ALCAM: A Novel Surface Marker on EpCAMlow Circulating Tumor Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient-Derived Tumor Cell lines
2.2. Naïve Neutrophil Isolation
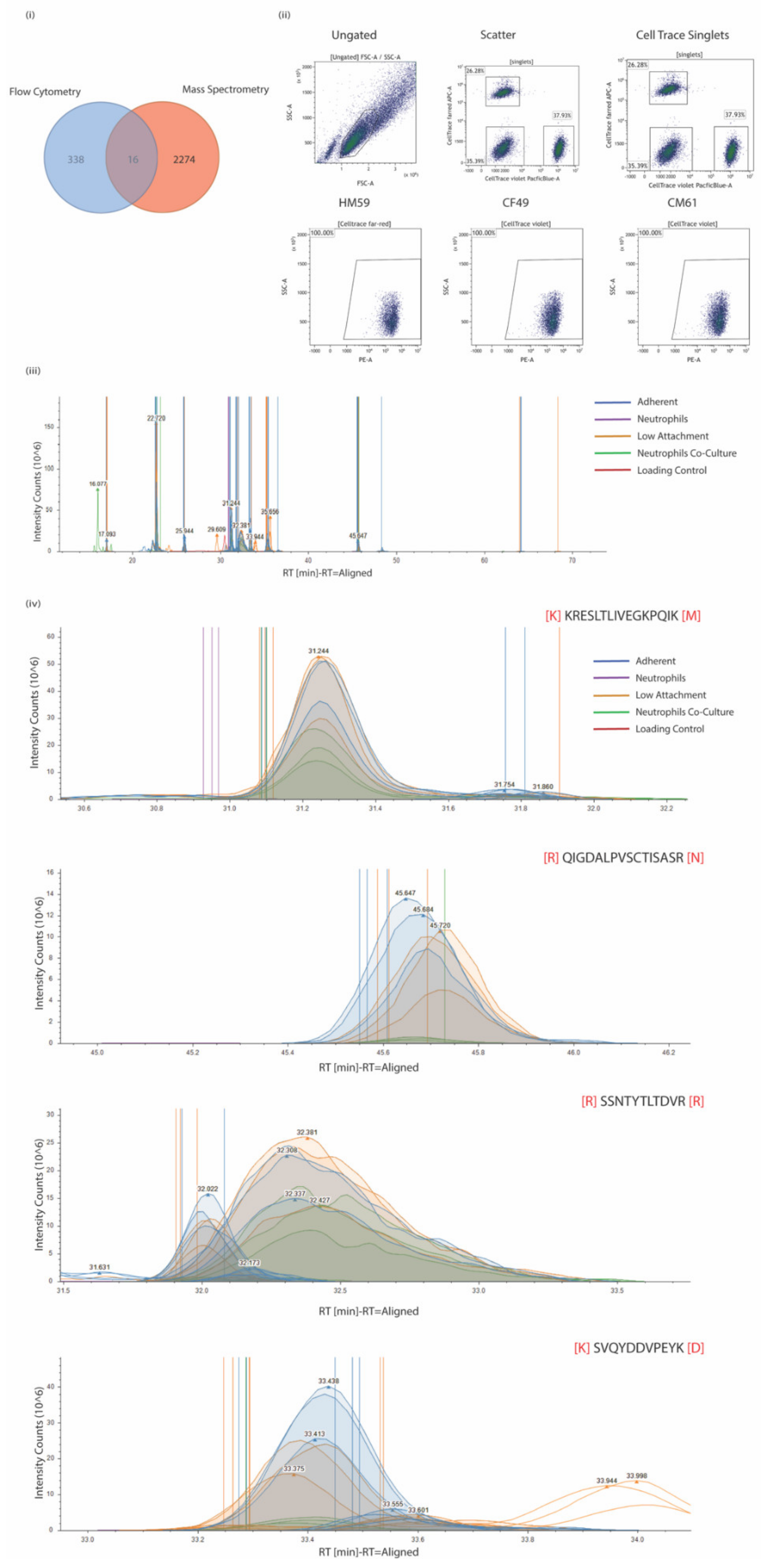
2.3. Flow Cytometric Surface Protein Characterization
2.4. Karyotyping
2.5. Preparation of Surface Proteome
2.6. Preparation of Samples for LC-MS
2.7. Transmission Electron Microscopy
2.8. LC-MS/MS Analysis
2.9. Proteome Discoverer Label-Free Quantitation Analysis
2.10. GEMM Animal Experiments
2.11. CTC and DTC Isolation from KPC Mice
2.12. HEK 293 and CTC Cell Line Staining
2.13. CTC Immunostaining and Enumeration
2.14. Enrichment Analyses
2.15. Generation of a Mouse–Human Hybrid TMA
2.16. Histochemical Validation of Selected Markers
3. Results
3.1. CTCs Express Multiple Surface Proteins That Might Play a Role in Pancreatic Cancer
3.2. Variation in Biomarkers on Surface Could Be Attributed to the Genetic Makeup of the CTCs
3.3. CTCs Express Differing Levels of Biomarkers on the Surface When Grown in Different Culture Conditions
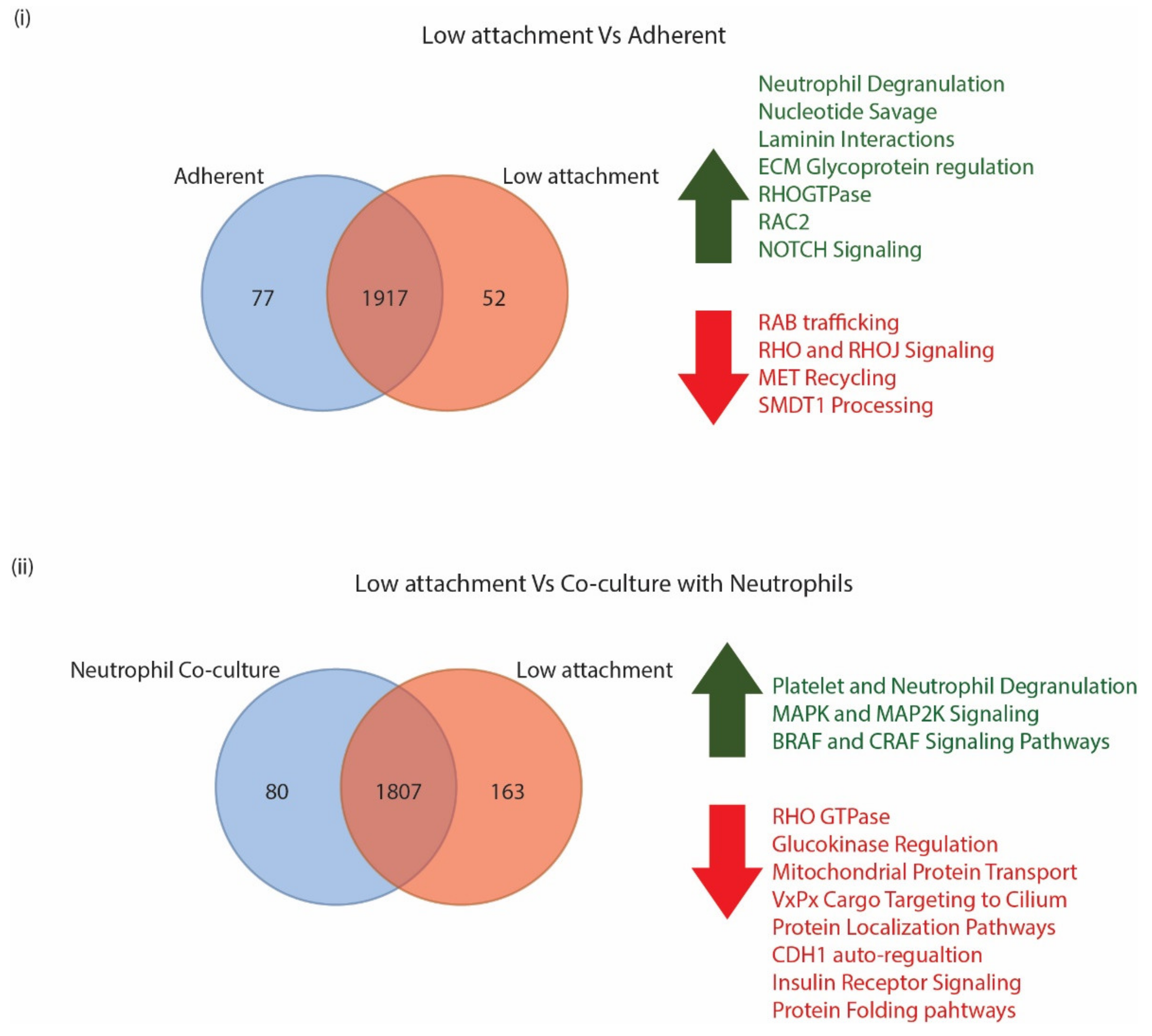
3.3.1. CTC Surface Markers Differ in Cells Grown in Adherent or Low Attachment Conditions
3.3.2. CTC Surface Markers Are Differentially Regulated on Exposure to Naïve Neutrophils
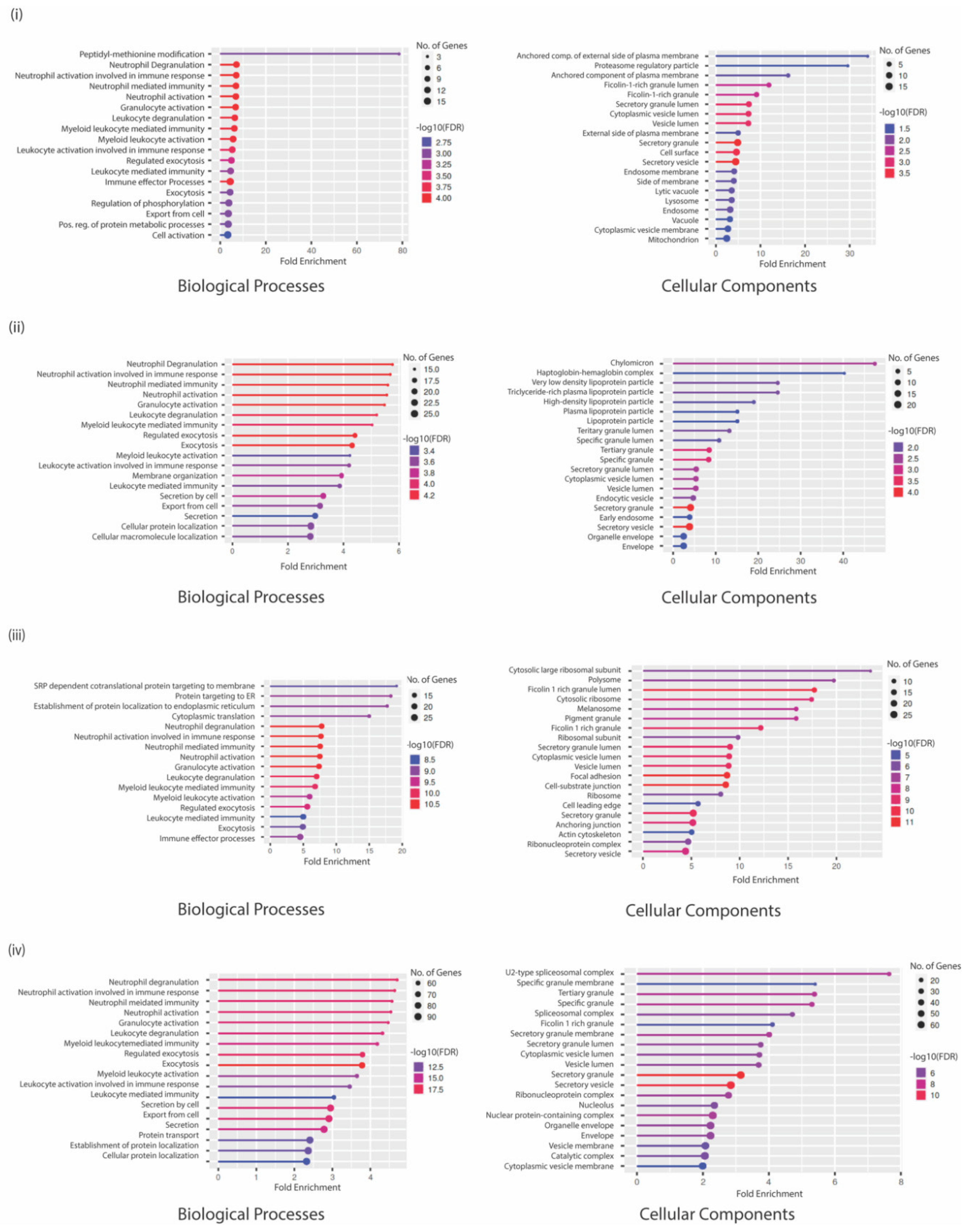
3.4. GO Analysis Indicates a Unique Repertoire of Enrichments in Different CTC Culture Conditions
3.5. ALCAM Is a Reliable Marker Expressed in PDAC CTCs
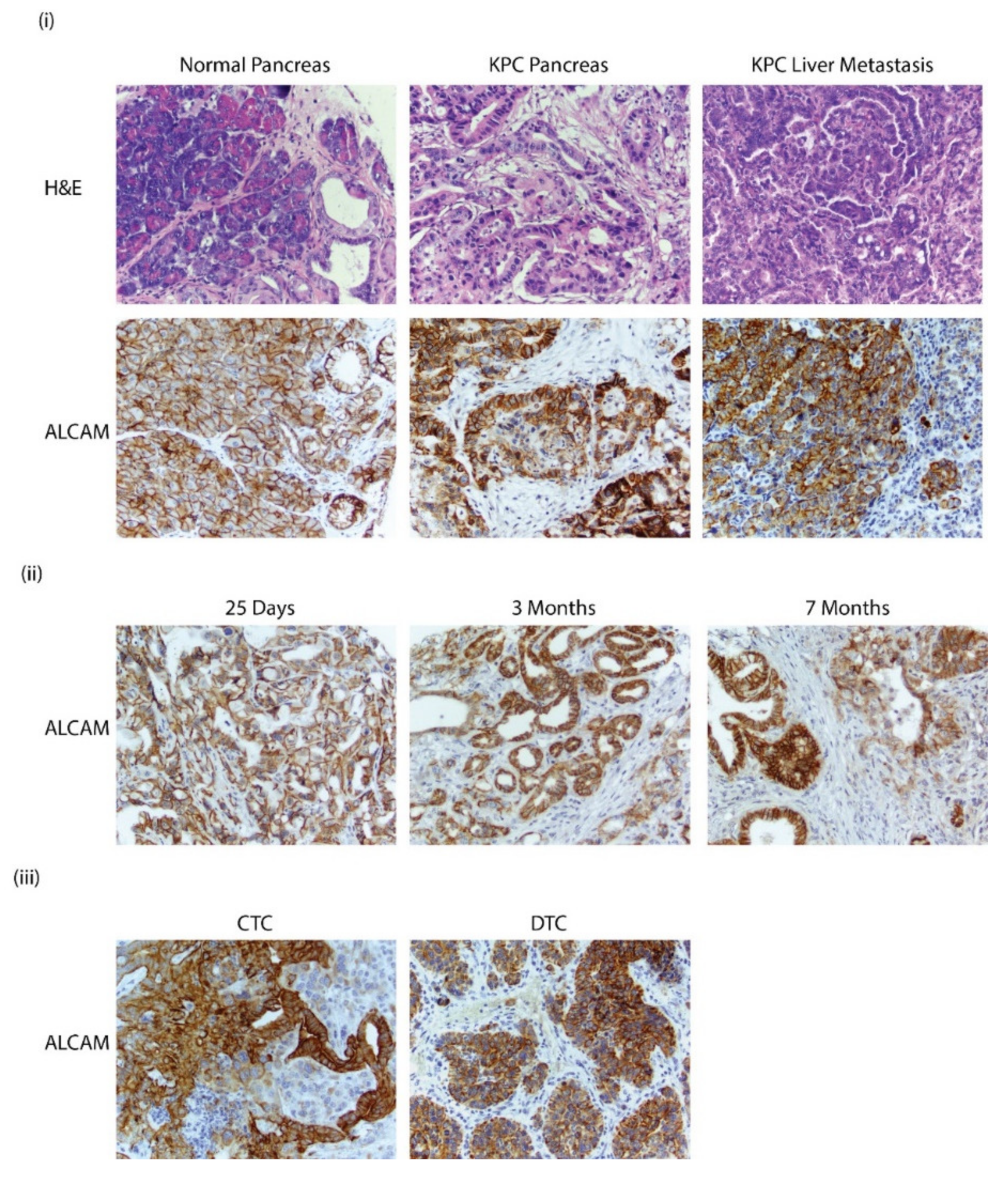
3.6. Alcam Is Expressed in PDAC GEMM Tumors, Distant Metastatic Sites, CTCs, and DTCs
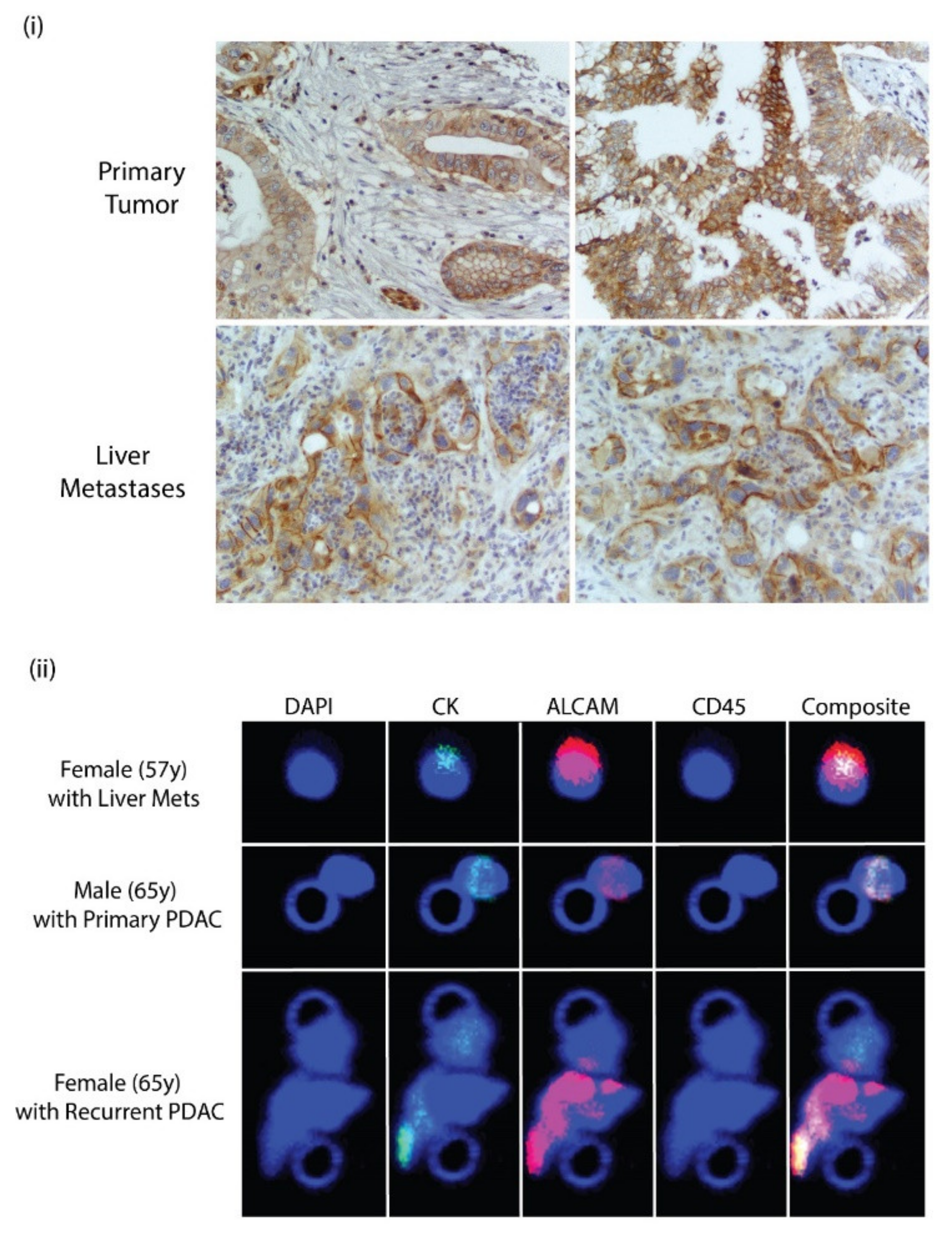
3.7. ALCAM Is Expressed in Diseased Pancreas and Related Metastases and on the Surface of CTCs Isolated from PDAC Patients
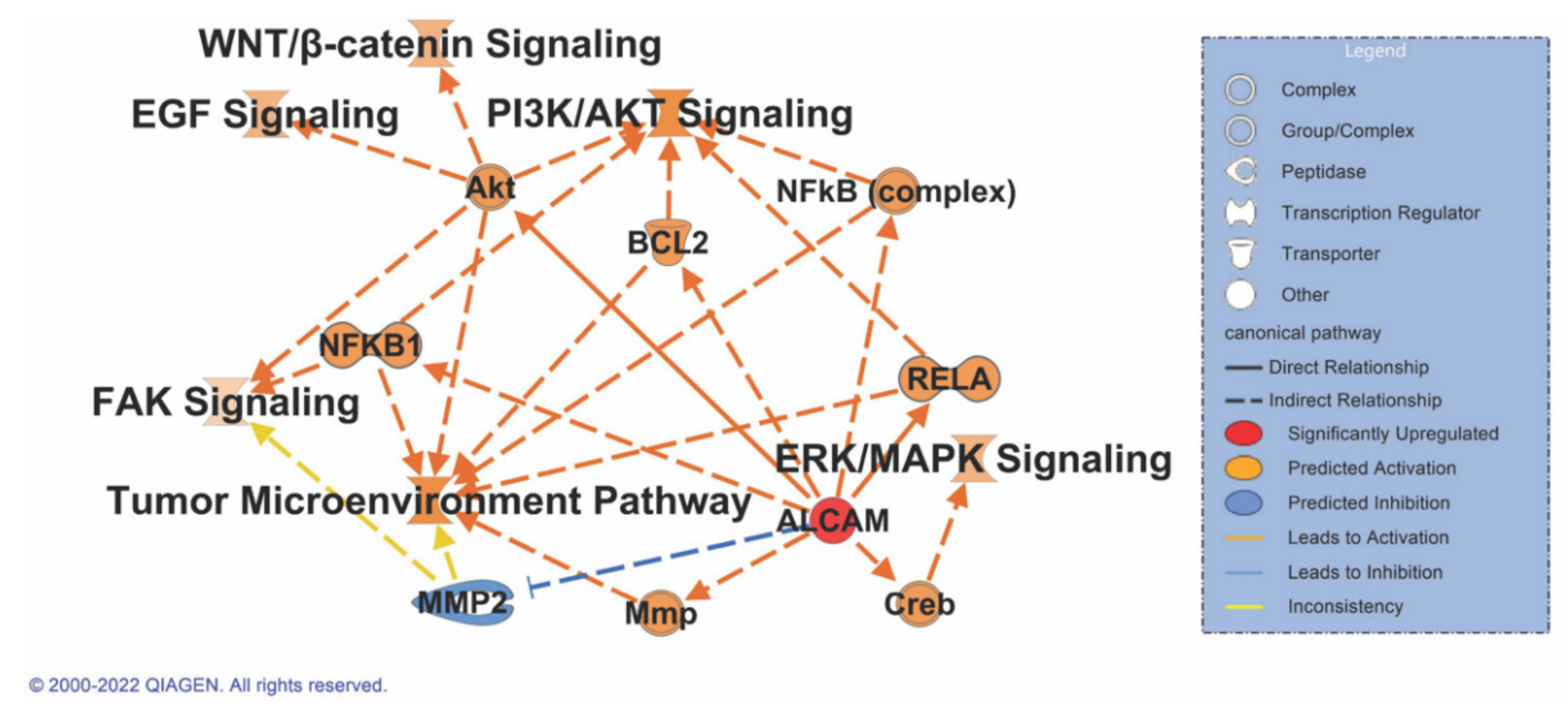
3.8. ALCAM Correlates with Multiple Signaling Pathways in Pancreatic Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menon, R.; Shahin, H. Extracellular vesicles in spontaneous preterm birth. Am. J. Reprod. Immunol. 2020, 85, e13353. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov-Markov, S.; Perales-Paton, J.; Bockorny, B.; Dopazo, A.; Munoz, M.; Banos, N.; Bonilla, V.; Menendez, C.; Duran, Y.; Huang, L.; et al. Discovery of New Targets to Control Metastasis in Pancreatic Cancer by Single-cell Transcriptomics Analysis of Circulating Tumor Cells. Mol. Cancer Ther. 2020, 19, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ma, Y.; Dong, X.; Zhang, Z.; Tian, X.; Zhao, X.; Yang, Y. Molecular characterization of circulating tumor cells in pancreatic ductal adenocarcinoma: Potential diagnostic and prognostic significance in clinical practice. Hepatobiliary Surg. Nutr. 2021, 10, 796–810. [Google Scholar] [CrossRef]
- Arnoletti, J.P.; Reza, J.; Rosales, A.; Monreal, A.; Fanaian, N.; Whisner, S.; Srivastava, M.; Rivera-Otero, J.; Yu, G.; Phanstiel Iv, O.; et al. Pancreatic Ductal Adenocarcinoma (PDAC) circulating tumor cells influence myeloid cell differentiation to support their survival and immunoresistance in portal vein circulation. PLoS ONE 2022, 17, e0265725. [Google Scholar] [CrossRef]
