, 110912. [Google Scholar] [CrossRef]
- Han, Y.B.; Tian, M.; Wang, X.X.; Fan, D.H.; Li, W.Z.; Wu, F.; Liu, L. Berberine ameliorates obesity-induced chronic inflammation through suppression of ER stress and promotion of macrophage M2 polarization at least partly via downregulating lncRNA Gomafu. Int. Immunopharmacol. 2020, 86, 106741. [Google Scholar] [CrossRef]
- Jeong, H.W.; Hsu, K.C.; Lee, J.W.; Ham, M.; Huh, J.Y.; Shin, H.J.; Kim, W.S.; Kim, J.B. Berberine suppresses proinflammatory responses through AMPK activation in macrophages. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E955–E964. [Google Scholar] [CrossRef]
- Yang, J.; Ma, X.J.; Li, L.; Wang, L.; Chen, Y.G.; Liu, J.; Luo, Y.; Zhuang, Z.J.; Yang, W.J.; Zang, S.F.; et al. Berberine ameliorates non-alcoholic steatohepatitis in ApoE(-/-) mice. Exp. There. Med. 2017, 14, 4134–4140. [Google Scholar] [CrossRef] [Green Version]
- Kochumon, S.; Wilson, A.; Chandy, B.; Shenouda, S.; Tuomilehto, J.; Sindhu, S.; Ahmad, R. Palmitate Activates CCL4 Expression in Human Monocytic Cells via TLR4/MyD88 Dependent Activation of NF-kappaB/MAPK/PI3K Signaling Systems. Cell Physiol. Biochem. 2018, 46, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Fan, Z.; Li, L.; Lu, J.; Zhai, Y.; Zhao, J. The chemokine system and its role in obesity. J. Cell Physiol. 2019, 234, 3336–3346. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Yoon, J.H.; Ghim, J.; Yea, K.; Song, P.; Park, S.; Lee, A.; Hong, C.P.; Jang, M.S.; et al. CXCL12 secreted from adipose tissue recruits macrophages and induces insulin resistance in mice. Diabetologia 2014, 57, 1456–1465. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noh, J.-W.; Jun, M.-S.; Yang, H.-K.; Lee, B.-C. Cellular and Molecular Mechanisms and Effects of Berberine on Obesity-Induced Inflammation. Biomedicines 2022, 10, 1739. https://doi.org/10.3390/biomedicines10071739
Noh J-W, Jun M-S, Yang H-K, Lee B-C. Cellular and Molecular Mechanisms and Effects of Berberine on Obesity-Induced Inflammation. Biomedicines. 2022; 10(7):1739. https://doi.org/10.3390/biomedicines10071739
Chicago/Turabian StyleNoh, Ji-Won, Min-Soo Jun, Hee-Kwon Yang, and Byung-Cheol Lee. 2022. "Cellular and Molecular Mechanisms and Effects of Berberine on Obesity-Induced Inflammation" Biomedicines 10, no. 7: 1739. https://doi.org/10.3390/biomedicines10071739





