Interplay between Artificial Intelligence and Biomechanics Modeling in the Cardiovascular Disease Prediction
Abstract
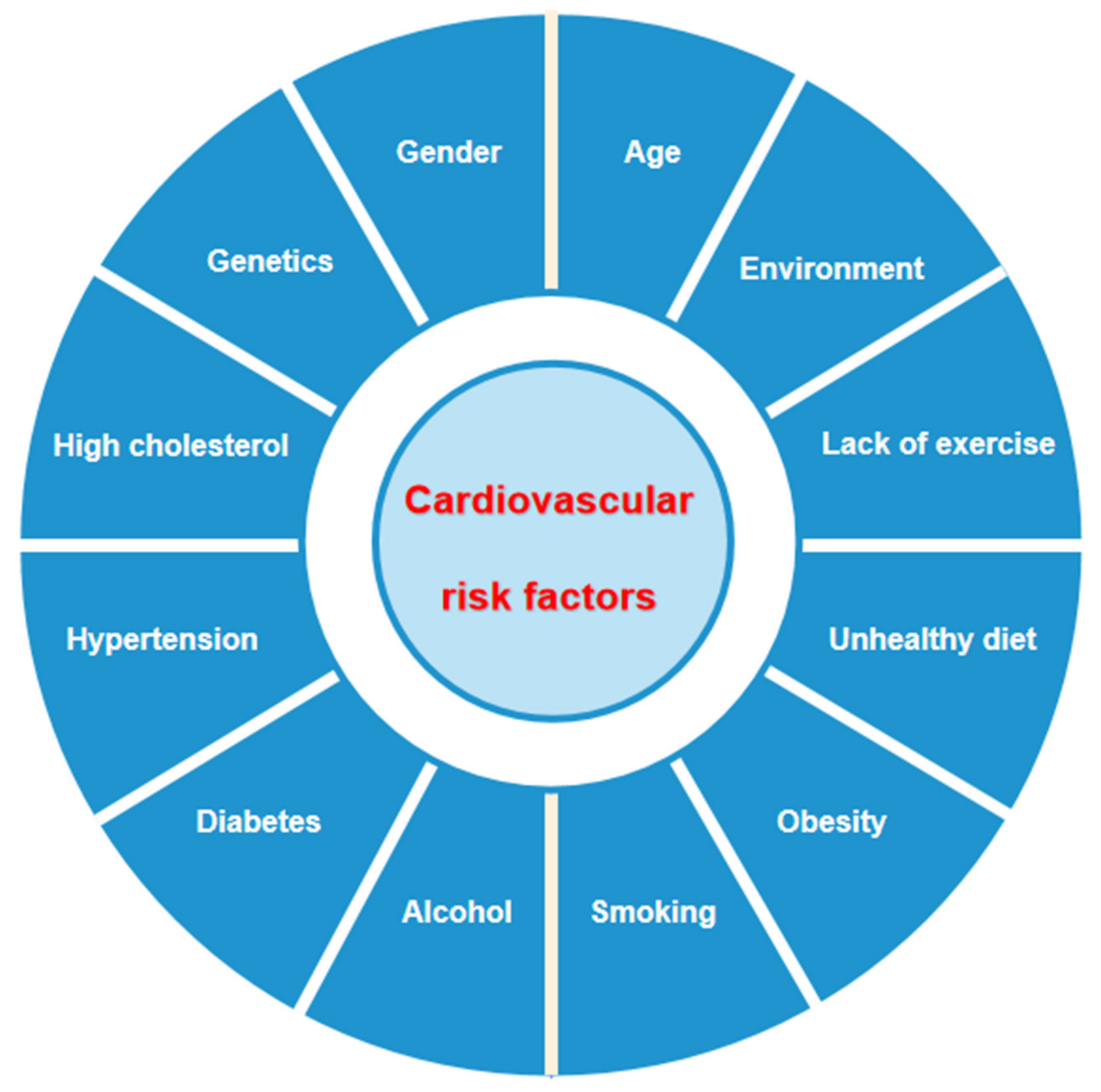
1. Introduction
2. Overview of Artificial Intelligence
3. Application of Artificial Intelligence in the Prediction of Cardiovascular Disease
3.1. Prediction of Cardiovascular Morbidity or Mortality
3.2. Prediction of Cardiovascular Biomechanics Modeling
3.2.1. Traditional Computational Modeling and Simulation
3.2.2. ML−Based Hemodynamics with Vascular Geometries, Equations and Methods
Geometric Modeling
Governing Equation (ML−Based Partial Differential Equation)
A ML−Based Surrogate for Computational Fluid Dynamics
4. Challenges and Future Prospects
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Tsao, C.W. Heart Disease and Stroke Statistics—2020 Update: A Report From the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Leong, D.; Mckee, M.; Anand, S.S.; Schwalm, J.D.; Teo, K.; Mente, A.; Yusuf, S. Reducing the Global Burden of Cardiovascular Disease, Part 1: The Epidemiology and Risk Factors. Circ. Res. 2017, 121, 677. [Google Scholar] [CrossRef] [PubMed]
- Yichong, L.; Limin, W.; Yong, J.; Mei, Z. GW24-e1907Association between changes in status of multiple cardiovascular risk factors and carotid atherosclerosis progression: A population-based cohort study. Heart 2013, 99, E126–E127. [Google Scholar]
- Heb, A.; Prt, B.; Ee, B.; Edm, C.; Raf, C.; Alb, D.; Hmk, E.; Kcf, F.; Mre, G.; Hw, H. Ten things to know about ten cardiovascular disease risk factors—ScienceDirect. Am. J. Prev. Cardiol. 2021, 5, 100149. [Google Scholar]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Group G-N-JGBoCDW. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019 Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- Timmis, A.; Vardas, P.; Townsend, N.; Torbica, A.; Katus, H.; De Smedt, D.; Gale, C.P.; Maggioni, A.P.; Petersen, S.E.; Huculeci, R. European Society of Cardiology: Cardiovascular disease statistics 2021. Eur. Heart J. 2022, 43, 716–799. [Google Scholar] [CrossRef]
- Andersson, C.; Johnson, A.D.; Benjamin, E.J.; Levy, D.; Vasan, R.S. 70-year legacy of the Framingham Heart Study. Nat. Rev. Cardiol. 2019, 16, 1–12. [Google Scholar] [CrossRef]
- Pitchai, B.; Khin, M.-U.; Gowraganahalli, J. Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol. Res. Off. J. Ital. Pharmacol. Soc. 2016, 113, 600–609. [Google Scholar]
- Briganti, G.; Le Moine, O. Artificial intelligence in medicine: Today and tomorrow. Front. Med. 2020, 7, 27. [Google Scholar] [CrossRef]
- Lüscher, T.F.; Lyon, A.; Amstein, R.; Maisel, A. Artificial Intelligence: The Pathway to the Future of Cardiovascular Medicine; Oxford University Press: Oxford, UK, 2022. [Google Scholar]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef]
- Kohli, P.S.; Arora, S. Application of machine learning in disease prediction. In Proceedings of the 2018 4th International conference on computing communication and automation (ICCCA), Greater Noida, India, 14–15 December 2018; pp. 1–4. [Google Scholar]
- Kaelin, V.C.; Valizadeh, M.; Salgado, Z.; Parde, N.; Khetani, M.A. Artificial intelligence in rehabilitation targeting the participation of children and youth with disabilities: Scoping review. J. Med. Internet Res. 2021, 23, e25745. [Google Scholar] [CrossRef]
- Ialongo, C.; Pieri, M.; Bernardini, S. Artificial neural network for total laboratory automation to improve the management of sample dilution: Smart automation for clinical laboratory timeliness. Slas Technol. Transl. Life Sci. Innov. 2017, 22, 44–49. [Google Scholar] [CrossRef]
- Mehta, M.C.; Katz, I.T.; Jha, A.K. Transforming global health with AI. N. Engl. J. Med. 2020, 382, 791–793. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Ayodele, T.O. Types of machine learning algorithms. New Adv. Mach. Learn. 2010, 3, 19–48. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Kutz, J.N. Deep learning in fluid dynamics. J. Fluid Mech. 2017, 814, 1–4. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, C.C. Neural networks and deep learning. Springer 2018, 10, 978-3. [Google Scholar]
- Abedi, V.; Goyal, N.; Tsivgoulis, G.; Hosseinichimeh, N.; Hontecillas, R.; Bassaganya-Riera, J.; Elijovich, L.; Metter, J.E.; Alexandrov, A.W.; Liebeskind, D.S. Novel screening tool for stroke using artificial neural network. Stroke 2017, 48, 1678–1681. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liang, L.; Sun, W. Estimation of in vivo constitutive parameters of the aortic wall using a machine learning approach. Comput. Methods Appl. Mech. Eng. 2019, 347, 201–217. [Google Scholar] [CrossRef]
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: A multicenter study. JACC Cardiovasc. Imaging 2018, 11, 1654–1663. [Google Scholar] [CrossRef]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Physics-Informed Neural Networks: A Deep Learning Framework for Solving Forward and Inverse Problems Involving Nonlinear Partial Differential Equations. J. Comput. Phys. 2018, 378, 686–707. [Google Scholar] [CrossRef]
- Shameer, K.; Johnson, K.W.; Glicksberg, B.S.; Dudley, J.T.; Sengupta, P.P. Machine learning in cardiovascular medicine: Are we there yet? Heart 2018, 104, 1156–1164. [Google Scholar] [CrossRef]
- Arzani, A.; Wang, J.-X.; Sacks, M.S.; Shadden, S.C. Machine Learning for Cardiovascular Biomechanics Modeling: Challenges and Beyond. Ann. Biomed. Eng. 2022, 50, 615–627. [Google Scholar] [CrossRef]