- Moutinho-Ribeiro, P.; Macedo, G.; Melo, S.A. Pancreatic Cancer Diagnosis and Management: Has the Time Come to Prick the Bubble? Front. Endocrinol. 2018, 9, 779. [Google Scholar] [CrossRef]
- DiPardo, B.J.; Winograd, P.; Court, C.M.; Tomlinson, J.S. Pancreatic cancer circulating tumor cells: Applications for personalized oncology. Expert Rev. Mol. Diagn. 2018, 18, 809–820. [Google Scholar] [CrossRef]
- Martini, V.; Timme-Bronsert, S.; Fichtner-Feigl, S.; Hoeppner, J.; Kulemann, B. Circulating Tumor Cells in Pancreatic Cancer: Current Perspectives. Cancers 2019, 11, 1659. [Google Scholar] [CrossRef]
- Xie, Z.B.; Yao, L.; Jin, C.; Fu, D.L. Circulating tumor cells in pancreatic cancer patients: Efficacy in diagnosis and value in prognosis. Discov. Med. 2016, 22, 121–128. [Google Scholar]
- Van der Toom, E.E.; Groot, V.P.; Glavaris, S.A.; Gemenetzis, G.; Chalfin, H.J.; Wood, L.D.; Wolfgang, C.L.; de la Rosette, J.; de Reijke, T.M.; Pienta, K.J. Analogous detection of circulating tumor cells using the AccuCyte((R))—CyteFinder((R)) system and ISET system in patients with locally advanced and metastatic prostate cancer. Prostate 2018, 78, 300–307. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Scheidmann, M.C.; Aceto, N. Beyond Enumeration: Functional and Computational Analysis of Circulating Tumor Cells to Investigate Cancer Metastasis. Front. Med. 2018, 5, 34. [Google Scholar] [CrossRef]
- Paoletti, C.; Miao, J.; Dolce, E.M.; Darga, E.P.; Repollet, M.I.; Doyle, G.V.; Gralow, J.R.; Hortobagyi, G.N.; Smerage, J.B.; Barlow, W.E.; et al. Circulating Tumor Cell Clusters in Patients with Metastatic Breast Cancer: A SWOG S0500 Translational Medicine Study. Clin. Cancer Res. 2019, 25, 6089–6097. [Google Scholar] [CrossRef]
- O’Toole, S.A.; Spillane, C.; Huang, Y.; Fitzgerald, M.C.; Ffrench, B.; Mohamed, B.; Ward, M.; Gallagher, M.; Kelly, T.; O’Brien, C.; et al. Circulating tumour cell enumeration does not correlate with Miller-Payne grade in a cohort of breast cancer patients undergoing neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2020, 181, 571–580. [Google Scholar] [CrossRef]
- Ankeny, J.S.; Court, C.M.; Hou, S.; Li, Q.; Song, M.; Wu, D.; Chen, J.F.; Lee, T.; Lin, M.; Sho, S.; et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br. J. Cancer 2016, 114, 1367–1375. [Google Scholar] [CrossRef]
- Yu, J.; Gemenetzis, G.; Kinny-Koster, B.; Habib, J.R.; Groot, V.P.; Teinor, J.; Yin, L.; Pu, N.; Hasanain, A.; van Oosten, F.; et al. Pancreatic circulating tumor cell detection by targeted single-cell next-generation sequencing. Cancer Lett. 2020, 493, 245–253. [Google Scholar] [CrossRef]
- Liu, H.; Sun, B.; Wang, S.; Liu, C.; Lu, Y.; Li, D.; Liu, X. Circulating Tumor Cells as a Biomarker in Pancreatic Ductal Adenocarcinoma. Cell. Physiol. Biochem. 2017, 42, 373–382. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Atwal, S.K.; Spoerke, J.M.; Savage, H.; Pandita, A.; Yeh, R.F.; Pirzkall, A.; Fine, B.M.; Amler, L.C.; Chen, D.S.; et al. Molecular biomarker analyses using circulating tumor cells. PLoS ONE 2010, 5, e12517. [Google Scholar] [CrossRef]
- Miltenyi, S.; Muller, W.; Weichel, W.; Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 1990, 11, 231–238. [Google Scholar] [CrossRef]
