Systems Genetic Identification of Mitochondrion-Associated Alzheimer’s Disease Genes and Implications for Disease Risk Prediction
Abstract
:1. Introduction
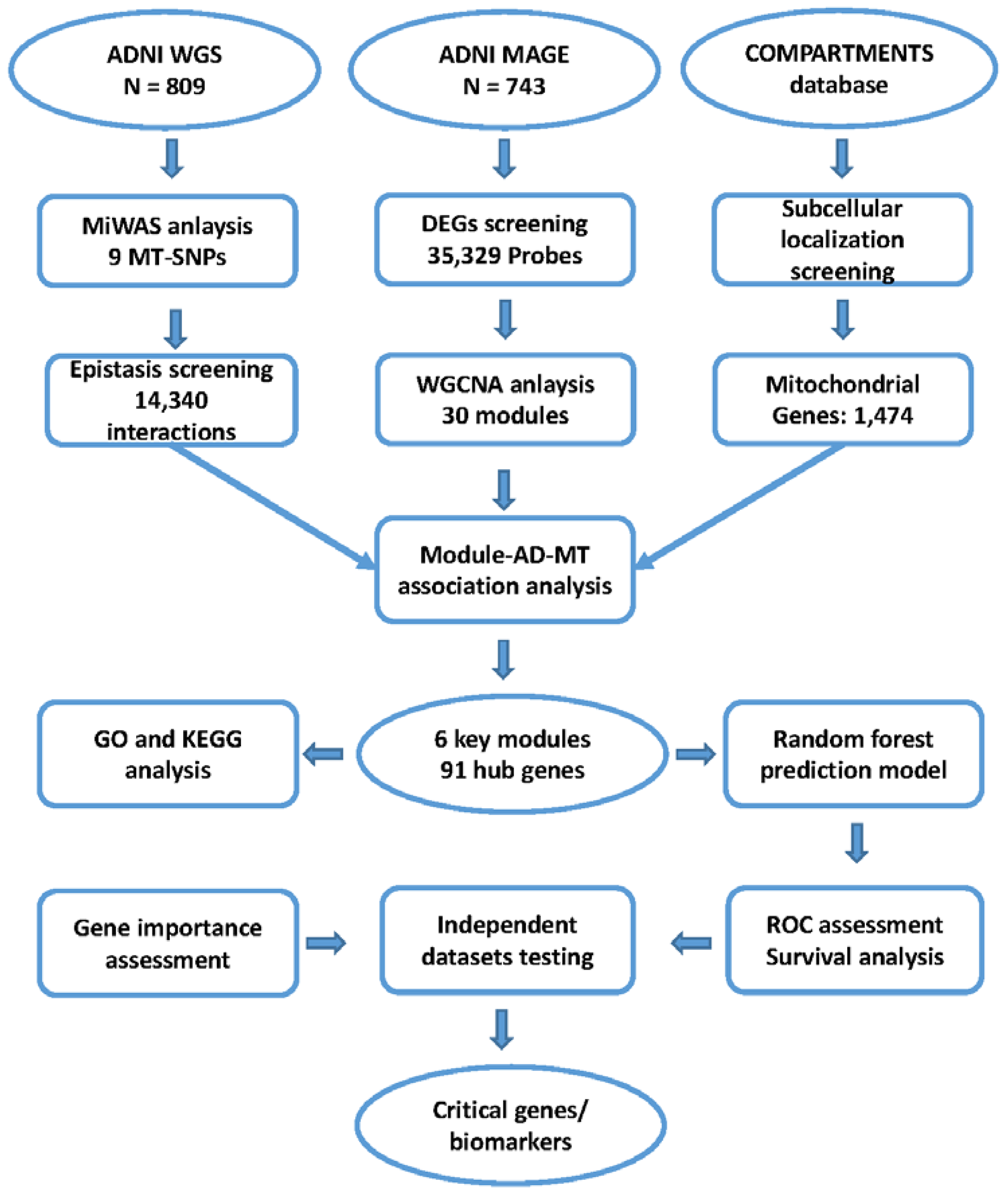
2. Materials and Methods
2.1. Study Subjects
2.2. Genotyping and Quality Control
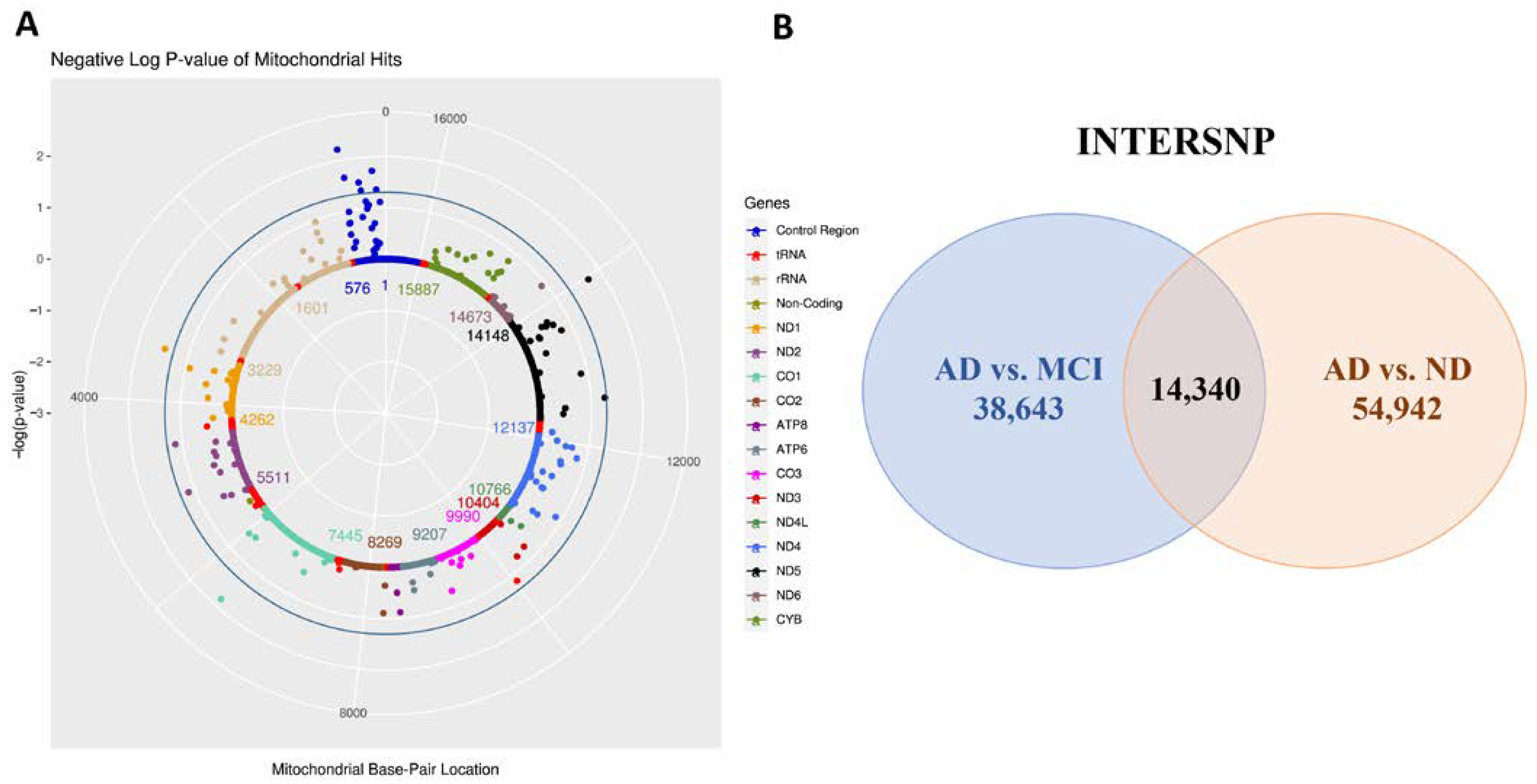
2.3. Mitochondrial Genome-Wide Association Study
2.4. Epistasis Screening
2.5. Microarray Data Processing and Differentially Expressed Gene Analysis
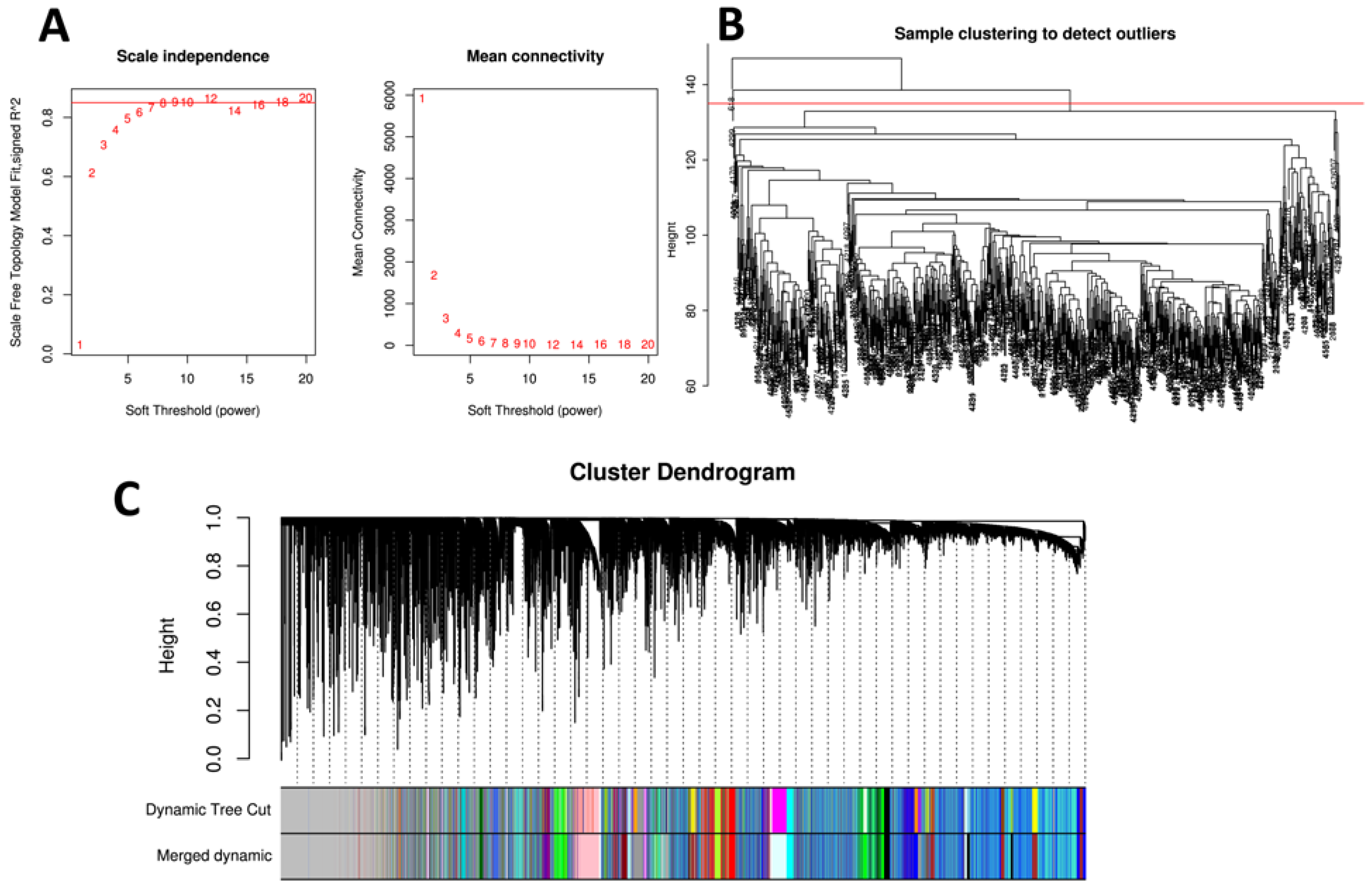
2.6. Weighted Co-Expression Network Analysis
2.7. Identification of Key Modules and Hub Genes
2.8. Functional Enrichment Analysis of Key Modules
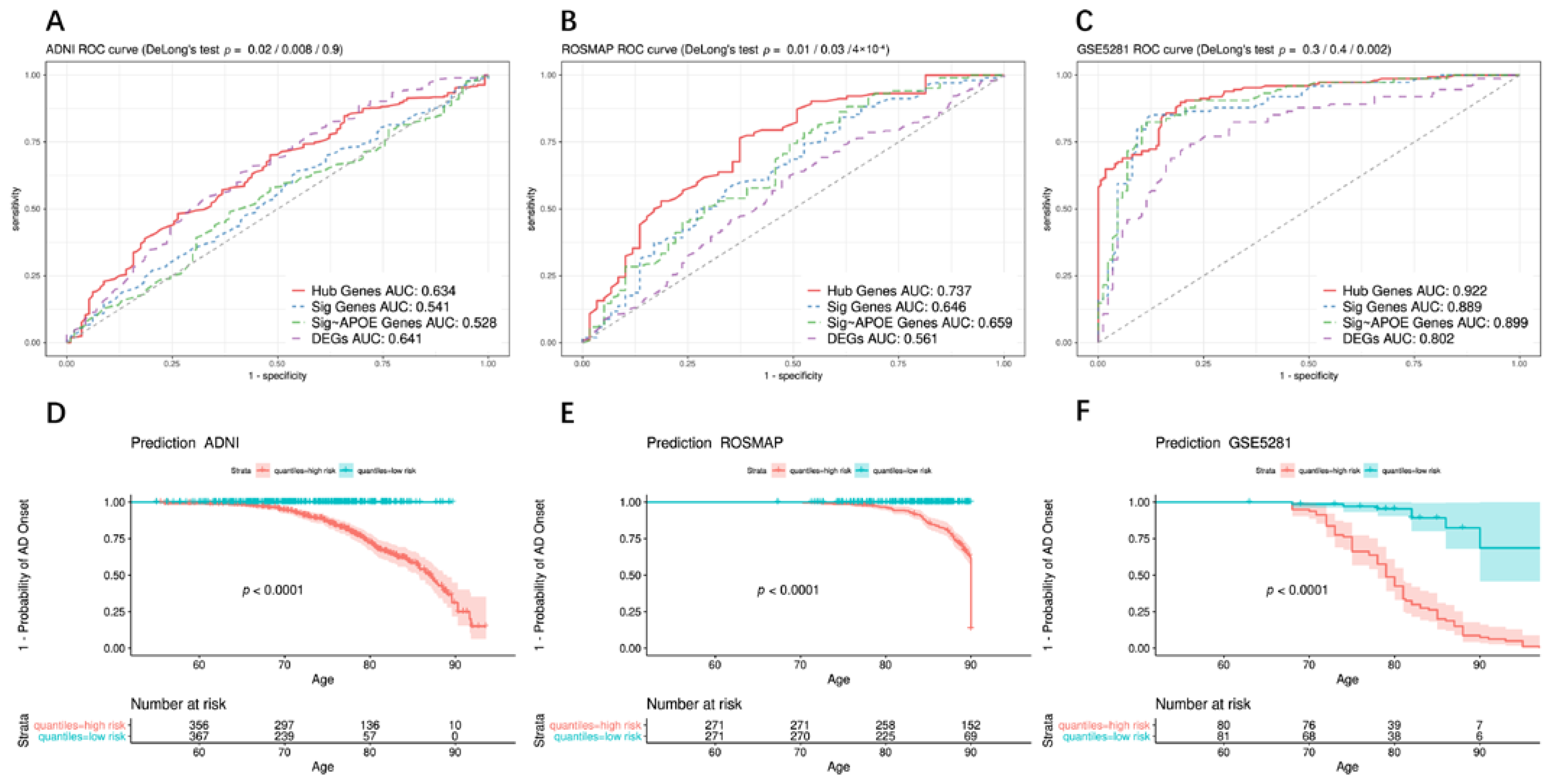
2.9. Construction and Evaluation of the Predictive Model
3. Results
3.1. Mitochondrial Genome-Wide Association Study
3.2. Mitochondrial Epistasis Screening
3.3. Identification of Differentially Expressed Genes
3.4. Weighted Gene Co-Expression Network Analysis and Detection of Key Modules
3.5. Functional Enrichment Analysis of Key Modules
3.6. Identification of Hub Genes
3.7. Construction and Evaluation of the Predictive Model
3.8. Identification of Critical Genes
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheltens, P.; Blennow, K.; Breteler, M.M.B.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s Disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic Meta-Analysis of Diagnosed Alzheimer’s Disease Identifies New Risk Loci and Implicates Aβ, Tau, Immunity and Lipid Processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáez, M.E.; González-Pérez, A.; Hernández-Olasagarre, B.; Beà, A.; Moreno-Grau, S.; de Rojas, I.; Monté-Rubio, G.; Orellana, A.; Valero, S.; Comella, J.X.; et al. Genome Wide Meta-Analysis Identifies Common Genetic Signatures Shared by Heart Function and Alzheimer’s Disease. Sci. Rep. 2019, 9, 16665. [Google Scholar] [CrossRef]
- Ebanks, B.; Ingram, T.L.; Chakrabarti, L. ATP Synthase and Alzheimer’s Disease: Putting a Spin on the Mitochondrial Hypothesis. Aging 2020, 12, 16647–16662. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Reiman, E.M.; Beach, T.G.; Serrano, G.E.; Sabbagh, M.N.; Nielsen, M.; Caselli, R.J.; Shi, J. Effect of ApoE Isoforms on Mitochondria in Alzheimer Disease. Neurology 2020, 94, e2404–e2411. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Gleichmann, M.; Cheng, A. Mitochondria in Neuroplasticity and Neurological Disorders. Neuron 2008, 60, 748–766. [Google Scholar] [CrossRef] [Green Version]