- Diller, G.-P.; Kempny, A.; Babu-Narayan, S.V.; Henrichs, M.; Brida, M.; Uebing, A.; Lammers, A.E.; Baumgartner, H.; Li, W.; Wort, S.J. Machine learning algorithms estimating prognosis and guiding therapy in adult congenital heart disease: Data from a single tertiary centre including 10 019 patients. Eur. Heart J. 2019, 40, 1069–1077. [Google Scholar] [CrossRef]
- D’Agostino Sr, R.B.; Vasan, R.S.; Pencina, M.J.; Wolf, P.A.; Cobain, M.; Massaro, J.M.; Kannel, W.B. General cardiovascular risk profile for use in primary care: The Framingham Heart Study. Circulation 2008, 117, 743–753. [Google Scholar] [CrossRef]
- Conroy, R.M.; Pyörälä, K.; Fitzgerald, A.E.; Sans, S.; Menotti, A.; De Backer, G.; De Bacquer, D.; Ducimetiere, P.; Jousilahti, P.; Keil, U. Estimation of ten-year risk of fatal cardiovascular disease in Europe: The SCORE project. Eur. Heart J. 2003, 24, 987–1003. [Google Scholar] [CrossRef]
- Goff, D.C.; Lloyd-Jones, D.M.; Bennett, G.; Coady, S.; D’agostino, R.B.; Gibbons, R.; Greenland, P.; Lackland, D.T.; Levy, D.; O’donnell, C.J. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014, 63, 2935–2959. [Google Scholar] [CrossRef] [PubMed]
- Kakadiaris, I.A.; Vrigkas, M.; Yen, A.A.; Kuznetsova, T.; Budoff, M.; Naghavi, M. Machine learning outperforms ACC/AHA CVD risk calculator in MESA. J. Am. Heart Assoc. 2018, 7, e009476. [Google Scholar] [CrossRef]
- Dimopoulos, A.C.; Nikolaidou, M.; Caballero, F.F.; Engchuan, W.; Sanchez-Niubo, A.; Arndt, H.; Ayuso-Mateos, J.L.; Haro, J.M.; Chatterji, S.; Georgousopoulou, E.N. Machine learning methodologies versus cardiovascular risk scores, in predicting disease risk. BMC Med. Res. Methodol. 2018, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alaa, A.M.; Bolton, T.; Di Angelantonio, E.; Rudd, J.H.; Van der Schaar, M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLoS ONE 2019, 14, e0213653. [Google Scholar] [CrossRef]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.H.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: A 5-year multicentre prospective registry analysis. Eur. Heart J. 2017, 38, 500–507. [Google Scholar] [CrossRef]
- van Rosendael, A.R.; Maliakal, G.; Kolli, K.K.; Beecy, A.; Al’Aref, S.J.; Dwivedi, A.; Singh, G.; Panday, M.; Kumar, A.; Ma, X. Maximization of the usage of coronary CTA derived plaque information using a machine learning based algorithm to improve risk stratification; insights from the CONFIRM registry. J. Cardiovasc. Comput. Tomogr. 2018, 12, 204–209. [Google Scholar] [CrossRef]
- Tourassi, G.D.; Armato, S.G.; Lessmann, N.; Isgum, I.; Setio, A.; Vos, B.D.; Ciompi, F.; Jong, P.D.; Oudkerk, M.; Mali, W. Deep convolutional neural networks for automatic coronary calcium scoring in a screening study with low-dose chest CT. Med. Imaging Comput. Aided Diagnosis. SPIE 2016, 9785, 978511. [Google Scholar]
- Zreik, M.; Lessmann, N.; van Hamersvelt, R.W.; Wolterink, J.M.; Voskuil, M.; Viergever, M.A.; Leiner, T.; Išgum, I. Deep learning analysis of the myocardium in coronary CT angiography for identification of patients with functionally significant coronary artery stenosis. Med. Image Anal. 2018, 44, 72–85. [Google Scholar] [CrossRef]
- Gessert, N.; Lutz, M.; Heyder, M.; Latus, S.; Leistner, D.M.; Abdelwahed, Y.S.; Schlaefer, A. Automatic plaque detection in IVOCT pullbacks using convolutional neural networks. IEEE Trans. Med. Imaging 2018, 38, 426–434. [Google Scholar] [CrossRef]
- Narula, S.; Shameer, K.; Omar, A.S.; Dudley, J.T.; Sengupta, P.P. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J. Am. Coll. Cardiol. 2016, 68, 2287–2295. [Google Scholar] [CrossRef]
- Sengupta, P.P.; Huang, Y.M.; Bansal, M.; Ashrafi, A.; Dudley, J.T. Cognitive Machine-Learning Algorithm for Cardiac Imaging: A Pilot Study for Differentiating Constrictive Pericarditis From Restrictive Cardiomyopathy. Circ. Cardiovasc. Imaging 2016, 9, e004330. [Google Scholar] [CrossRef]
- Gjoreski, M.; Gradišek, A.; Budna, B.; Gams, M.; Poglajen, G. Machine learning and end-to-end deep learning for the detection of chronic heart failure from heart sounds. IEEE Access 2020, 8, 20313–20324. [Google Scholar] [CrossRef]
- Zack, C.J.; Senecal, C.; Kinar, Y.; Metzger, Y.; Bar-Sinai, Y.; Widmer, R.J.; Lennon, R.; Singh, M.; Bell, M.R.; Lerman, A. Leveraging machine learning techniques to forecast patient prognosis after percutaneous coronary intervention. Cardiovasc. Interv. 2019, 12, 1304–1311. [Google Scholar] [CrossRef]
- JérMe, A.; Nicolas, A.; Pascal, A.; Ivan, P.; Olivier, M.; Myriem, B.; Sophie, P.; Philippe, M.; Cyril, F.; Alessandro, P. A Comparison of a Machine Learning Model with EuroSCORE II in Predicting Mortality after Elective Cardiac Surgery: A Decision Curve Analysis. PLoS ONE 2017, 12, e0169772. [Google Scholar]
- Wolterink, J.M.; Leiner, T.; de Vos, B.D.; van Hamersvelt, R.W.; Viergever, M.A.; Išgum, I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med. Image Anal. 2016, 34, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Kelm, M.B.; Mittal, S.; Zheng, Y.; Tsymbal, A.; Comaniciu, D. Detection, Grading and Classification of Coronary Stenoses in Computed Tomography Angiography; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Hu, H.; Liu, Z.; Chen, G.; Yuan, D.; Zheng, T. Analysis of aortic wall stress and morphology in patients with type B aortic dissection. Med. Nov. Technol. Dev. 2021, 11, 100081. [Google Scholar] [CrossRef]
