EGF-Dependent Activation of ELK1 Contributes to the Induction of CLDND1 Expression Involved in Tight Junction Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Luciferase Reporter and Expression Constructs
2.3. Transfection and Luciferase Activity Assay
2.4. Chromatin Immunoprecipitation (ChIP) Assay
2.5. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.6. Western Blotting
2.7. Statistical Analysis
3. Results
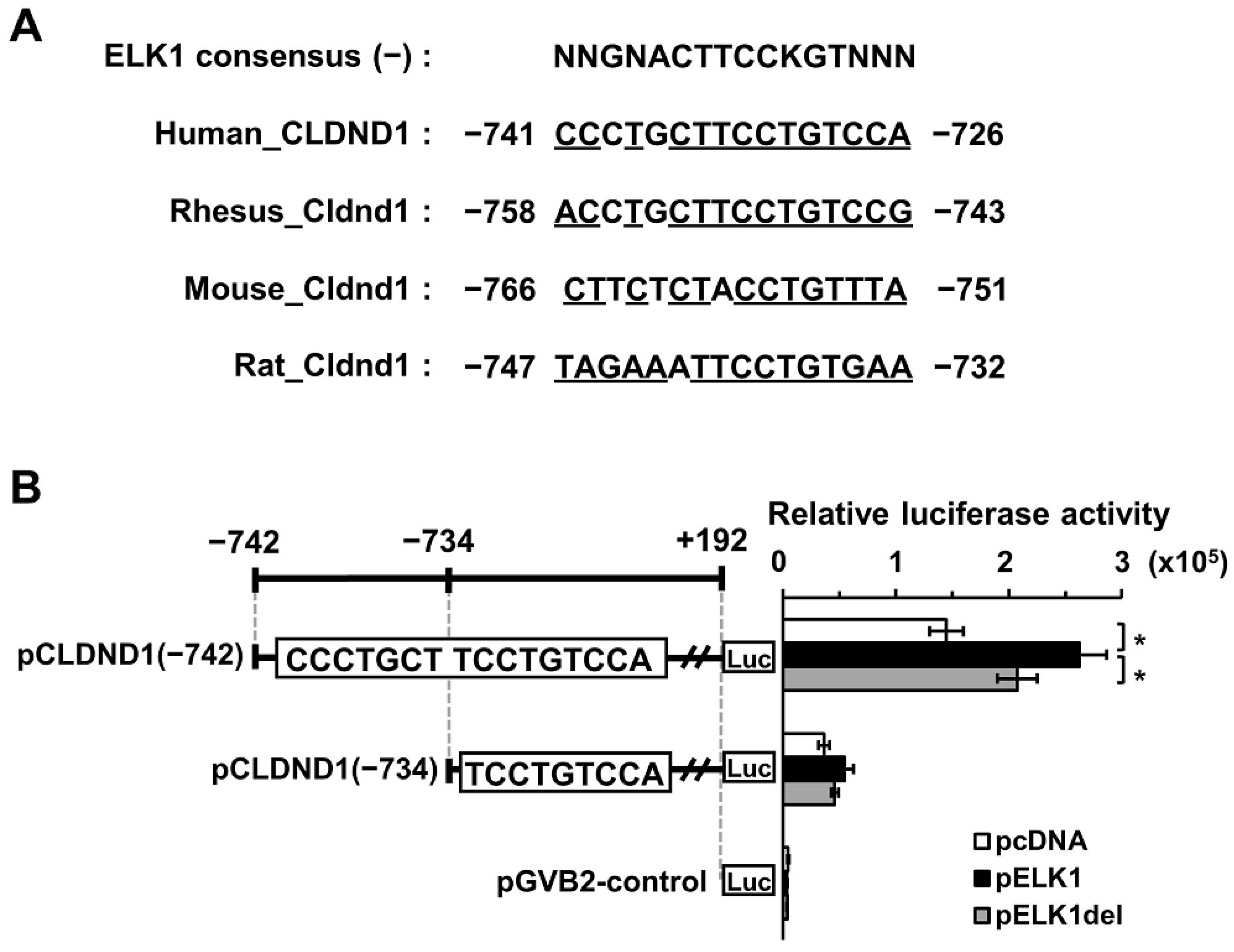
3.1. Effect of ELK1 on the Enhancer Region of CLDND1
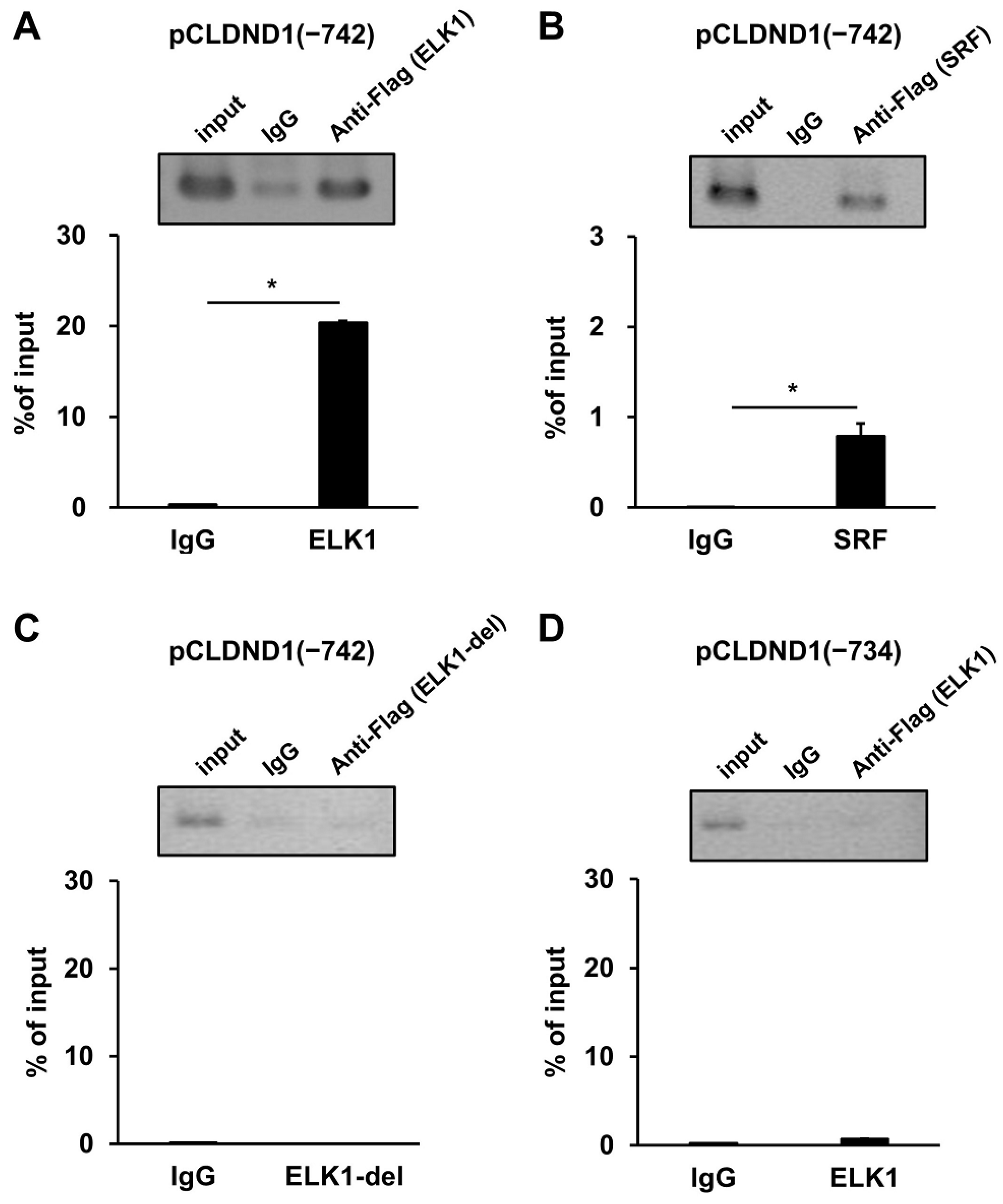
3.2. Binding of ELK1 and SRF to the Enhancer Region of CLDND1
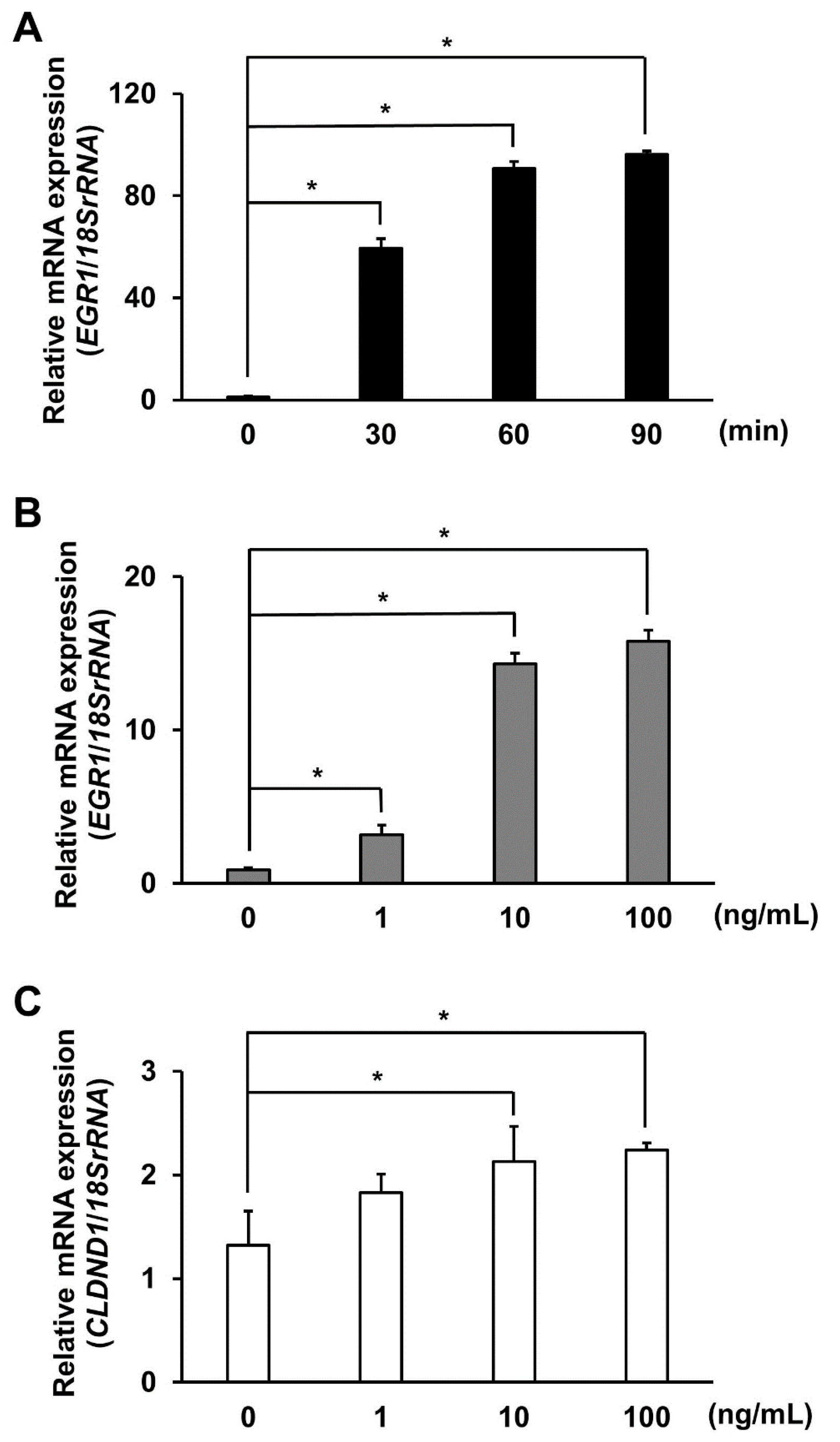
3.3. Effect of ELK1 Overexpression on CLDND1 Expression
3.4. Effect of EGF Stimulation on CLDND1 Expression
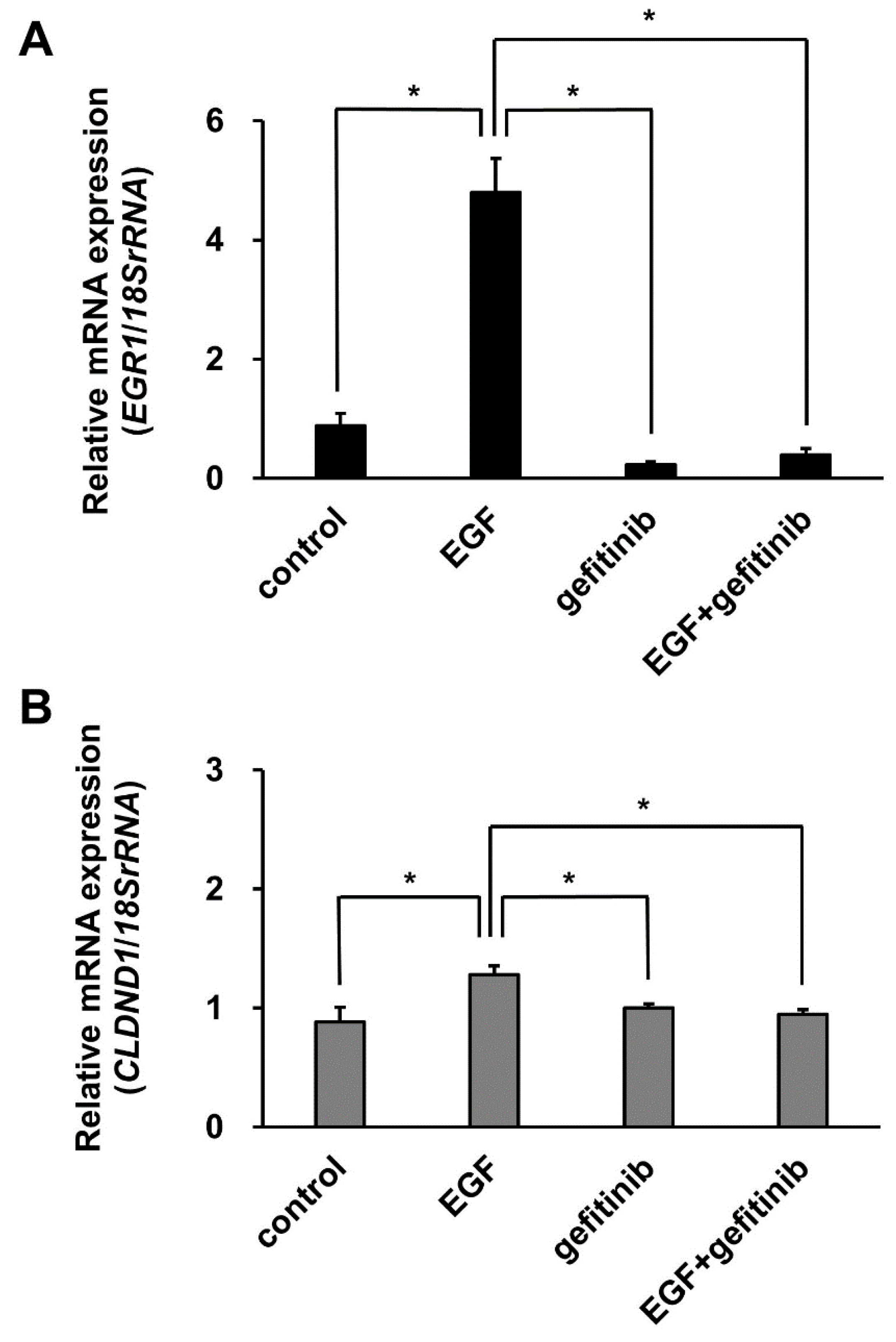
3.5. Effect of EGF Stimulation on the Binding of the ELK1 Complex to the CLDND1 Enhancer Region
3.6. Effect of Gefitinib Treatment on CLDND1 Expression Induced by EGF Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef]
- Suzuki, H.; Tani, K.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J. Mol. Biol. 2015, 427, 291–297. [Google Scholar] [CrossRef]
- Tsukita, S.; Yamazaki, Y.; Katsuno, T.; Tamura, A.; Tsukita, S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 2008, 27, 6930–6938. [Google Scholar] [CrossRef]
- Cavallaro, U.; Dejana, E. Adhesion molecule signalling: Not always a sticky business. Nat. Rev. Mol. Cell Biol. 2011, 12, 189–197. [Google Scholar] [CrossRef]
- Günzel, D.; Yu, A.S.L. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef]