- Lautrup, S.; Lou, G.; Aman, Y.; Nilsen, H.; Tao, J.; Fang, E.F. Microglial Mitophagy Mitigates Neuroinflammation in Alzheimer’s Disease. Neurochem. Int. 2019, 129, 104469. [Google Scholar] [CrossRef]
- Schapira, A.H.V. Mitochondrial Diseases. Lancet 2012, 379, 1825–1834. [Google Scholar] [CrossRef]
- da Cunha, F.M.; Torelli, N.Q.; Kowaltowski, A.J. Mitochondrial Retrograde Signaling: Triggers, Pathways, and Outcomes. Oxid. Med. Cell Longev. 2015, 2015, 482582. [Google Scholar] [CrossRef] [Green Version]
- Onyango, I.G.; Dennis, J.; Khan, S.M. Mitochondrial Dysfunction in Alzheimer’s Disease and the Rationale for Bioenergetics Based Therapies. Aging Dis. 2016, 7, 201–214. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.-Y.; Liu, Y.; Chen, L.; Luo, J.; Zhang, C.; Shen, X.-F. Identification of the Potential Biomarkers in Patients with Glioma: A Weighted Gene Co-Expression Network Analysis. Carcinogenesis 2020, 41, 743–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.-Y.; Chao, F.; Zhuo, Z.; Ma, Z.; Li, W.; Chen, G. Identification of Hub Genes in Prostate Cancer Using Robust Rank Aggregation and Weighted Gene Co-Expression Network Analysis. Aging 2019, 11, 4736–4756. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C.; Aisen, P.S.; Beckett, L.A.; Donohue, M.C.; Gamst, A.C.; Harvey, D.J.; Jack, C.R.; Jagust, W.J.; Shaw, L.M.; Toga, A.W.; et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): Clinical Characterization. Neurology 2010, 74, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, D.A.; Buchman, A.S.; Boyle, P.A.; Barnes, L.L.; Wilson, R.S.; Schneider, J.A. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimer’s Dis. 2018, 64, S161–S189. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Walker, D.G.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Gene Expression Profiles in Anatomically and Functionally Distinct Regions of the Normal Aged Human Brain. Physiol. Genom. 