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975. [Google Scholar] [CrossRef]
- Schmidt, T.G.; Skerra, A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007, 2, 1528–1535. [Google Scholar] [CrossRef]
- Xiong, K.; Wei, W.; Jin, Y.; Wang, S.; Zhao, D.; Wang, S.; Gao, X.; Qiao, C.; Yue, H.; Ma, G.; et al. Biomimetic Immuno-Magnetosomes for High-Performance Enrichment of Circulating Tumor Cells. Adv. Mater. 2016, 28, 7929–7935. [Google Scholar] [CrossRef]
- Sequist, L.V.; Nagrath, S.; Toner, M.; Haber, D.A.; Lynch, T.J. The CTC-chip: An exciting new tool to detect circulating tumor cells in lung cancer patients. J. Thorac. Oncol. 2009, 4, 281–283. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Gottert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [Google Scholar] [CrossRef]
- Jan, Y.J.; Chen, J.F.; Zhu, Y.; Lu, Y.T.; Chen, S.H.; Chung, H.; Smalley, M.; Huang, Y.W.; Dong, J.; Chen, L.C.; et al. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 78–93. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [CrossRef]
- Yoon, H.J.; Shanker, A.; Wang, Y.; Kozminsky, M.; Jin, Q.; Palanisamy, N.; Burness, M.L.; Azizi, E.; Simeone, D.M.; Wicha, M.S.; et al. Tunable Thermal-Sensitive Polymer-Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv. Mater. 2016, 28, 4891–4897. [Google Scholar] [CrossRef]
- Loeian, M.S.; Mehdi Aghaei, S.; Farhadi, F.; Rai, V.; Yang, H.W.; Johnson, M.D.; Aqil, F.; Mandadi, M.; Rai, S.N.; Panchapakesan, B. Liquid biopsy using the nanotube-CTC-chip: Capture of invasive CTCs with high purity using preferential adherence in breast cancer patients. Lab Chip 2019, 19, 1899–1915. [Google Scholar] [CrossRef]
- Harouaka, R.A.; Zhou, M.D.; Yeh, Y.T.; Khan, W.J.; Das, A.; Liu, X.; Christ, C.C.; Dicker, D.T.; Baney, T.S.; Kaifi, J.T.; et al. Flexible micro spring array device for high-throughput enrichment of viable circulating tumor cells. Clin. Chem. 2014, 60, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853. [Google Scholar] [CrossRef]
- Zhou, M.D.; Hao, S.; Williams, A.J.; Harouaka, R.A.; Schrand, B.; Rawal, S.; Ao, Z.; Brenneman, R.; Gilboa, E.; Lu, B.; et al. Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells. Sci. Rep. 2014, 4, 7392. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.N.; Jayachandran, G.; Hardy, M.R.; Venkata Subramanian, A.M.; Meng, X.; Reuben, J.M. Antigen-agnostic microfluidics-based circulating tumor cell enrichment and downstream molecular characterization. PLoS ONE 2020, 15, e0241123. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal. Chem. 2010, 82, 6629–6635. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Guan, G.; Bhagat, A.A. ClearCell(R) FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells. Cytom. Part A 2018, 93, 1251–1254. [Google Scholar] [CrossRef]
- Hyun, K.A.; Kwon, K.; Han, H.; Kim, S.I.; Jung, H.I. Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients. Biosens. Bioelectron. 2013, 40, 206–212. [Google Scholar] [CrossRef]
- Konigsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; de Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011, 50, 700–710. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, W.H.; Song, Z.; Lou, H.; Kang, X. Comparison of circulating tumor cell (CTC) detection rates with epithelial cell adhesion molecule (EpCAM) and cell surface vimentin (CSV) antibodies in different solid tumors: A retrospective study. PeerJ 2021, 9, e10777. [Google Scholar] [CrossRef]
- Kurihara, T.; Itoi, T.; Sofuni, A.; Itokawa, F.; Tsuchiya, T.; Tsuji, S.; Ishii, K.; Ikeuchi, N.; Tsuchida, A.; Kasuya, K.; et al. Detection of circulating tumor cells in patients with pancreatic cancer: A preliminary result. J. Hepato-Biliary-Pancreat. Surg. 2008, 15, 189–195. [Google Scholar] [CrossRef]
- Khoja, L.; Backen, A.; Sloane, R.; Menasce, L.; Ryder, D.; Krebs, M.; Board, R.; Clack, G.; Hughes, A.; Blackhall, F.; et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br. J. Cancer 2012, 106, 508–516. [Google Scholar] [CrossRef]
- De Albuquerque, A.; Kubisch, I.; Ernst, D.; Breier, G.; Stamminger, G.; Fersis, N.; Stolzel, U.; Boese-Landgraf, J.; Eichler, A.; Kaul, S. Development of a molecular multimarker assay for the analysis of circulating tumor cells in adenocarcinoma patients. Clin. Lab. 2012, 58, 373–384. [Google Scholar]
- Bidard, F.C.; Huguet, F.; Louvet, C.; Mineur, L.; Bouche, O.; Chibaudel, B.; Artru, P.; Desseigne, F.; Bachet, J.B.; Mathiot, C.; et al. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: The ancillary CirCe 07 study to the LAP 07 trial. Ann. Oncol. 2013, 24, 2057–2061. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Ning, N.; Chen, Q.; Yang, Z.; Guo, Y.; Xu, D.; Zhang, D.; Zhan, T.; Cui, W. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. Int. J. Cancer 2015, 136, 1228–1233. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodriguez-Garrote, M.; Lisa, E.; Mendia, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Goldmann, U.; Muller, S.; van Oostrum, M.; Muller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Williams, A.; Rawal, S.; Ao, Z.; Torres-Munoz, J.; Balic, M.; Zhou, M.D.; Zheng, S.Y.; Tai, Y.C.; Cote, R.J.; Datar, R. Clinical Translation of a Novel Microfilter Technology Capture, Characterization and Culture of Circulating Tumor Cells. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 220–223. [Google Scholar]
- Lin, H.K.; Zheng, S.; Williams, A.J.; Balic, M.; Groshen, S.; Scher, H.I.; Fleisher, M.; Stadler, W.; Datar, R.H.; Tai, Y.C.; et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 2010, 16, 5011–5018. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Ram Makena, M.; Gatla, H.; Verlekar, D.; Sukhavasi, S.; Pandey, M.K.; Pramanik, K.C. Wnt/beta-Catenin Signaling: The Culprit in Pancreatic Carcinogenesis and Therapeutic Resistance. Int. J. Mol. Sci. 2019, 20, 4242. [Google Scholar] [CrossRef]
- Zeng, G.; Germinaro, M.; Micsenyi, A.; Monga, N.K.; Bell, A.; Sood, A.; Malhotra, V.; Sood, N.; Midda, V.; Monga, D.K.; et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia 2006, 8, 279–289. [Google Scholar] [CrossRef]