- Rangel, L.B.A.; Agarwal, R.; D’Souza, T.; Pizer, E.S.; Alò, P.L.; Lancaster, W.D.; Gregoire, L.; Schwartz, D.R.; Cho, K.R.; Morin, P.J. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res. 2003, 9, 2567–2575. [Google Scholar]
- Kominsky, S.L.; Vali, M.; Korz, D.; Gabig, T.G.; Weitzman, S.A.; Argani, P.; Sukumar, S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol. 2004, 164, 1627–1633. [Google Scholar] [CrossRef]
- Ushiku, T.; Shinozaki-Ushiku, A.; Maeda, D.; Morita, S.; Fukayama, M. Distinct expression pattern of claudin-6, a primitive phenotypic tight junction molecule, in germ cell tumours and visceral carcinomas. Histopathology 2012, 61, 1043–1056. [Google Scholar] [CrossRef]
- Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Goto, K.; Kawahara, I.; Nishiguchi, Y.; Kishi, S.; Mori, S.; Ohmori, H.; Kondoh, M.; et al. Anti-claudin-4 extracellular domain antibody enhances the antitumoral effects of chemotherapeutic and antibody drugs in colorectal cancer. Oncotarget 2018, 9, 37367–37378. [Google Scholar] [CrossRef]
- Kuwada, M.; Chihara, Y.; Luo, Y.; Li, X.; Nishiguchi, Y.; Fujiwara, R.; Sasaki, T.; Fujii, K.; Ohmori, H.; Fujimoto, K.; et al. Pro-chemotherapeutic effects of antibody against extracellular domain of claudin-4 in bladder cancer. Cancer Lett. 2015, 369, 212–221. [Google Scholar] [CrossRef]
- Nishiguchi, Y.; Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Ohmori, H.; Kishi, S.; Mori, S.; Goto, K.; Yasui, W.; Sho, M.; et al. Targeting claudin-4 enhances CDDP-chemosensitivity in gastric cancer. Oncotarget 2019, 10, 2189–2202. [Google Scholar] [CrossRef]