2007, 28, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhang, D.-F.; Luo, R.; Wu, Y.; Zhou, H.; Kong, L.-L.; Bi, R.; Yao, Y.-G. A Systematic Integrated Analysis of Brain Expression Profiles Reveals YAP1 and Other Prioritized Hub Genes as Important Upstream Regulators in Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 215–229. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. MtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1.23.1–1.23.26. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Gomez, D.; Leipzig, J.; Shen, L.; Lott, M.; Stassen, A.P.M.; Wallace, D.C.; Wiggs, J.L.; Falk, M.J.; van Oven, M.; Gai, X. Phy-Mer: A Novel Alignment-Free and Reference-Independent Mitochondrial Haplogroup Classifier. Bioinformatics 2015, 31, 1310–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridge, P.G.; Wadsworth, M.E.; Miller, J.B.; Saykin, A.J.; Green, R.C.; Kauwe, J.S.K. Assembly of 809 Whole Mitochondrial Genomes with Clinical, Imaging, and Fluid Biomarker Phenotyping. Alzheimer Dement. 2018, 14, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, B.; Torres, M.; Jiang, X.; McKean-Cowdin, R.; Nousome, D.; Kim, S.-J.; Mehta, H.H.; Yen, K.; Cohen, P.; Varma, R. A Mitochondrial Genome-Wide Association Study of Cataract in a Latino Population. Transl. Vis. Sci. Technol. 2020, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A Tutorial on Conducting Genome-Wide Association Studies: Quality Control and Statistical Analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Herold, C.; Steffens, M.; Brockschmidt, F.F.; Baur, M.P.; Becker, T. INTERSNP: Genome-Wide Interaction Analysis Guided by a Priori Information. Bioinformatics 2009, 25, 3275–3281. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.; Schneider, J.A.; De Jager, P.L.; Bennett, D.A.; Zhang, H.-Y. Genome-Wide Interaction Analysis of Pathological Hallmarks in Alzheimer’s Disease. Neurobiol. Aging 2020, 93, 61–68. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Cordell, H.J. Epistasis: What It Means, What It Doesn’t Mean, and Statistical Methods to Detect It in Humans. Hum. Mol. Genet. 