- Oliveira-Cunha, M.; Newman, W.G.; Siriwardena, A.K. Epidermal growth factor receptor in pancreatic cancer. Cancers 2011, 3, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Troiani, T.; Martinelli, E.; Capasso, A.; Morgillo, F.; Orditura, M.; De Vita, F.; Ciardiello, F. Targeting EGFR in pancreatic cancer treatment. Curr. Drug Targets 2012, 13, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.T.; Navas, C.; Martin-Serrano, G.; Grana-Castro, O.; Lechuga, C.G.; Martin-Diaz, L.; Djurec, M.; Li, J.; Morales-Cacho, L.; Esteban-Burgos, L.; et al. Complete Regression of Advanced Pancreatic Ductal Adenocarcinomas upon Combined Inhibition of EGFR and C-RAF. Cancer Cell 2019, 35, 573–587.e576. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Herrmann, D.; Evans, T.J.; Morton, J.P.; Timpson, P. Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine. Gut 2019, 68, 742–758. [Google Scholar] [CrossRef]
- Kanteti, R.; Mirzapoiazova, T.; Riehm, J.J.; Dhanasingh, I.; Mambetsariev, B.; Wang, J.; Kulkarni, P.; Kaushik, G.; Seshacharyulu, P.; Ponnusamy, M.P.; et al. Focal adhesion kinase a potential therapeutic target for pancreatic cancer and malignant pleural mesothelioma. Cancer Biol. Ther. 2018, 19, 316–327. [Google Scholar] [CrossRef]
- Furukawa, T. Impacts of activation of the mitogen-activated protein kinase pathway in pancreatic cancer. Front. Oncol. 2015, 5, 23. [Google Scholar] [CrossRef]
- Feig, C.; Gopinathan, A.; Neesse, A.; Chan, D.S.; Cook, N.; Tuveson, D.A. The pancreas cancer microenvironment. Clin. Cancer Res. 2012, 18, 4266–4276. [Google Scholar] [CrossRef]
- Ho, W.J.; Jaffee, E.M.; Zheng, L. The tumour microenvironment in pancreatic cancer—Clinical challenges and opportunities. Nat. Rev. Clin. Oncol 2020, 17, 527–540. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef]
- Bojic, S.; Hallam, D.; Alcada, N.; Ghareeb, A.; Queen, R.; Pervinder, S.; Buck, H.; Amitai Lange, A.; Figueiredo, G.; Rooney, P.; et al. CD200 Expression Marks a Population of Quiescent Limbal Epithelial Stem Cells with Holoclone Forming Ability. Stem Cells 2018, 36, 1723–1735. [Google Scholar] [CrossRef]
- Reduzzi, C.; Vismara, M.; Gerratana, L.; Silvestri, M.; De Braud, F.; Raspagliesi, F.; Verzoni, E.; Di Cosimo, S.; Locati, L.D.; Cristofanilli, M.; et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin. Cancer Biol. 2020, 60, 344–350. [Google Scholar] [CrossRef]
- Fujiwara, K.; Ohuchida, K.; Sada, M.; Horioka, K.; Ulrich, C.D., 3rd; Shindo, K.; Ohtsuka, T.; Takahata, S.; Mizumoto, K.; Oda, Y.; et al. CD166/ALCAM expression is characteristic of tumorigenicity and invasive and migratory activities of pancreatic cancer cells. PLoS ONE 2014, 9, e107247. [Google Scholar] [CrossRef]
- Kahlert, C.; Weber, H.; Mogler, C.; Bergmann, F.; Schirmacher, P.; Kenngott, H.G.; Matterne, U.; Mollberg, N.; Rahbari, N.N.; Hinz, U.; et al. Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br. J. Cancer 2009, 101, 457–464. [Google Scholar] [CrossRef]
- Tachezy, M.; Zander, H.; Marx, A.H.; Stahl, P.R.; Gebauer, F.; Izbicki, J.R.; Bockhorn, M. ALCAM (CD166) expression and serum levels in pancreatic cancer. PLoS ONE 2012, 7, e39018. [Google Scholar] [CrossRef][Green Version]
- Xu, X.; Rao, G.S.; Groh, V.; Spies, T.; Gattuso, P.; Kaufman, H.L.; Plate, J.; Prinz, R.A. Major histocompatibility complex class I-related chain A/B (MICA/B) expression in tumor tissue and serum of pancreatic cancer: Role of uric acid accumulation in gemcitabine-induced MICA/B expression. BMC Cancer 2011, 11, 194. [Google Scholar] [CrossRef]