- Luo, Y.; Kishi, S.; Sasaki, T.; Ohmori, H.; Fujiwara-Tani, R.; Mori, S.; Goto, K.; Nishiguchi, Y.; Mori, T.; Kawahara, I.; et al. Targeting claudin-4 enhances chemosensitivity in breast cancer. Cancer Sci. 2020, 111, 1840–1850. [Google Scholar] [CrossRef]
- Sahin, U.; Türeci, Ö.; Manikhas, G.; Lordick, F.; Rusyn, A.; Vynnychenko, I.; Dudov, A.; Bazin, I.; Bondarenko, I.; Melichar, B.; et al. FAST: A randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first-line treatment of advanced CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma. Ann. Oncol. 2021, 32, 609–619. [Google Scholar] [CrossRef]
- Lee, J.W.; Hsiao, W.T.; Chen, H.Y.; Hsu, L.P.; Chen, P.R.; Lin, M.D.; Chiu, S.J.; Shih, W.L.; Hsu, Y.C. Upregulated claudin-1 expression confers resistance to cell death of nasopharyngeal carcinoma cells. Int. J. Cancer 2010, 126, 1353–1366. [Google Scholar] [CrossRef]
- Achari, C.; Winslow, S.; Larsson, C. Down regulation of CLDND1 induces apoptosis in breast cancer cells. PLoS ONE 2015, 10, e0130300. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef]
- Ohnishi, M.; Ochiai, H.; Matsuoka, K.; Akagi, M.; Nakayama, Y.; Shima, A.; Uda, A.; Matsuoka, H.; Kamishikiryo, J.; Michihara, A.; et al. Claudin domain containing 1 contributing to endothelial cell adhesion decreases in presence of cerebellar hemorrhage. J. Neurosci. Res. 2017, 95, 2051–2058. [Google Scholar] [CrossRef]
- Matsuoka, H.; Shima, A.; Uda, A.; Ezaki, H.; Michihara, A. The retinoic acid receptor-related orphan receptor a positively regulates tight junction protein claudin domain-containing 1 mRNA expression in human brain endothelial cells. J. Biochem. 2017, 161, 441–450. [Google Scholar] [CrossRef]