2002, 11, 2463–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, J.X.; Pletscher-Frankild, S.; Tsafou, K.; Stolte, C.; O’Donoghue, S.I.; Schneider, R.; Jensen, L.J. Compartments: Unification and Visualization of Protein Subcellular Localization Evidence. Database 2014, 2014, bau012. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Falah, M.; Farhadi, M.; Kamrava, S.K.; Mahmoudian, S.; Daneshi, A.; Balali, M.; Asghari, A.; Houshmand, M. Association of Genetic Variations in the Mitochondrial DNA Control Region with Presbycusis. Clin. Interv. Aging 2017, 12, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Brandon, M.C.; Lott, M.T.; Nguyen, K.C.; Spolim, S.; Navathe, S.B.; Baldi, P.; Wallace, D.C. MITOMAP: A Human Mitochondrial Genome Database--2004 Update. Nucleic Acids Res. 2005, 33, D611–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunnon, K.; Keohane, A.; Pidsley, R.; Newhouse, S.; Riddoch-Contreras, J.; Thubron, E.B.; Devall, M.; Soininen, H.; Kłoszewska, I.; Mecocci, P.; et al. Mitochondrial Genes Are Altered in Blood Early in Alzheimer’s Disease. Neurobiol. Aging 2017, 53, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.M.; Meng, Q.; Perez de Acha, O.; Mustapic, M.; Cheng, A.; Eren, E.; Kundu, G.; Piao, Y.; Munk, R.; Wood, W.H.; et al. Mitochondrial RNA in Alzheimer’s Disease Circulating Extracellular Vesicles. Front. Cell Dev. Biol. 2020, 8, 581882. [Google Scholar] [CrossRef]
- Saman, S.; Lee, N.C.Y.; Inoyo, I.; Jin, J.; Li, Z.; Doyle, T.; McKee, A.C.; Hall, G.F. Proteins Recruited to Exosomes by Tau Overexpression Implicate Novel Cellular Mechanisms Linking Tau Secretion with Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 40, S47–S70. [Google Scholar] [CrossRef]
- Wang, H.; Bennett, D.A.; De Jager, P.L.; Zhang, Q.-Y.; Zhang, H.-Y. Genome-Wide Epistasis Analysis for Alzheimer’s Disease and Implications for Genetic Risk Prediction. Alzheimer’s Res. Ther. 2021, 13, 55. [Google Scholar] [CrossRef]
- Nho, K.; Nudelman, K.; Allen, M.; Hodges, A.; Kim, S.; Risacher, S.L.; Apostolova, L.G.; Lin, K.; Lunnon, K.; Wang, X.; et al. Genome-Wide Transcriptome Analysis Identifies Novel Dysregulated Genes Implicated in Alzheimer’s Pathology. Alzheimer Dement. 2020, 16, 1213–1223. [Google Scholar] [CrossRef]