- Gong, X.; Liang, Z.; Wang, Y.; Zhang, C.; Xie, S.; Fan, Y. Comparative study on hemodynamic environments around patient-specific carotid atherosclerotic plaques with different symmetrical features. Med. Nov. Technol. Dev. 2021, 11, 100079. [Google Scholar] [CrossRef]
- Arzani, A. Coronary artery plaque growth: A two-way coupled shear stress-driven model. Int. J. Numer. Methods Biomed. Eng. 2020, 36, e3293. [Google Scholar] [CrossRef]
- Hoogendoorn, A.; Kok, A.M.; Hartman, E.M.J.; Nisco, G.D.; Wentzel, J.J. Multidirectional wall shear stress promotes advanced coronary plaque development—Comparing five shear stress metrics. Cardiovasc. Res. 2019, 116, 1136–1146. [Google Scholar] [CrossRef]
- Domanin, M.; Gallo, D.; Vergara, C.; Biondetti, P.; Forzenigo, L.V.; Morbiducci, U. Prediction of long term restenosis risk after surgery in the carotid bifurcation by hemodynamic and geometric analysis. Ann. Biomed. Eng. 2019, 47, 1129–1140. [Google Scholar] [CrossRef]
- Gallo, D.; Steinman, D.A.; Morbiducci, U. An insight into the mechanistic role of the common carotid artery on the hemodynamics at the carotid bifurcation. Ann. Biomed. Eng. 2015, 43, 68–81. [Google Scholar] [CrossRef]
- Pouch, A.M.; Wang, H.; Takabe, M.; Jackson, B.M.; Gorman, J.H.; Gorman, R.C.; Yushkevich, P.A.; Sehgal, C.M. Fully automatic segmentation of the mitral leaflets in 3D transesophageal echocardiographic images using multi-atlas joint label fusion and deformable medial modeling. Med. Image Anal. 2014, 18, 118–129. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Zhang, M.; Tupin, S.; Qiao, A.; Liu, Y.; Ohta, M.; Anzai, H. Prediction of 3D Cardiovascular hemodynamics before and after coronary artery bypass surgery via deep learning. Commun. Biol. 2021, 4, 99. [Google Scholar] [CrossRef]
- Feiger, B.; Gounley, J.; Adler, D.; Leopold, J.A.; Randles, A. Accelerating massively parallel hemodynamic models of coarctation of the aorta using neural networks. Sci. Rep. 2020, 10, 9508. [Google Scholar]
- Zarins, C.K.; Giddens, D.P.; Bharadvaj, B.K.; Sottiurai, V.S.; Mabon, R.F.; Glagov, S. Carotid bifurcation atherosclerosis. Quantitative correlation of plaque localization with flow velocity profiles and wall shear stress. Circ. Res. 1983, 53, 502–514. [Google Scholar] [CrossRef]
- Stone, P.H.; Coskun, A.U.; Kinlay, S.; Clark, M.E.; Sonka, M.; Wahle, A.; Ilegbusi, O.J.; Yeghiazarians, Y.; Popma, J.J.; Orav, J.E.; et al. Effect of Endothelial Shear Stress on the Progression of Coronary Artery Disease, Vascular Remodeling, and In-Stent Restenosis in Humans: In Vivo 6-Month Follow-Up Study. Circ. J. Am. Heart Assoc. 2003, 108, 438–444. [Google Scholar] [CrossRef]
- Toba, T.; Otake, H.; Choi, G.; Kim, H.J.; Onishi, H.; Sugizaki, Y.; Takeshige, R.; Nagasawa, A.; Nagano, Y.; Tsukiyama, Y.; et al. Wall Shear Stress and Plaque Vulnerability: Computational Fluid Dynamics Analysis Derived from cCTA and OCT. JACC Cardiovasc. Imaging 2020, 14, 315–317. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Farghadan, A.; McConnell, D.; Barker, A.J.; Wentzel, J.J.; Budoff, M.J.; Arzani, A. The Story of Wall Shear Stress in Coronary Artery Atherosclerosis: Biochemical Transport and Mechanotransduction. J. Biomech. Eng. 2020, 143, 4049026. [Google Scholar] [CrossRef]
- Lee, S.W.; Antiga, L.; Steinman, D.A. Correlations Among Indicators of Disturbed Flow at the Normal Carotid Bifurcation. J. Biomech. Eng. 2009, 131, 061013. [Google Scholar] [CrossRef]
- Liu, X.; Pu, F.; Fan, Y.; Deng, X.; Li, D.; Li, S. A numerical study on the flow of blood and the transport of LDL in the human aorta: The physiological significance of the helical flow in the aortic arch. Am. J. Physiol. 2009, 297, 163–170. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Liang, Y.; Deng, X.; Fan, Y. Spatiotemporal changes of local hemodynamics and plaque components during atherosclerotic progression in rabbit. Comput. Methods Programs Biomed. 2022, 220, 106814. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.P.; Ferrara, D.E.; Sorescu, D.C.; Guldberg, R.E.; Taylor, W.R.R.; Giddens, D.P. Hemodynamic Shear Stresses in Mouse Aortas: Implications for Atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Himburg, H.A.; Grzybowski, D.M.; Hazel, A.L.; LaMack, J.A.; Li, X.-M.; Friedman, M.H. Spatial comparison between wall shear stress measures and porcine arterial endothelial permeability. Am. J. Physiol. -Heart Circ. Physiol. 2004, 286, H1916–H1922. [Google Scholar] [CrossRef] [PubMed]
- Gibson, C.M.; Díaz, L.; Kandarpa, K.; Sacks, F.M.; Pasternak, R.C.; Sandor, T.; Feldman, C.; Stone, P.H. Relation of vessel wall shear stress to atherosclerosis progression in human coronary arteries. Arterioscler. Thromb. J. Vasc. Biol. 1993, 13, 310–315. [Google Scholar] [CrossRef]
- Martin, C.; Sun, W.; Elefteriades, J. Patient-specific finite element analysis of ascending aorta aneurysms. Am. J. Physiol. Heart Circ. Physiol. 2015, 308, H1306–H1316. [Google Scholar] [CrossRef]
- Lopes, D.; Agujetas, R.; Puga, H.; Teixeira, J.; Lima, R.; Alejo, J.; Ferrera, C. Analysis of finite element and finite volume methods for fluid-structure interaction simulation of blood flow in a real stenosed artery. Int. J. Mech. Sci. 2021, 207, 106650. [Google Scholar] [CrossRef]
- Taylor, C.A.; Hughes, T.J.R.; Zarins, C.K. Finite element modeling of blood flow in arteries. Comput. Methods Appl. Mech. Eng. 1998, 158, 155–196. [Google Scholar] [CrossRef]