- Matsuoka, H.; Michihara, A. Identification of the RORα transcriptional network contributes to the search for therapeutic targets in atherosclerosis. Biol. Pharm. Bull. 2021, 44, 1607–1616. [Google Scholar] [CrossRef]
- Wang, Y.; Kumar, N.; Crumbley, C.; Griffin, P.R.; Burris, T.P. A second class of nuclear receptors for oxysterols: Regulation of RORα and RORγ activity by 24S-hydroxycholesterol (cerebrosterol). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2010, 1801, 917–923. [Google Scholar] [CrossRef]
- Wang, Y.; Solt, L.A.; Burris, T.P. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor α. J. Biol. Chem. 2010, 285, 15668–15673. [Google Scholar] [CrossRef]
- Shima, A.; Matsuoka, H.; Miya, K.; Michihara, A. Lovastatin suppresses the transcriptional regulation of CLDND1 in human hepatoma cells. BPB Rep. 2020, 3, 113–118. [Google Scholar] [CrossRef]
- Matsuoka, H.; Tamura, A.; Kinehara, M.; Shima, A.; Uda, A.; Tahara, H.; Michihara, A. Levels of tight junction protein CLDND1 are regulated by microRNA-124 in the cerebellum of stroke-prone spontaneously hypertensive rats. Biochem. Biophys. Res. Commun. 2018, 498, 817–823. [Google Scholar] [CrossRef]
- Shima, A.; Matsuoka, H.; Yamaoka, A.; Michihara, A. Transcription of CLDND1 in human brain endothelial cells is regulated by the myeloid zinc finger 1. Clin. Exp. Pharmacol. Physiol. 2021, 48, 260–269. [Google Scholar] [CrossRef]