- Martín-Maestro, P.; Gargini, R.; García, E.; Simón, D.; Avila, J.; García-Escudero, V. Mitophagy Failure in APP and Tau Overexpression Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2019, 70, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Solleiro-Villavicencio, H.; Rivas-Arancibia, S. Effect of Chronic Oxidative Stress on Neuroinflammatory Response Mediated by CD4+T Cells in Neurodegenerative Diseases. Front. Cell Neurosci. 2018, 12, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, E.-J.; Fischer, N.; Efferth, T. Phytochemicals as Inhibitors of NF-ΚB for Treatment of Alzheimer’s Disease. Pharmacol. Res. 2018, 129, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Manczak, M.; Kandimalla, R.; Yin, X.; Reddy, P.H. Hippocampal Mutant APP and Amyloid Beta-Induced Cognitive Decline, Dendritic Spine Loss, Defective Autophagy, Mitophagy and Mitochondrial Abnormalities in a Mouse Model of Alzheimer’s Disease. Hum. Mol. Genet. 2018, 27, 1332–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaillant-Beuchot, L.; Mary, A.; Pardossi-Piquard, R.; Bourgeois, A.; Lauritzen, I.; Eysert, F.; Kinoshita, P.F.; Cazareth, J.; Badot, C.; Fragaki, K.; et al. Accumulation of Amyloid Precursor Protein C-Terminal Fragments Triggers Mitochondrial Structure, Function, and Mitophagy Defects in Alzheimer’s Disease Models and Human Brains. Acta Neuropathol. 2021, 141, 39–65. [Google Scholar] [CrossRef]
- Chen, X.; Stern, D.; Yan, S.D. Mitochondrial Dysfunction and Alzheimer’s Disease. Curr. Alzheimer Res. 2006, 3, 515–520. [Google Scholar] [CrossRef]
- Yao, J.; Du, H.; Yan, S.; Fang, F.; Wang, C.; Lue, L.-F.; Guo, L.; Chen, D.; Stern, D.M.; Gunn Moore, F.J.; et al. Inhibition of Amyloid-Beta (Abeta) Peptide-Binding Alcohol Dehydrogenase-Abeta Interaction Reduces Abeta Accumulation and Improves Mitochondrial Function in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2011, 31, 2313–2320. [Google Scholar] [CrossRef]
- Xiao, X.; Chen, Q.; Zhu, X.; Wang, Y. ABAD/17β-HSD10 Reduction Contributes to the Protective Mechanism of Huperzine a on the Cerebral Mitochondrial Function in APP/PS1 Mice. Neurobiol. Aging 2019, 81, 77–87. [Google Scholar] [CrossRef]