- Samady, H.; Eshtehardi, P.; McDaniel, M.C.; Suo, J.P.; Dhawan, S.S.; Maynard, C.; Timmins, L.H.; Quyyumi, A.A.; Giddens, D.P. Coronary Artery Wall Shear Stress Is Associated With Progression and Transformation of Atherosclerotic Plaque and Arterial Remodeling in Patients With Coronary Artery Disease. Circulation 2011, 124, 779–788. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Zhang, P.; Feng, C.; Sun, A.; Kang, H.; Deng, X.; Fan, Y. Numerical simulation of haemodynamics and low-density lipoprotein transport in the rabbit aorta and their correlation with atherosclerotic plaque thickness. J. R. Soc. Interface 2017, 14, 20170140. [Google Scholar] [CrossRef]
- Stone, P.H.; Saito, S.; Takahashi, S.; Makita, Y.; Nakamura, S.; Kawasaki, T.; Takahashi, A.; Katsuki, T.-A.; Nakamura, S.; Namiki, A.; et al. Prediction of Progression of Coronary Artery Disease and Clinical Outcomes Using Vascular Profiling of Endothelial Shear Stress and Arterial Plaque Characteristics: The PREDICTION Study. Circulation 2012, 126, 172–181. [Google Scholar] [CrossRef]
- Peiffer, V.; Sherwin, S.J.; Weinberg, P.D. Computation in the rabbit aorta of a new metric—The transverse wall shear stress—To quantify the multidirectional character of disturbed blood flow. J. Biomech. 2013, 46, 2651–2658. [Google Scholar] [CrossRef]
- Amirhossein, A.; Shadden, C.S. Wall shear stress fixed points in cardiovascular fluid mechanics. J. Biomech. 2018, 73, 145–152. [Google Scholar]
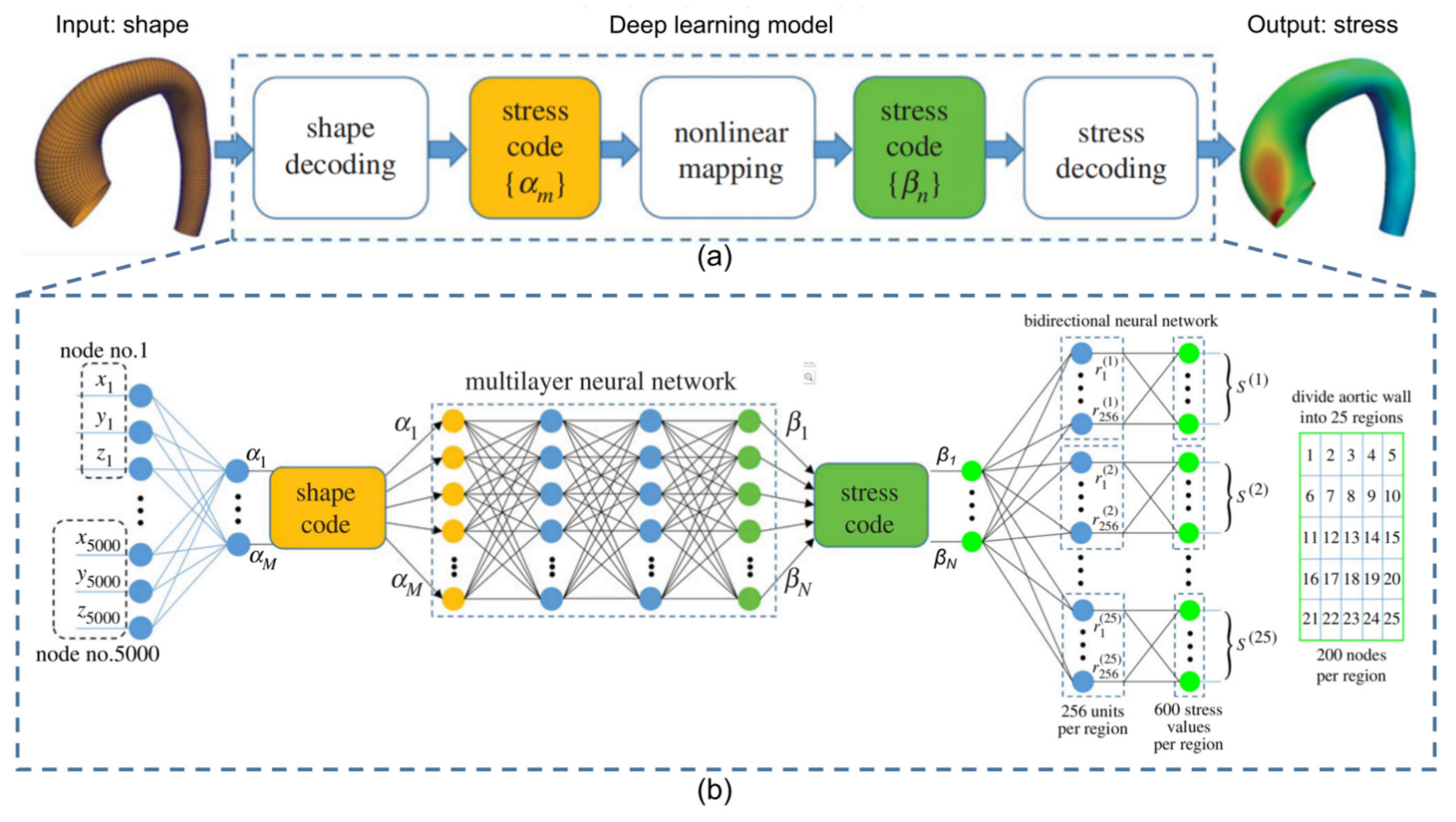
- Liang, L.; Minliang, L.; Caitlin, M.; Wei, S. A deep learning approach to estimate stress distribution: A fast and accurate surrogate of finite-element analysis. J. R. Soc. Interface 2018, 15, 20170844. [Google Scholar] [CrossRef]
- Mao, W.; Caballero, A.; Mckay, R.; Primiano, C.; Sun, W. Fully-coupled fluid-structure interaction simulation of the aortic and mitral valves in a realistic 3D left ventricle model. PLoS ONE 2017, 12, e0184729. [Google Scholar] [CrossRef]
- Wittek, A.; Derwich, W.; Karatolios, K.; Fritzen, C.P.; Blase, C. A finite element updating approach for identification of the anisotropic hyperelastic properties of normal and diseased aortic walls from 4D ultrasound strain imaging. J. Mech. Behav. Biomed. Mater. 2016, 58, 122–138. [Google Scholar] [CrossRef]
- Schmid, P.J.; Sesterhenn, J. Dynamic Mode Decomposition of Numerical and Experimental Data. J. Fluid Mech. 2010, 656, 5–28. [Google Scholar] [CrossRef]
- Dowell, E.H.; Hall, K.C.; Romanowski, M.C. Eigenmode Analysis in Unsteady Aerodynamics: Reduced Order Models. Appl. Mech. Rev. 1997, 50, 371. [Google Scholar] [CrossRef]
- Weinberg, E.J.; Mofrad, M. Transient, Three-dimensional, Multiscale Simulations of the Human Aortic Valve. Cardiovasc. Eng. 2007, 7, 140–155. [Google Scholar] [CrossRef]
- Liang, L.; Liu, M.; Martin, C.; Elefteriades, J.A.; Sun, W. A machine learning approach to investigate the relationship between shape features and numerically predicted risk of ascending aortic aneurysm. Biomech. Modeling Mechanobiol. 2017, 16, 1519–1533. [Google Scholar] [CrossRef]
- Raissi, M. Deep Hidden Physics Models: Deep Learning of Nonlinear Partial Differential Equations. J. Mach. Learn. Res. 2018. [Google Scholar]
- Chi, Z.; Beile, L.; Deyu, L.; Yubo, F. Application of multiscale coupling models in the numerical study of circulation system. Med. Nov. Technol. Devices 2022, 14, 100117. [Google Scholar] [CrossRef]
- Sun, L.; Gao, H.; Pan, S.; Wang, J.X. Surrogate Modeling for Fluid Flows Based on Physics-Constrained Deep Learning Without Simulation Data. Comput. Methods Appl. Mech. Eng. 2019, 361, 112732. [Google Scholar] [CrossRef]
- Raissi, M.; Wang, Z.; Triantafyllou, M.S.; Karniadakis, G.E. Deep learning of vortex-induced vibrations. J. Fluid Mech. 2018, 861, 119–137. [Google Scholar] [CrossRef]
- Huang, J.; Liu, H.; Cai, W. Online in situ prediction of 3-D flame evolution from its history 2-D projections via deep learning. J. Fluid Mech. 2019, 875, R2. [Google Scholar] [CrossRef]
- Peng, G.; Alber, M.; Tepole, A.B.; Cannon, W.R.; Kuhl, E. Multiscale Modeling Meets Machine Learning: What Can We Learn? Arch. Comput. Methods Eng. 2020, 28, 1017–1037. [Google Scholar] [CrossRef]
- Miyanawala, T.P.; Jaiman, R.K. An Efficient Deep Learning Technique for the Navier-Stokes Equations: Application to Unsteady Wake Flow Dynamics. arXiv 2017, arXiv:1710.09099. [Google Scholar]
- Guo, X.; Wei, L.; Iorio, F. Convolutional Neural Networks for Steady Flow Approximation. In Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, San Francisco, CA, USA, 13–17 August, 2016; pp. 481–490. [Google Scholar]
- Ribeiro, M.D.; Rehman, A.; Ahmed, S.; Dengel, A. DeepCFD: Efficient Steady-State Laminar Flow Approximation with Deep Convolutional Neural Networks. arXiv 2020, arXiv:2004.08826. [Google Scholar]
- Tompson, J.; Schlachter, K.; Sprechmann, P.; Perlin, K. Accelerating Eulerian Fluid Simulation With Convolutional Networks. In Proceedings of the International Conference on Machine Learning. PMLR, Sydney, Australia, 6–11 August, 2017; pp. 3424–3433. [Google Scholar]
- Umberto, M.; Kok, A.M.; Kwak, B.R.; Stone, P.H.; Steinman, D.A.; Wentzel, J.J. Atherosclerosis at arterial bifurcations: Evidence for the role of haemodynamics and geometry. Thromb. Haemost. 2016, 115, 484–492. [Google Scholar]
- Arzani, A.; Shadden, S.C. Characterization of the transport topology in patient-specific abdominal aortic aneurysm models. Phys. Fluids 2012, 24, 81901. [Google Scholar] [CrossRef]
- Shavelle, D.M. Mathematical Modeling of Blood Flow to Evaluate the Hemodynamic Significance of Peripheral Vascular Lesions. Angiol. Vasc. Surg. 2021, 6, 76. [Google Scholar] [CrossRef]