- Duan, Q.; Li, H.; Gao, C.; Zhao, H.; Wu, S.; Wu, H.; Wang, C.; Shen, Q.; Yin, T. High glucose promotes pancreatic cancer cells to escape from immune surveillance via AMPK-Bmi1-GATA2-MICA/B pathway. J. Exp. Clin. Cancer Res. 2019, 38, 192. [Google Scholar] [CrossRef]
- Wang, V.M.; Ferreira, R.M.M.; Almagro, J.; Evan, T.; Legrave, N.; Zaw Thin, M.; Frith, D.; Carvalho, J.; Barry, D.J.; Snijders, A.P.; et al. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat. Cell Biol. 2019, 21, 1425–1435. [Google Scholar] [CrossRef]
- Lu, W.; Fei, A.; Jiang, Y.; Chen, L.; Wang, Y. Tetraspanin CD9 interacts with alpha-secretase to enhance its oncogenic function in pancreatic cancer. Am. J. Transl. Res. 2020, 12, 5525–5537. [Google Scholar]
- Wei, Q.; Zhang, J.; Li, Z.; Wei, L.; Ren, L. Serum Exo-EphA2 as a Potential Diagnostic Biomarker for Pancreatic Cancer. Pancreas 2020, 49, 1213–1219. [Google Scholar] [CrossRef]
- Fan, J.; Wei, Q.; Koay, E.J.; Liu, Y.; Ning, B.; Bernard, P.W.; Zhang, N.; Han, H.; Katz, M.H.; Zhao, Z.; et al. Chemoresistance Transmission via Exosome-Mediated EphA2 Transfer in Pancreatic Cancer. Theranostics 2018, 8, 5986–5994. [Google Scholar] [CrossRef]
- Chen, Q.; Pu, N.; Yin, H.; Zhang, J.; Zhao, G.; Lou, W.; Wu, W. CD73 acts as a prognostic biomarker and promotes progression and immune escape in pancreatic cancer. J. Cell. Mol. Med. 2020, 24, 8674–8686. [Google Scholar] [CrossRef]
- Zhou, L.; Jia, S.; Chen, Y.; Wang, W.; Wu, Z.; Yu, W.; Zhang, M.; Ding, G.; Cao, L. The distinct role of CD73 in the progression of pancreatic cancer. J. Mol. Med. 2019, 97, 803–815. [Google Scholar] [CrossRef]
- Nguyen, A.M.; Zhou, J.; Sicairos, B.; Sonney, S.; Du, Y. Upregulation of CD73 Confers Acquired Radioresistance and is Required for Maintaining Irradiation-selected Pancreatic Cancer Cells in a Mesenchymal State. Mol. Cell. Proteom. 2020, 19, 375–389. [Google Scholar] [CrossRef]
- Dias, J.; Sandberg, J.K.; Leeansyah, E. Extensive Phenotypic Analysis, Transcription Factor Profiling, and Effector Cytokine Production of Human MAIT Cells by Flow Cytometry. Methods Mol. Biol. 2017, 1514, 241–256. [Google Scholar] [CrossRef]
- Magill, L.; Adriani, M.; Berthou, V.; Chen, K.; Gleizes, A.; Hacein-Bey-Abina, S.; Hincelin-Mery, A.; Mariette, X.; Pallardy, M.; Spindeldreher, S.; et al. Low Percentage of Signal Regulatory Protein α/β+ Memory B Cells in Blood Predicts Development of Anti-drug Antibodies (ADA) in Adalimumab-Treated Rheumatoid Arthritis Patients. Front. Immunol. 2018, 9, 2865. [Google Scholar] [CrossRef]
- Ronnberg, E.; Boey, D.Z.H.; Ravindran, A.; Safholm, J.; Orre, A.C.; Al-Ameri, M.; Adner, M.; Dahlen, S.E.; Dahlin, J.S.; Nilsson, G. Immunoprofiling Reveals Novel Mast Cell Receptors and the Continuous Nature of Human Lung Mast Cell Heterogeneity. Front. Immunol. 2021, 12, 804812. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, A.L.; Joo, H.J.; Han, S.; Kim, S.H.; Kim, H.Y.; Lim, J.S.; Lee, M.S.; Choi, H.K.; Yang, Y. Development of suspension cell culture model to mimic circulating tumor cells. Oncotarget 2018, 9, 622–640. [Google Scholar] [CrossRef]