- Lim, Y.-A.; Grimm, A.; Giese, M.; Mensah-Nyagan, A.G.; Villafranca, J.E.; Ittner, L.M.; Eckert, A.; Götz, J. Inhibition of the Mitochondrial Enzyme ABAD Restores the Amyloid-β-Mediated Deregulation of Estradiol. PLoS ONE 2011, 6, e28887. [Google Scholar] [CrossRef] [Green Version]
- Pluvinage, J.V.; Haney, M.S.; Smith, B.A.H.; Sun, J.; Iram, T.; Bonanno, L.; Li, L.; Lee, D.P.; Morgens, D.W.; Yang, A.C.; et al. CD22 Blockade Restores Homeostatic Microglial Phagocytosis in Ageing Brains. Nature 2019, 568, 187–192. [Google Scholar] [CrossRef]
- Bu, X.-L.; Sun, P.-Y.; Fan, D.-Y.; Wang, J.; Sun, H.-L.; Cheng, Y.; Zeng, G.-H.; Chen, D.-W.; Li, H.-Y.; Yi, X.; et al. Associations of Plasma Soluble CD22 Levels with Brain Amyloid Burden and Cognitive Decline in Alzheimer’s Disease. Sci. Adv. 2022, 8, eabm5667. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.N.; Theiss, A.L. Gut Bacteria Signaling to Mitochondria in Intestinal Inflammation and Cancer. Gut Microbes 2020, 11, 285–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Wang, T.; Jin, F. Alzheimer’s Disease and Gut Microbiota. Sci. China Life Sci. 2016, 59, 1006–1023. [Google Scholar] [CrossRef] [Green Version]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A.; Ojala, J.; Haapasalo, A.; Soininen, H.; Hiltunen, M. Impaired Autophagy and APP Processing in Alzheimer’s Disease: The Potential Role of Beclin 1 Interactome. Prog. Neurobiol. 2013, 106, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.-Q.; Ren, C.; Xia, Z.-F.; Yao, Y.-M. Organelle-Specific Autophagy in Inflammatory Diseases: A Potential Therapeutic Target Underlying the Quality Control of Multiple Organelles. Autophagy 2021, 17, 385–401. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Chen, C.; Shen, H.; He, B.Z.; Yu, D.; Jiang, S.; Zhao, S.; Gao, Z.; Zhu, Z.; Chen, X.; et al. GenTree, an Integrated Resource for Analyzing the Evolution and Function of Primate-Specific Coding Genes. Genome Res. 2019, 29, 682–696. [Google Scholar] [CrossRef]
- Murphy, M.P.; Hartley, R.C. Mitochondria as a Therapeutic Target for Common Pathologies. Nat. Rev. Drug Discov. 2018, 17, 865–886. [Google Scholar] [CrossRef] [Green Version]

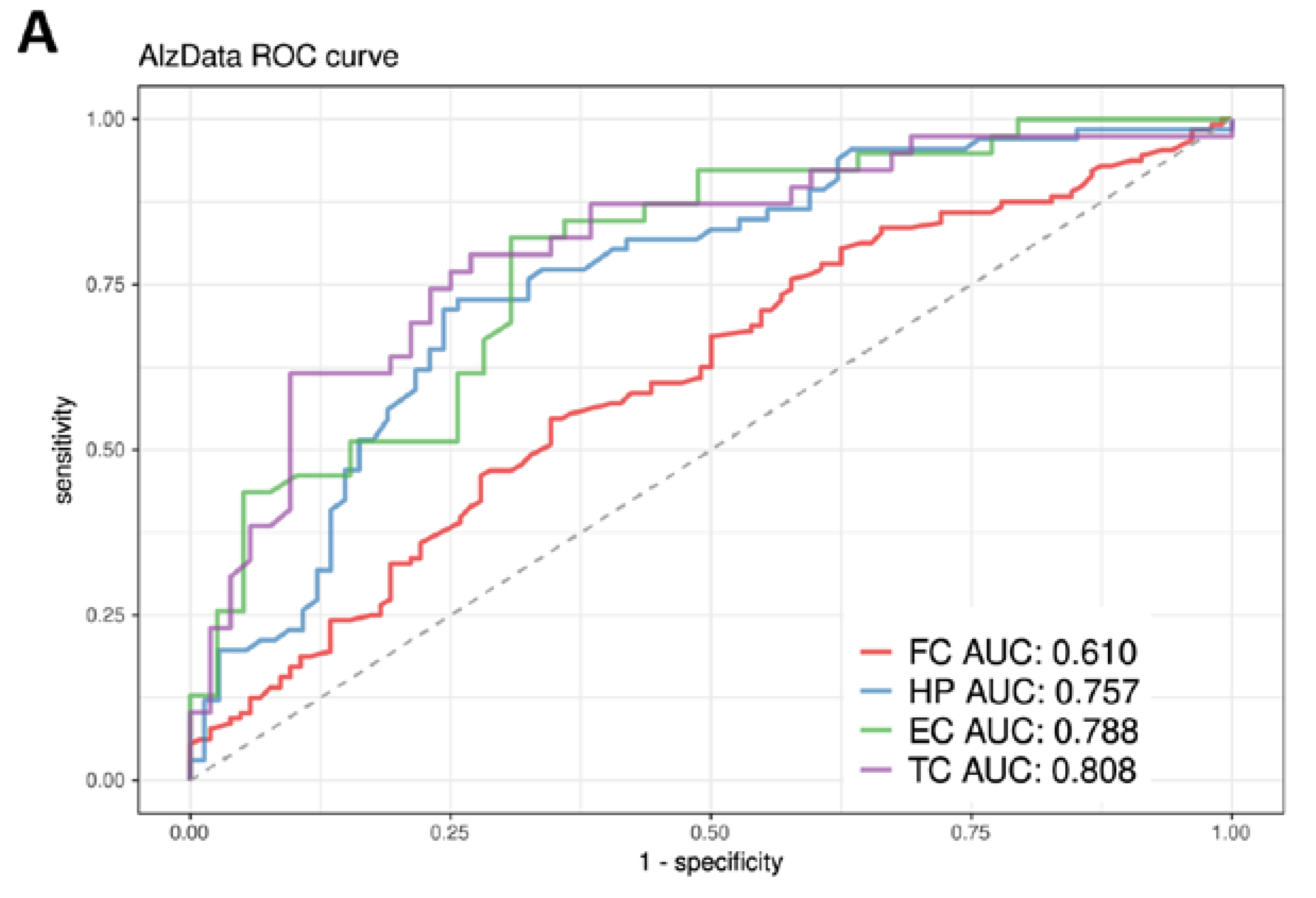
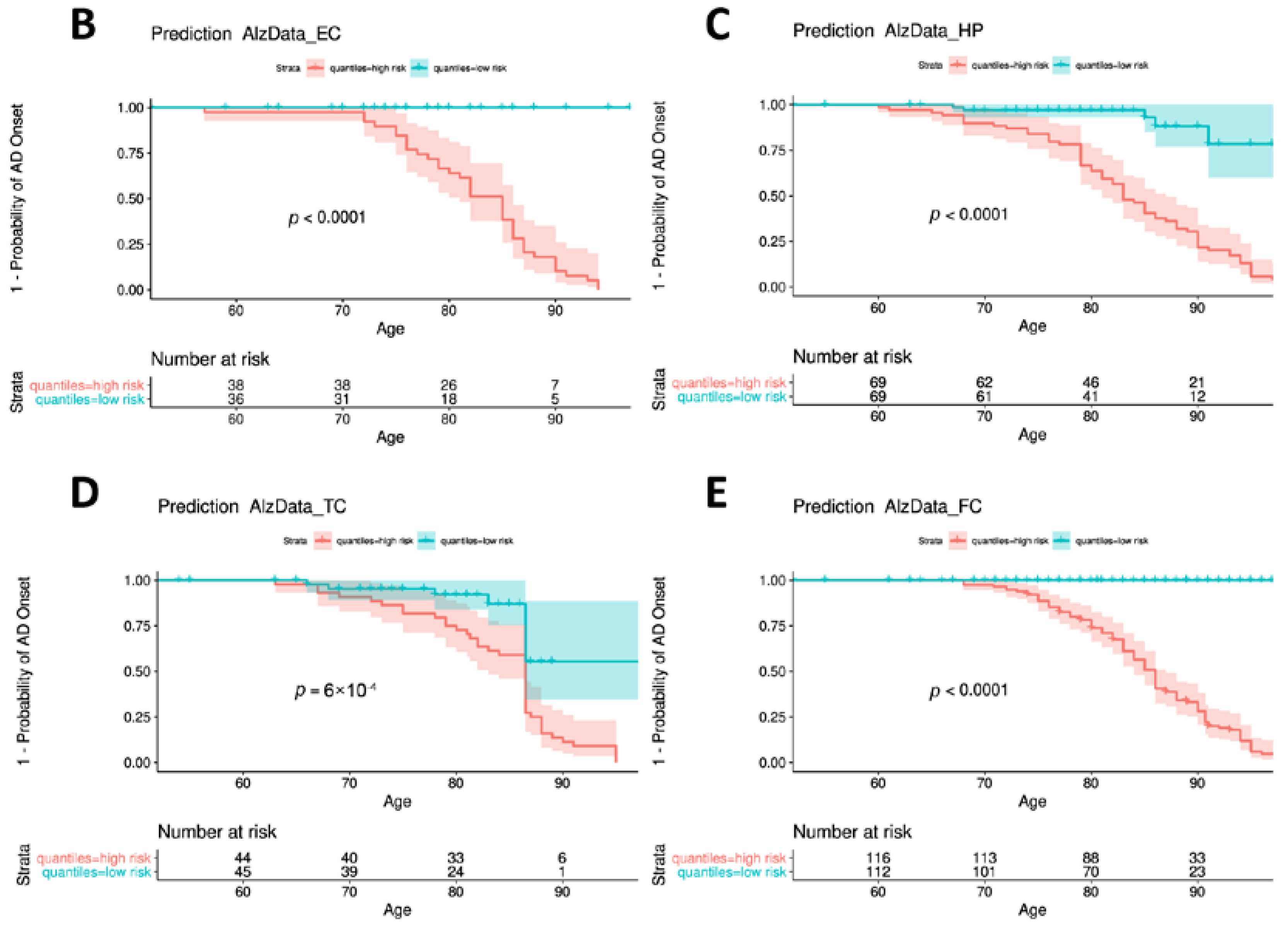