- Shima, A.; Matsuoka, H.; Hamashima, T.; Yamaoka, A.; Koga, Y.; Michihara, A. Transcription of CLDND1 is regulated mainly by the competitive action of MZF1 and SP1 that binds to the enhancer of the promoter region. BPB Rep. 2020, 3, 190–195. [Google Scholar] [CrossRef]
- De La Brousse, F.C.; McKnight, S.L. Glimpses of allostery in the control of eukaryotic gene expression. Trends Genet. 1993, 9, 151–154. [Google Scholar] [CrossRef]
- Sharrocks, A.D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001, 2, 827–837. [Google Scholar] [CrossRef]
- Yang, R.; Li, X.; Wu, Y.; Zhang, G.; Liu, X.; Li, Y.; Bao, Y.; Yang, W.; Cui, H. EGFR activates GDH1 transcription to promote glutamine metabolism through MEK/ERK/ELK1 pathway in glioblastoma. Oncogene 2020, 39, 2975–2986. [Google Scholar] [CrossRef]
- Cavigelli, M.; Dolfi, F.; Claret, F.X.; Karin, M. Induction of c-fos expression through JNK-mediated TCF/Elk1 phosphorylation. EMBO J. 1995, 14, 5957–5964. [Google Scholar] [CrossRef]
- Enslen, H.; Raingeaud, J.; Davis, R.J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 1998, 273, 1741–1748. [Google Scholar] [CrossRef]
- Whitmarsh, A.J.; Shore, P.; Sharrocks, A.D.; Davis, R.J. Integration of MAP kinase signal transduction pathways at the serum response element. Science 1995, 269, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Buchwalter, G.; Gross, C.; Wasylyk, B. Ets ternary complex transcription factors. Gene 2004, 324, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, C.; Gonzalez-Angulo, A.M.; Liu, P.; Hayashi, N.; Lluch, A.; Ferrer-Lozano, J.; Hortobágyi, G.N. High ERK Protein Expression Levels Correlate with Shorter Survival in Triple-Negative Breast Cancer Patients. Oncologist 2012, 17, 766–774. [Google Scholar] [CrossRef]