- Vardhan, M.; Gounley, J.; Chen, S.J.; Chi, E.C.; Randles, A. Non-invasive characterization of complex coronary lesions. Sci. Rep. 2021, 11, 1–15. [Google Scholar]
- Berhane, H.; Scott, M.B.; Elbaz, M.S.M.; Jarvis, K.; McCarthy, P.M.; Carr, J.C.; Malaisrie, C.; Avery, R.J.; Barker, A.J.; Robinson, J.D.; et al. Fully automated 3D aortic segmentation of 4D flow MRI for hemodynamic analysis using deep learning. Magn. Reson. Med. 2020, 84, 2204–2218. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, S.; Huang, J.; Metaxas, D.; Axel, L. Sparse Deformable Models with Application to Cardiac Motion Analysis; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Zhang, S.; Zhan, Y.; Metaxas, D.N. Deformable segmentation via sparse representation and dictionary learning. Med. Image Anal. 2012, 16, 1385–1396. [Google Scholar] [CrossRef]
- Litjens, G.J.S.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Laak, J.v.d.; Ginneken, B.v.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Liang, L.; Mao, W.; Sun, W. A feasibility study of deep learning for predicting hemodynamics of human thoracic aorta. J. Biomech. 2019, 99, 109544. [Google Scholar] [CrossRef]
- Luo, Y.; Fan, Z.; Baek, S.; Lu, J. Machine learning–aided exploration of relationship between strength and elastic properties in ascending thoracic aneurysm. Int. J. Numer. Methods Biomed. Eng. 2018, 34, e2977.2971–e2977.2912. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, D.; Georgescu, B.; Nguyen, H.V.; Comaniciu, D. 3D Deep Learning for Efficient and Robust Landmark Detection in Volumetric Data. In Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; Volume 9349, pp. 565–572. [Google Scholar]
- Moeskops, P.; Wolterink, J.M.; Velden, B.H.M.v.d.; Gilhuijs, K.G.A.; Leiner, T.; Viergever, M.A.; Išgum, I. Deep Learning for Multi-Task Medical Image Segmentation in Multiple Modalities. arXiv 2016, arXiv:1704.03379. [Google Scholar]
- Groenendyk, J.W.; Mehta, N.N. Applying the ordinal model of atherosclerosis to imaging science: A brief review. Open Heart 2018, 5, e000861. [Google Scholar] [CrossRef]
- Hitchner, E.; Zayed, M.A.; Lee, G.K.; Morrison, D.E.; Lane, B.; Zhou, W. Intravascular ultrasound as a clinical adjunct for carotid plaque characterization. J. Vasc. Surg. 2014, 59 3, 774–780. [Google Scholar] [CrossRef][Green Version]
- Zacharatos, H.; Hassan, A.E.; Qureshi, A.I. Intravascular Ultrasound: Principles and Cerebrovascular Applications. Am. J. Neuroradiol. 2010, 31, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Randoux, B.; Marro, B.; Koskas, F.; Duyme, M.; Sahel, M.; Zouaoui, A.; Marsault, C. Carotid artery stenosis: Prospective comparison of CT, three-dimensional gadolinium-enhanced MR, and conventional angiography. Radiology 2001, 220 1, 179–185. [Google Scholar] [CrossRef]
- Loizou, P.C.; Pattichis, C.S.; Pantziaris, M.; Nicolaides, A.N. An integrated system for the segmentation of atherosclerotic carotid plaque ultrasound video. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2014, 61, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Iyer, K.; Najarian, C.; Fattah, A.A.; Arthurs, C.J.; Soroushmehr, S.M.R.; Subban, V.; Sankardas, M.A.; Nadakuditi, R.R.; Nallamothu, B.K.; Figueroa, C.A. AngioNet: A convolutional neural network for vessel segmentation in X-ray angiography. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Prabhu, D.; Kolluru, C.; Gharaibeh, Y.; Wilson, D.L. Automated plaque characterization using deep learning on coronary intravascular optical coherence tomographic images. Biomed. Opt. Express 2019, 10, 6497. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lan, Y.; Chen, S.; Zhong, Y.; Che, W. Lumen contour segmentation in IVOCT based on N-type CNN. IEEE Access 2019, 7, 135573–135581. [Google Scholar] [CrossRef]
- Athanasiou, L.S.; Bourantas, C.V.; Rigas, G.; Sakellarios, A.I.; Exarchos, T.P.; Siogkas, P.K.; Ricciardi, A.; Naka, K.K.; Papafaklis, M.I.; Michalis, L.K.; et al. Methodology for fully automated segmentation and plaque characterization in intracoronary optical coherence tomography images. J. Biomed. Opt. 2014, 19, 026009. [Google Scholar] [CrossRef]
- Abdolmanafi, A.; Duong, L.; Dahdah, N.; Cheriet, F. Deep feature learning for automatic tissue classification of coronary artery using optical coherence tomography. Biomed. Opt. Express 2017, 8 (Pt 2), 1203–1220. [Google Scholar] [CrossRef]
- Karimi-Bidhendi, S.; Arafati, A.; Cheng, A.L.; Wu, Y.; Kheradvar, A.; Jafarkhani, H. Fully-automated deep-learning segmentation of pediatric cardiovascular magnetic resonance of patients with complex congenital heart diseases. J. Cardiovasc. Magn. Reson. 2020, 22, 1–24. [Google Scholar] [CrossRef]
- Kong, F.; Wilson, N.; Shadden, S. A deep-learning approach for direct whole-heart mesh reconstruction. Med. Image Anal. 2021, 74, 102222. [Google Scholar] [CrossRef]
- Weng, W.; Zhu, X. INet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access 2021, 9, 16591–16603. [Google Scholar] [CrossRef]
- Liang, L.; Kong, F.; Martin, C.K.; Pham, T.; Wang, Q.; Duncan, J.S.; Sun, W. Machine learning-based 3-D geometry reconstruction and modeling of aortic valve deformation using 3-D computed tomography images. Int. J. Numer. Methods Biomed. Eng. 2017, 33, e2827. [Google Scholar] [CrossRef]