- May, A.N.; Crawford, B.D.; Nedelcu, A.M. In Vitro Model-Systems to Understand the Biology and Clinical Significance of Circulating Tumor Cell Clusters. Front. Oncol. 2018, 8, 63. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Iwagishi, R.; Tanaka, R.; Seto, M.; Takagi, T.; Norioka, N.; Ueyama, T.; Kawamura, T.; Takagi, J.; Ogawa, Y.; Shirakabe, K. Negatively charged amino acids in the stalk region of membrane proteins reduce ectodomain shedding. J. Biol. Chem. 2020, 295, 12343–12352. [Google Scholar] [CrossRef] [PubMed]
- Micciche, F.; Da Riva, L.; Fabbi, M.; Pilotti, S.; Mondellini, P.; Ferrini, S.; Canevari, S.; Pierotti, M.A.; Bongarzone, I. Activated leukocyte cell adhesion molecule expression and shedding in thyroid tumors. PLoS ONE 2011, 6, e17141. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, J.; Lin, J.; Pan, Q.; Yu, Y.; Sun, F. Cluster of differentiation 166 (CD166) regulated by phosphatidylinositide 3-Kinase (PI3K)/AKT signaling to exert its anti-apoptotic role via yes-associated protein (YAP) in liver cancer. J. Biol. Chem. 2014, 289, 6921–6933. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gu, Z.; Ni, P.; Qiao, Y.; Chen, C.; Liu, X.; Lin, J.; Chen, N.; Fan, Q. NF-kappaB P50/P65 hetero-dimer mediates differential regulation of CD166/ALCAM expression via interaction with micoRNA-9 after serum deprivation, providing evidence for a novel negative auto-regulatory loop. Nucleic Acids Res. 2011, 39, 6440–6455. [Google Scholar] [CrossRef]
- Bosserhoff, A.K.; Hofmeister, S.; Ruedel, A.; Schubert, T. DCC is expressed in a CD166-positive subpopulation of chondrocytes in human osteoarthritic cartilage and modulates CRE activity. Int. J. Clin. Exp. Pathol. 2014, 7, 1947–1956. [Google Scholar]
- Charles Jacob, H.K.; Charles Richard, J.L.; Signorelli, R.; Kashuv, T.; Lavania, S.; Vaish, U.; Boopathy, R.; Middleton, A.; Boone, M.M.; Sundaram, R.; et al. Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 2727. [Google Scholar] [CrossRef]
- Amantini, C.; Morelli, M.B.; Nabissi, M.; Piva, F.; Marinelli, O.; Maggi, F.; Bianchi, F.; Bittoni, A.; Berardi, R.; Giampieri, R.; et al. Expression Profiling of Circulating Tumor Cells in Pancreatic Ductal Adenocarcinoma Patients: Biomarkers Predicting Overall Survival. Front. Oncol. 2019, 9, 874. [Google Scholar] [CrossRef]
- Hong, X.; Michalski, C.W.; Kong, B.; Zhang, W.; Raggi, M.C.; Sauliunaite, D.; De Oliveira, T.; Friess, H.; Kleeff, J. ALCAM is associated with chemoresistance and tumor cell adhesion in pancreatic cancer. J. Surg. Oncol. 2010, 101, 564–569. [Google Scholar] [CrossRef]
- Ni, C.; Zhang, Z.; Zhu, X.; Liu, Y.; Qu, D.; Wu, P.; Huang, J.; Xu, A.X. Prognostic value of CD166 expression in cancers of the digestive system: A systematic review and meta-analysis. PLoS ONE 2013, 8, e70958. [Google Scholar] [CrossRef]
- Tachezy, M.; Zander, H.; Marx, A.H.; Gebauer, F.; Rawnaq, T.; Kaifi, J.T.; Sauter, G.; Izbicki, J.R.; Bockhorn, M. ALCAM (CD166) expression as novel prognostic biomarker for pancreatic neuroendocrine tumor patients. J. Surg. Res. 2011, 170, 226–232. [Google Scholar] [CrossRef]