- Ng, K.Y.; Chan, L.H.; Chai, S.; Tong, M.; Guan, X.Y.; Lee, N.P.; Yuan, Y.; Xie, D.; Lee, T.K.; Dusetti, N.J.; et al. TP53INP1 downregulation activates a p73-dependent DUSP10/ERK signaling pathway to promote metastasis of hepatocellular carcinoma. Cancer Res. 2017, 77, 4602–4612. [Google Scholar] [CrossRef]
- Yu, Y.; Luk, F.; Yang, J.L.; Walsh, W.R. Ras/Raf/MEK/ERK pathway is associated with lung metastasis of osteosarcoma in an orthotopic mouse model. Anticancer Res. 2011, 31, 1147–1152. [Google Scholar]
- Wakeling, A.E.; Guy, S.P.; Woodburn, J.R.; Ashton, S.E.; Curry, B.J.; Barker, A.J.; Gibson, K.H. ZD1839 (Iressa): An orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002, 62, 5749–5754. [Google Scholar] [PubMed]
- Recondo, G.; Facchinetti, F.; Olaussen, K.A.; Besse, B.; Friboulet, L. Making the first move in EGFR-driven or ALK-driven NSCLC: First-generation or next-generation TKI? Nat. Rev. Clin. Oncol. 2018, 15, 694–708. [Google Scholar] [CrossRef]
- Shore, P.; Whitmarsh, A.J.; Bhaskaran, R.; Davis, R.J.; Waltho, J.P.; Sharrocks, A.D. Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 1996, 16, 3338–3349. [Google Scholar] [CrossRef][Green Version]
- Gille, H.; Kortenjann, M.; Thomae, O.; Moomaw, C.; Slaughter, C.; Cobb, M.H.; Shaw, P.E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995, 14, 951–962. [Google Scholar] [CrossRef]
- Tsunoda, T.; Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef]