- Zheng, Y.; John, M.; Liao, R.; Nöttling, A.; Boese, J.M.; Kempfert, J.; Walther, T.; Brockmann, G.; Comaniciu, D. Automatic Aorta Segmentation and Valve Landmark Detection in C-Arm CT for Transcatheter Aortic Valve Implantation. IEEE Trans. Med. Imaging 2012, 31, 2307–2321. [Google Scholar] [CrossRef]
- Oktay, O.; Ferrante, E.; Kamnitsas, K.; Heinrich, M.P.; Bai, W.; Caballero, J.; Cook, S.A.; de Marvao, A.; Dawes, T.J.W.; O’Regan, D.P.; et al. Anatomically Constrained Neural Networks (ACNNs): Application to Cardiac Image Enhancement and Segmentation. IEEE Trans. Med. Imaging 2018, 37, 384–395. [Google Scholar] [CrossRef]
- Kong, F.; Shadden, S.C. Automating Model Generation for Imagebased Cardiac Flow Simulation. J. Biomech. Eng. 2020, 142, 111011. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.P.K.; Lamata, P.; Montana, G. Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation. arXiv 2016, arXiv:1608.03974. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; González Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Jordanski, M.; Radovic, M.D.; Milosevic, Z.; Filipovic, N.D.; Obradovic, Z. Machine Learning Approach for Predicting Wall Shear Distribution for Abdominal Aortic Aneurysm and Carotid Bifurcation Models. IEEE J. Biomed. Health Inform. 2018, 22, 537–544. [Google Scholar] [CrossRef]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden fluid mechanics: Learning velocity and pressure fields from flow visualizations. Science 2020, 367, 1026–1030. [Google Scholar] [CrossRef]
- Madani, A.; Bakhaty, A.; Kim, J.; Mubarak, Y.; Mofrad, M. Bridging Finite Element and Machine Learning Modeling: Stress Prediction of Arterial Walls in Atherosclerosis. J. Biomech. Eng. 2019, 141, 084502. [Google Scholar] [CrossRef]
- Fukami, K.; Fukagata, K.; Taira, K. Super-resolution reconstruction of turbulent flows with machine learning. J. Fluid Mech. 2019, 870, 106–120. [Google Scholar] [CrossRef]
- Arzani, A.; Wang, J.-X.; D’Souza, R.M. Uncovering near-wall blood flow from sparse data with physics-informed neural networks. Phys. Fluids 2021, 33, 071905. [Google Scholar] [CrossRef]
- Chorin, A.J. Numerical solution of the Navier-Stokes equations. Math. Comput. 1968, 22, 745–762. [Google Scholar] [CrossRef]
- Rudy, S.H.; Brunton, S.L.; Proctor, J.L.; Kutz, J.N. Data-driven discovery of partial differential equations. Sci. Adv. 2017, 3, e1602614. [Google Scholar] [CrossRef] [PubMed]
- Hayden, S. Learning partial differential equations via data discovery and sparse optimization. Proc. R. Society. Math. Phys. Eng. Sci. 2017, 473, 20160446. [Google Scholar]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Machine learning of linear differential equations using Gaussian processes. J. Comput. Phys. 2017, 348, 683–693. [Google Scholar] [CrossRef]
- Raissi, M.; Karniadakis, G.E. Hidden Physics Models: Machine Learning of Nonlinear Partial\n Differential Equations. J. Comput. Phys. 2018, 357, 125–141. [Google Scholar] [CrossRef]
- Parussini, L.; Venturi, D.; Perdikaris, P.; Karniadakis, G.E. Multi-fidelity Gaussian process regression for prediction of random fields. J. Comput. Phys. 2017, 336, 36–50. [Google Scholar] [CrossRef]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Physics Informed Deep Learning (Part I): Data-driven Solutions of Nonlinear Partial Differential Equations. arXiv 2017, arXiv:1711.10561. [Google Scholar]
- Karniadakis, G.E.; Kevrekidis, I.G.; Lu, L.; Perdikaris, P.; Wang, S.; Yang, L. Physics-informed machine learning. Nat. Rev. Phys. 2021, 3, 422–440. [Google Scholar] [CrossRef]
- Lee, S.; You, D. Data-driven prediction of unsteady flow over a circular cylinder using deep learning. J. Fluid Mech. 2019, 879, 217–254. [Google Scholar] [CrossRef]
- Gaidzik, F.; Stucht, D.; Roloff, C.; Speck, O.; Janiga, G. Transient flow prediction in an idealized aneurysm geometry using data assimilation. Comput. Biol. Med. 2019, 115, 103507. [Google Scholar] [CrossRef]
- Funke, S.W.; Nordaas, M.; Evju, Y.; Alnaes, M.S.; Mardal, K.A. Variational data assimilation for transient blood flow simulations—Cerebral aneurysms as an illustrative example. Int. J. Numer. Methods Biomed. Eng. 2018, 35, e3152. [Google Scholar] [CrossRef]
- Habibi, M.; D’Souza, R.M.; Dawson, S.; Arzani, A. Integrating multi-fidelity blood flow data with reduced-order data assimilation. Comput. Biol. Med. 2021, 135, 104566. [Google Scholar] [CrossRef]
- Adriaan, C.; Young-Hak, K.; Mariusz, K.; Christian, T.; Jakob, D.G.; Akira, K.; Lubbers, M.L.; Joost, D.; Lucian, I.; Saikiran, R. Diagnostic Accuracy of a Machine-Learning Approach to Coronary Computed Tomographic Angiography–Based Fractional Flow Reserve. Circ. Cardiovasc. Imaging 2018, 11, e007217. [Google Scholar]
- Itu, L.; Rapaka, S.; Passerini, T.; Georgescu, B.; Schwemmer, C.; Schoebinger, M.; Flohr, T.; Sharma, P.; Comaniciu, D. A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. J. Appl. Physiol. 2016, 121, 42–52. [Google Scholar] [CrossRef]
- Hu, X.; Yang, M.; Han, L.; Du, Y. Diagnostic performance of machine-learning-based computed fractional flow reserve (FFR) derived from coronary computed tomography angiography for the assessment of myocardial ischemia verified by invasive FFR. Int. J. Cardiovasc. Imaging 2018, 34, 1987–1996. [Google Scholar] [CrossRef]
- Liang, L.; Liu, M.; Sun, W. A Deep Learning Approach to Estimate Chemically-Treated Collagenous Tissue Nonlinear Anisotropic Stress-Strain Responses from Microscopy Images. Acta Biomater. 2017, 63, 227–235. [Google Scholar] [CrossRef]