| ADNI | AD (n = 175) | ND (n = 634) | Diff (p) * |
| Sex (F/M) | 70 F, 105 M | 293 F, 341 M | 0.15 |
| Age, y (SD) | 73.95(7.66) | 73.04 (6.90) | 0.13 |
| ROSMAP | AD (n = 220) | ND (n = 322) | Diff (p) * |
| Sex (F/M) | 147 F, 73 M | 128 F, 194 M | <0.0001 |
| Age, y (SD) | 88(3.51) | 85.5 (5.00) | <0.0001 |
| GSE5281 | AD (n = 87) | ND (n = 74) | Diff (p) * |
| Sex (F/M) | 37 F, 50 M | 21 F, 53 M | 0.07 |
| Age, y (SD) | 79.8 (6.91) | 79.5 (8.92) | 0.8 |
| AlzData (EC) | AD (n = 39) | ND (n = 39) | Diff (p) * |
| Sex (F/M) | 18 F, 21 M | 17 F, 22 M | 1.00 |
| Age, y (SD) | 82.4 (7.38) | 78 (11.1) | 0.04 |
| AlzData (HP) | AD (n = 74) | ND (n = 66) | Diff (p) * |
| Sex (F/M) | 45 F, 29 M | 23 F, 43 M | 0.002 |
| Age, y (SD) | 83.1 (9.44) | 80.2 (9.68) | 0.07 |
| AlzData (TC) | AD (n = 52) | ND (n = 39) | Diff (p) * |
| Sex (F/M) | 14 F, 20 M * | 18 F, 21 M* | 0.81 |
| Age, y (SD) | 83.1 (9.44) | 80.2 (9.68) | 0.07 |
| AlzData (FC) | AD (n = 104) | ND (n = 128) | Diff (p) * |
| Sex (F/M) | 44 F, 46 M * | 55 F, 65 M * | 0.68 |
| Age, y (SD) | 84.7 (7.53) | 81.7 (10.60) | 0.01 |
| Gene | Describe | Compartment * | GO BP * | BioSystems Pathway * | AD Literature (PMID) | MT Literature (PMID) |
|---|---|---|---|---|---|---|
| RTN3 | Reticulon 3 | plasma membrane; endoplasmic reticulum | GO:0006915; GO:0016032; GO:0071786; etc. | Pathways of neurodegeneration-multiple disease (Alzheimer disease); Transmission across Chemical Synapses (Neuronal System) | 23827971; 29356939; 28733667; etc. | 32048886; 17191123; 17031492; etc. |
| RASSF2 | Ras Association Domain Family Member 2 | nucleus | GO:0001501; GO:0001503; GO:0006468; etc. | Hippo signaling pathway-multiple species | —— | 22674380; etc. |
| TCL1A | TCL1 Family AKT Coactivator A | nucleus | GO:0007275; GO:0008284; GO:0010918; etc. | PI3K/Akt Signaling | —— | 26041471; 10983986; 30282833; etc. |
| BCL11A | BAF Chromatin Remodeling Complex Subunit BCL11A | nucleus | GO:0000122; GO:0006357; GO:0010976; etc. | —— | 30180184; 33911114; etc. | 33091040; 27838552; etc. |
| RANBP10 | RAN Binding Protein 10 | cytosol | GO:0005515; GO:0031267 | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) | 28659384; 28744327; etc. | —— |