- Boros, J.; O’Donnell, A.; Donaldson, I.J.; Kasza, A.; Zeef, L.; Sharrocks, A.D. Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res. 2009, 37, 7368–7380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cruzalegui, F.H.; Cano, E.; Treisman, R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene 1999, 18, 7948–7957. [Google Scholar] [CrossRef] [PubMed]
- Gregg, J.; Fraizer, G. Transcriptional regulation of EGR1 by EGF and the ERK Signaling pathway in prostate cancer cells. Genes Cancer 2011, 2, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Caputo, R.; Bianco, R.; Damiano, V.; Fontanini, G.; Cuccato, S.; De Placido, S.; Bianco, A.R.; Tortora, G. Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor. Clin. Cancer Res. 2001, 7, 1459–1465. [Google Scholar] [PubMed]
- Fang, H.; Wang, Y.; Xu, L.; Zhou, S.; Bai, J.; Wu, Y.; Qiao, J.; Jiang, X.; Zhu, D.; Ding, Y. EGFR inhibitor gefitinib regulates barrier function in human epidermal keratinocytes via the modulation of the expression of claudins. Int. J. Mol. Med. 2019, 43, 1522–1530. [Google Scholar] [CrossRef]
- Sladojevic, N.; Stamatovic, S.M.; Johnson, A.M.; Choi, J.; Hu, A.; Dithmer, S.; Blasig, I.E.; Keep, R.F.; Andjelkovic, A.V. Claudin-1-dependent destabilization of the blood–brain barrier in chronic stroke. J. Neurosci. 2019, 39, 743–757. [Google Scholar] [CrossRef]
- Fayein, N.A.; Stankoff, B.; Auffray, C.; Devignes, M.D. Characterization of tissue expression and full-length coding sequence of a novel human gene mapping at 3q12.1 and transcribed in oligodendrocytes. Gene 2002, 289, 119–129. [Google Scholar] [CrossRef]