- Hennigh, O. Lat-Net: Compressing Lattice Boltzmann Flow Simulations using Deep Neural Networks. arXiv 2017, arXiv:1705.09036. [Google Scholar]
- Zhou, Y.; He, Y.; Wu, J.; Cui, C.; Chen, M.; Sun, B. A method of parameter estimation for cardiovascular hemodynamics based on deep learning and its application to personalize a reduced-order model. Int. J. Numer. Methods Biomed. Eng. 2021, 38, e3533. [Google Scholar] [CrossRef]
- Sonoda, S.; Murata, N. Neural Network with Unbounded Activation Functions is Universal Approximator. Appl. Comput. Harmon. Anal. 2017, 43, 233–268. [Google Scholar] [CrossRef]
- Cai, S.; Mao, Z.; Wang, Z.; Yin, M.; Karniadakis, G.E. Physics-informed neural networks (PINNs) for fluid mechanics: A review. arXiv 2022, arXiv:2105.09506. [Google Scholar] [CrossRef]
- Jin, X.; Cai, S.; Li, H.; Karniadakis, G.E. NSFnets (Navier-Stokes flow nets): Physics-informed neural networks for the incompressible Navier-Stokes equations. J. Comput. Phys. 2021, 426, 109951. [Google Scholar] [CrossRef]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden Fluid Mechanics: A Navier-Stokes Informed Deep Learning Framework for Assimilating Flow Visualization Data. arXiv 2018, arXiv:1808.04327. [Google Scholar]
- Jagtap, A.D.; Kharazmi, E.; Karniadakis, G.E. Conservative physics-informed neural networks on discrete domains for conservation laws: Applications to forward and inverse problems. Comput. Methods Appl. Mech. Eng. 2020, 365, 113028. [Google Scholar] [CrossRef]
- Buoso, S.; Joyce, T.; Kozerke, S. Personalising left-ventricular biophysical models of the heart using parametric physics-informed neural networks. Med. Image Anal. 2021, 71, 102066. [Google Scholar] [CrossRef]
- Kissas, G.; Yang, Y.; Hwuang, E.; Witschey, W.R.; Detre, J.A.; Perdikaris, P. Machine learning in cardiovascular flows modeling: Predicting arterial blood pressure from non-invasive 4D flow MRI data using physics-informed neural networks. Comput. Methods Appl. Mech. Eng. 2019, 358, 112623. [Google Scholar] [CrossRef]
- Clauset, A.; Larremore, D.B.; Sinatra, R. Data-driven predictions in the science of science. Science 2017, 355, 477. [Google Scholar] [CrossRef]
- Karumuri, S.; Tripathy, R.; Bilionis, I.; Panchal, J. Simulator-free Solution of High-Dimensional Stochastic Elliptic Partial Differential Equations using Deep Neural Networks. J. Comput. Phys. 2019, 404, 109120. [Google Scholar] [CrossRef]
- Zhu, Y.; Zabaras, N.; Koutsourelakis, P.S.; Perdikaris, P. Physics-Constrained Deep Learning for High-dimensional Surrogate Modeling and Uncertainty Quantification without Labeled Data. J. Comput. Phys. 2019, 394, 56–81. [Google Scholar] [CrossRef]
- Geneva, N.; Zabaras, N. Modeling the Dynamics of PDE Systems with Physics-Constrained Deep Auto-Regressive Networks. J. Comput. Phys. 2019, 403, 109056. [Google Scholar] [CrossRef]
- Gao, H.; Sun, L.; Wang, J.X. Super-resolution and denoising of fluid flow using physics-informed convolutional neural networks without high-resolution labels. Phys. Fluids 2021, 33, 073603. [Google Scholar] [CrossRef]
- Wessels, H.; Weißenfels, C.; Wriggers, P. The Neural Particle Method—An Updated Lagrangian Physics Informed Neural Network for Computational Fluid Dynamics. arXiv 2020, arXiv:2003.10208. [Google Scholar] [CrossRef]
- Lucor, D.; Agrawal, A.; Sergent, A. Physics-Aware Deep Neural Networks for Surrogate Modeling of Turbulent Natural Convection. arXiv 2021, arXiv:2103.03565. [Google Scholar]
- Gasmi, C.F.; Tchelepi, H. Physics Informed Deep Learning for Flow and Transport in Porous Media. arXiv 2021, arXiv:2104.02629. [Google Scholar]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef]
- Siau, K.L.; Wang, W. Artificial Intelligence (AI) Ethics: Ethics of AI and Ethical AI. J. Database Manag. 2020, 31, 74–87. [Google Scholar] [CrossRef]
- Seetharam, K.; Shrestha, S.; Sengupta, P.P. Artificial Intelligence in Cardiovascular Medicine. Curr. Treat. Options Cardiovasc. Med. 2019, 21, 1–14. [Google Scholar] [CrossRef]
- Kagiyama, N.; Shrestha, S.; Farjo, P.D.; Sengupta, P.P. Artificial Intelligence: Practical Primer for Clinical Research in Cardiovascular Disease. J. Am. Heart Assoc. Cardiovasc. Cerebrovasc. Dis. 2019, 8, e012788. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Rodríguez, N.D.; Ser, J.D.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, Taxonomies, Opportunities and Challenges toward Responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Guidotti, R.; Monreale, A.; Turini, F.; Pedreschi, D.; Giannotti, F. A Survey of Methods for Explaining Black Box Models. ACM Comput. Surv. (CSUR) 2019, 51, 1–42. [Google Scholar] [CrossRef]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.-M.; von Tengg-Kobligk, H.; Summers, R.M.; Wiest, R. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol. Artif. Intell. 2020, 2, e190043. [Google Scholar] [CrossRef] [PubMed]




| Focus | Algorithm | Data Size | Input Variables | Performance (AUC) | Significant Discovery |
|---|---|---|---|---|---|
| CVD risk prediction [35] | ML (SVM) | 6459 | clinical data | 0.92 | ML algorithms significantly improved risk stratification while reducing adverse events. |
| CVD risk prediction [36] | ML (KNN, RF and DT) | 2020 | clinical data | Accu.: 0.83 Sens.: 0.89 Spec.: 0.46 | The RF gave the best results, while the k−NN gave the poorest results. |
| CVD risk prediction [37] | AutoPrognosis (SVM, RF, kNN, AdaBoost and GBM) | 423,604 | clinical data | 0.774 | ML model had better efficiency than traditional risk calculators. |