| REPS2 | RALBP1 Associated Eps Domain Containing 2 | cytosol | GO:0006897; GO:0007173; GO:0016197; etc. | EGF/EGFR Signaling Pathway | 32597797; etc. | —— |
| VCAN | Versican | extracellular | GO:0001501; GO:0007417; GO:0007155; etc. | Direct p53 effectors; Regulation of Wnt-mediated beta catenin signaling and target gene transcription; Spinal Cord Injury | 7793988; 29752348; 28724990; etc. | 30622695; 29060675; etc. |
| TMCC3 | Transmembrane And Coiled-Coil Domain Family 3 | Endoplasmic; reticulum | —— | —— | —— | —— |
| EPB41 | Erythrocyte Membrane Protein Band 4.1 | plasma membrane; nucleus; cytosol | GO:0007049; GO:0008360; GO:0030036; etc. | Syndecan-2-mediated signaling events; Neuronal System | 22815752; 24718034; etc. | —— |
| NEDD4L | NEDD4 Like E3 Ubiquitin Protein Ligase | Nucleus; cytosol | GO:0000122; GO:0000209; GO:0003254; etc. | Neurotrophic factor-mediated Trk receptor signaling; Ubiquitin mediated proteolysis TGF-beta Signaling Pathway | 32140098; 27686364; 28377502; etc. | 31959741; 32140098; etc. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Wang, H.; Bennett, D.A.; Zhang, Q.-Y.; Wang, G.; Zhang, H.-Y. Systems Genetic Identification of Mitochondrion-Associated Alzheimer’s Disease Genes and Implications for Disease Risk Prediction. Biomedicines 2022, 10, 1782. https://doi.org/10.3390/biomedicines10081782
Xu X, Wang H, Bennett DA, Zhang Q-Y, Wang G, Zhang H-Y. Systems Genetic Identification of Mitochondrion-Associated Alzheimer’s Disease Genes and Implications for Disease Risk Prediction. Biomedicines. 2022; 10(8):1782. https://doi.org/10.3390/biomedicines10081782
Chicago/Turabian StyleXu, Xuan, Hui Wang, David A. Bennett, Qing-Ye Zhang, Gang Wang, and Hong-Yu Zhang. 2022. "Systems Genetic Identification of Mitochondrion-Associated Alzheimer’s Disease Genes and Implications for Disease Risk Prediction" Biomedicines 10, no. 8: 1782. https://doi.org/10.3390/biomedicines10081782
APA StyleXu, X., Wang, H., Bennett, D. A., Zhang, Q.-Y., Wang, G., & Zhang, H.-Y. (2022). Systems Genetic Identification of Mitochondrion-Associated Alzheimer’s Disease Genes and Implications for Disease Risk Prediction. Biomedicines, 10(8), 1782. https://doi.org/10.3390/biomedicines10081782