- Jin, U.H.; Karki, K.; Kim, S.B.; Safe, S. Inhibition of pancreatic cancer Panc1 cell migration by omeprazole is dependent on aryl hydrocarbon receptor activation of JNK. Biochem. Biophys. Res. Commun. 2018, 501, 751–757. [Google Scholar] [CrossRef]
- Zhang, W.W.; Zhan, S.H.; Geng, C.X.; Sun, X.; Erkan, M.; Kleeff, J.; Xie, X.J. Activated leukocyte cell adhesion molecule regulates the interaction between pancreatic cancer cells and stellate cells. Mol. Med. Rep. 2016, 14, 3627–3633. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Sanders, A.J.; Ruge, F.; Dong, X.; Cui, Y.; Dou, Q.P.; Jia, S.; Hao, C.; Ji, J.; Jiang, W.G. Activated leukocyte cell adhesion molecule (ALCAM)/CD166 in pancreatic cancer, a pivotal link to clinical outcome and vascular embolism. Am. J. Cancer Res. 2021, 11, 5917–5932. [Google Scholar]

| Protein Marker | Gene Name | Description | CM61 (% + ve) | CF49 (% + ve) | HM59 (% + ve) | |
|---|---|---|---|---|---|---|
| 1 | CD85H | LILRA2 | Leukocyte Immunoglobulin-Like Receptor Subfamily A member 2 | 99.93 | 100 | 100 |
| 2 | CD166 | ALCAM | Activated Leukocyte Cell Adhesion Molecule | 99.83 | 99.95 | 99.98 |
| 3 | MICA/MICB | MICA/MICB | MHC Class I polypeptide-related sequence A/B | 99.90 | 99.93 | 99.86 |
| 4 | CD9 | CD9 | CD9 antigen | 99.73 | 99.79 | 99.93 |
| 5 | EPHA2 | EPHA2 | Ephrin Type A receptor 2 | 97.63 | 99.19 | 99.16 |
| 6 | CD252 | TNFSF4 | Tumor Necrosis Factor Ligand Superfamily member 4 | 93.08 | 95.88 | 97.67 |
| 7 | CD129 | IL9R | Interleukin-9 Receptor | 90.50 | 94.89 | 93.97 |
| 8 | CD73 | NT5E | 5′ Nucleotidase | 83.43 | 90.05 | 90.40 |
| 9 | CD263 | TNFRSF10C | Tumor Necrosis factor receptor superfamily member 10C | 67.75 | 78.85 | 83.37 |
| 10 | CD215 | IL15RA | Interleukin 15 receptor subunit alpha | 60.04 | 57.96 | 72.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Signorelli, R.; Giret, T.M.; Umland, O.; Hadisurya, M.; Lavania, S.; Charles Richard, J.L.; Middleton, A.; Boone, M.M.; Ergonul, A.B.; Tao, W.A.; et al. ALCAM: A Novel Surface Marker on EpCAMlow Circulating Tumor Cells. Biomedicines 2022, 10, 1983. https://doi.org/10.3390/biomedicines10081983
Signorelli R, Giret TM, Umland O, Hadisurya M, Lavania S, Charles Richard JL, Middleton A, Boone MM, Ergonul AB, Tao WA, et al. ALCAM: A Novel Surface Marker on EpCAMlow Circulating Tumor Cells. Biomedicines. 2022; 10(8):1983. https://doi.org/10.3390/biomedicines10081983
Chicago/Turabian StyleSignorelli, Rossana, Teresa Maidana Giret, Oliver Umland, Marco Hadisurya, Shweta Lavania, John Lalith Charles Richard, Ashley Middleton, Melinda Minucci Boone, Ayse Burcu Ergonul, Weiguo Andy Tao, and et al. 2022. "ALCAM: A Novel Surface Marker on EpCAMlow Circulating Tumor Cells" Biomedicines 10, no. 8: 1983. https://doi.org/10.3390/biomedicines10081983
APA StyleSignorelli, R., Giret, T. M., Umland, O., Hadisurya, M., Lavania, S., Charles Richard, J. L., Middleton, A., Boone, M. M., Ergonul, A. B., Tao, W. A., Amirian, H., Iliuk, A., Khan, A., Diaz, R., Cortes, D. B., Garcia-Buitrago, M., & Charles Jacob, H. K. (2022). ALCAM: A Novel Surface Marker on EpCAMlow Circulating Tumor Cells. Biomedicines, 10(8), 1983. https://doi.org/10.3390/biomedicines10081983