| CAD mortality prediction [38] | ML (LogitBoost) | 10,030 | clinical and CCTA data | 0.79 | The accuracy of the ML model was better compared to the traditional or CCTA severity scores alone. |
| CAD risk prediction [39] | ML (XGBoost) | 8844 | clinical and CCTA data | 0.771 | The risk score based on ML had greater prognostic accuracy than current CCTA integrated risk scores. |
| CAC identification [40] | CNN + RF | 50 | CCTA data | / | CAC could be automatically identified and classified in CCTA using CNN and RF algorithms. |
| Coronary artery stenosis identification [41] | CNN + CAE + SVM | 166 | FFR and CCTA data | 0.74 | The CNN could be used to automatically identify functionally significant coronary artery stenosis. |
| Obstructive disease prediction [27] | DL | 1638 | MPI data | 0.80/0.76 | The DL algorithm could automatically interpret MPI more accurately. |
| CHD Plaque detection [42] | CNN | 49 | IVOCT data | Accu.: 0.917 Sens.: 0.909 Spec.: 0.924 | It’s feasible to construct a DL−based clinical decision support system for plaque detection. |
| HCM discrimination [43] | ML (SVMs + RF) + ANN | 139 | STE data | 0.795 | The ML−based models had higher diagnostic sensitivity and specificity. |
| CP/ RCM discrimination [44] | ML (AMC, RF, SVM and kNN) | 94 | Clinical and STE data | 0.962 | The AMC gave the best results. |
| Prognosis prediction [31] | DL | 10,019 | Clinical and ECG data | Accu.: 0.906 | It was feasible to build a DL−based model to estimate the prognosis in ACHD. |
| CHF identification [45] | ML (RF) + DL | 947 | Clinical and heart sounds data | 0.893 | The heart sound−based detection methods for different CHF phases were proposed through ML and DL. |
| ACI identification [25] | ANN | 260 | clinical data | Spec.: 0.862 Sens.: 0.8 | ANN could be used for the recognition of ACI and differentiation of ACI from stroke intelligently. |
| Perioperative mortality prediction + Readmission [46] | ML (RF) | 11,709 | Perioperative clinical data | 0.9/0.88 | ML was more predictive in identifying postoperative mortality 180d after PCI and rehospitalization for CHF 30d after surgery. |
| Perioperative Mortality prediction [47] | ML (GBM, RF, Naïve Bayes, SVM) | 6520 | Perioperative clinical data | 0.795 | ML model was more accurate in predicting mortality after elective cardiac surgery than the traditional prediction model. |
| Algorithms | Name of Authors | Objectives | Training Set | Significant Discovery |
|---|---|---|---|---|
| ML | Jordanski et al. [125] | WSS | FEA results | Three ML models (MLR, MLP, GCRF) were developed for the calculation of WSS distribution, and the GCRF achieved the highest coefficient of determination (0.930–0.948) for the AAA model and (0.946–0.954) for carotid bifurcation model. |
| ML | Feiger et al. [58] | Pressure, WSS | LBM results | The 3D simulation−based ML model was developed to accurately predict pressure gradient across the stenosis and WSS for patients with coarctation of the aorta. |
| DL | Li et al. [57] | Velocity, Pressure gradient | FEA results | The hemodynamic prediction results of deep learning was in agreement with the conventional CFD method, but the calculation time was reduced 600−fold. |
| DL | Raissi et al. [126] | Velocity, Pressure | DNS results | A physics−informed deep−learning framework was capable of encoding the Navier−Stokes equations into the neural networks while being agnostic to the geometry or the initial and boundary conditions. |
| DNN | Madani et al. [127] | Stress | FEA results | The DNNs outperformed alternative prediction models and performance scales with the amount of training data. |
| DNN | liang et al. [102] | Pressure, Velocity | FEA results | The trained DNNs were capable of predicting the steady−state distributions of pressure and flow velocity inside the thoracic aorta with an average error of 1.9608% and 1.4269%. |
| CNN | Kai et al. [128] | Velocity | DNS results | The CNN model was found to reconstruct turbulent flows from extremely coarse flow field images with remarkable accuracy. |
| PINN | Arzani et al. [129] | WSS | N−S equations | PINN was used to obtain near−wall hemodynamics and WSS data from sparse velocity measurements and without knowledge of the inlet/outlet boundary conditions. |
| FC−NN | Sun et al. [86] | Velocity, Pressure | N−S equations | A physics−constrained deep neural network−based approach was developed for surrogate modeling of fluid flows without relying on any simulation data. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liu, X.; Deng, X.; Fan, Y. Interplay between Artificial Intelligence and Biomechanics Modeling in the Cardiovascular Disease Prediction. Biomedicines 2022, 10, 2157. https://doi.org/10.3390/biomedicines10092157
Li X, Liu X, Deng X, Fan Y. Interplay between Artificial Intelligence and Biomechanics Modeling in the Cardiovascular Disease Prediction. Biomedicines. 2022; 10(9):2157. https://doi.org/10.3390/biomedicines10092157
Chicago/Turabian StyleLi, Xiaoyin, Xiao Liu, Xiaoyan Deng, and Yubo Fan. 2022. "Interplay between Artificial Intelligence and Biomechanics Modeling in the Cardiovascular Disease Prediction" Biomedicines 10, no. 9: 2157. https://doi.org/10.3390/biomedicines10092157
APA StyleLi, X., Liu, X., Deng, X., & Fan, Y. (2022). Interplay between Artificial Intelligence and Biomechanics Modeling in the Cardiovascular Disease Prediction. Biomedicines, 10(9), 2157. https://doi.org/10.3390/biomedicines10092157