- Goto, K.; Sugiyama, T.; Matsumura, R.; Zhang, X.M.; Kimura, R.; Taira, A.; Arita, E.; Iwase, K.; Kobayashi, E.; Iwadate, Y.; et al. Identification of cerebral infarction-specific antibody markers from autoantibodies detected in patients with systemic lupus erythematosus. J. Mol. Biomark. Diagn. 2015, 6, 1. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuoka, H.; Yamaoka, A.; Hamashima, T.; Shima, A.; Kosako, M.; Tahara, Y.; Kamishikiryo, J.; Michihara, A. EGF-Dependent Activation of ELK1 Contributes to the Induction of CLDND1 Expression Involved in Tight Junction Formation. Biomedicines 2022, 10, 1792. https://doi.org/10.3390/biomedicines10081792
Matsuoka H, Yamaoka A, Hamashima T, Shima A, Kosako M, Tahara Y, Kamishikiryo J, Michihara A. EGF-Dependent Activation of ELK1 Contributes to the Induction of CLDND1 Expression Involved in Tight Junction Formation. Biomedicines. 2022; 10(8):1792. https://doi.org/10.3390/biomedicines10081792
Chicago/Turabian StyleMatsuoka, Hiroshi, Alice Yamaoka, Takahiro Hamashima, Akiho Shima, Marin Kosako, Yuma Tahara, Jun Kamishikiryo, and Akihiro Michihara. 2022. "EGF-Dependent Activation of ELK1 Contributes to the Induction of CLDND1 Expression Involved in Tight Junction Formation" Biomedicines 10, no. 8: 1792. https://doi.org/10.3390/biomedicines10081792
APA StyleMatsuoka, H., Yamaoka, A., Hamashima, T., Shima, A., Kosako, M., Tahara, Y., Kamishikiryo, J., & Michihara, A. (2022). EGF-Dependent Activation of ELK1 Contributes to the Induction of CLDND1 Expression Involved in Tight Junction Formation. Biomedicines, 10(8), 1792. https://doi.org/10.3390/biomedicines10081792






