An Artificial Intelligence Approach to Guiding the Management of Heart Failure Patients Using Predictive Models: A Systematic Review
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction and Quality Assessment
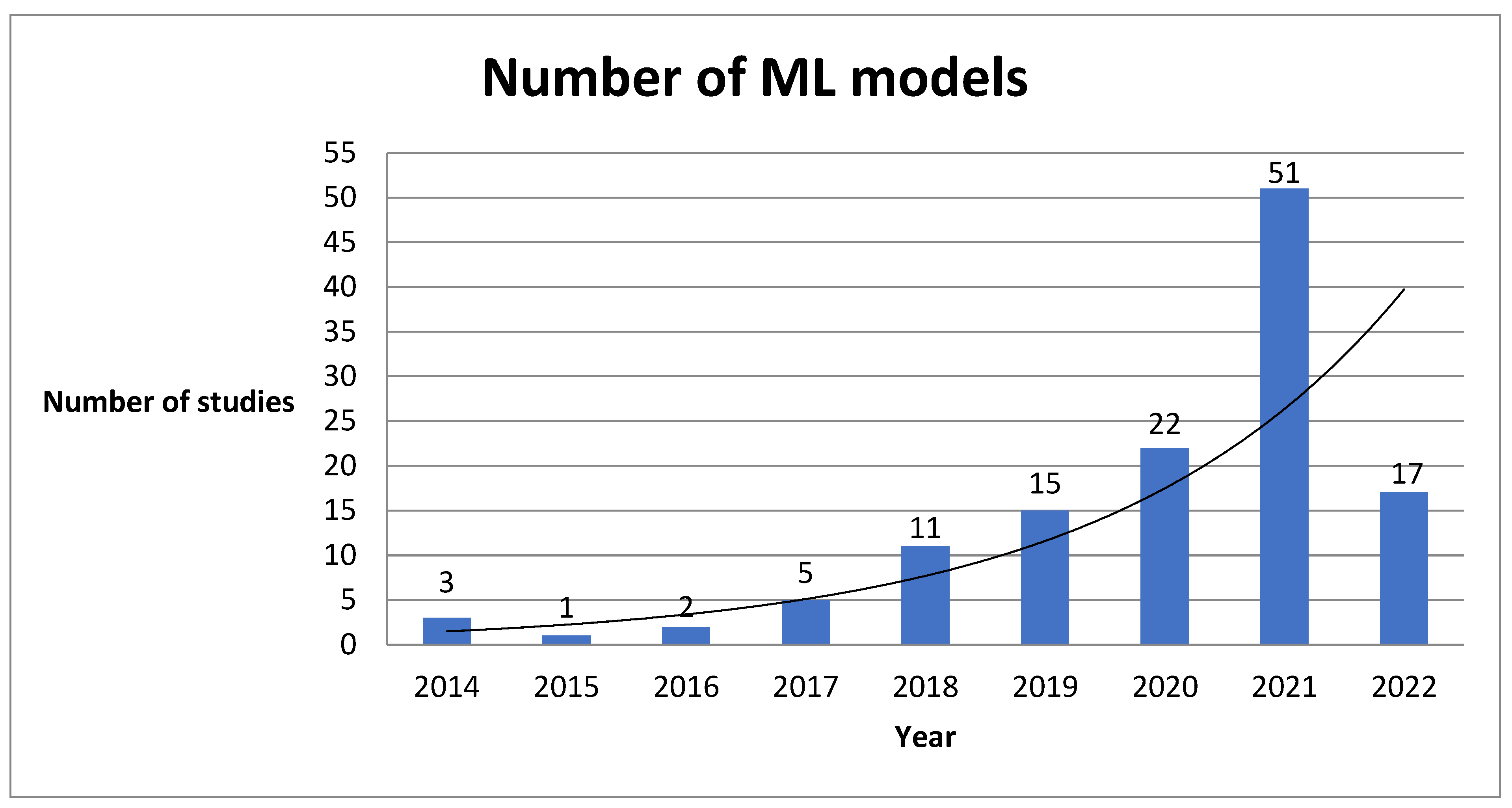
3. Results
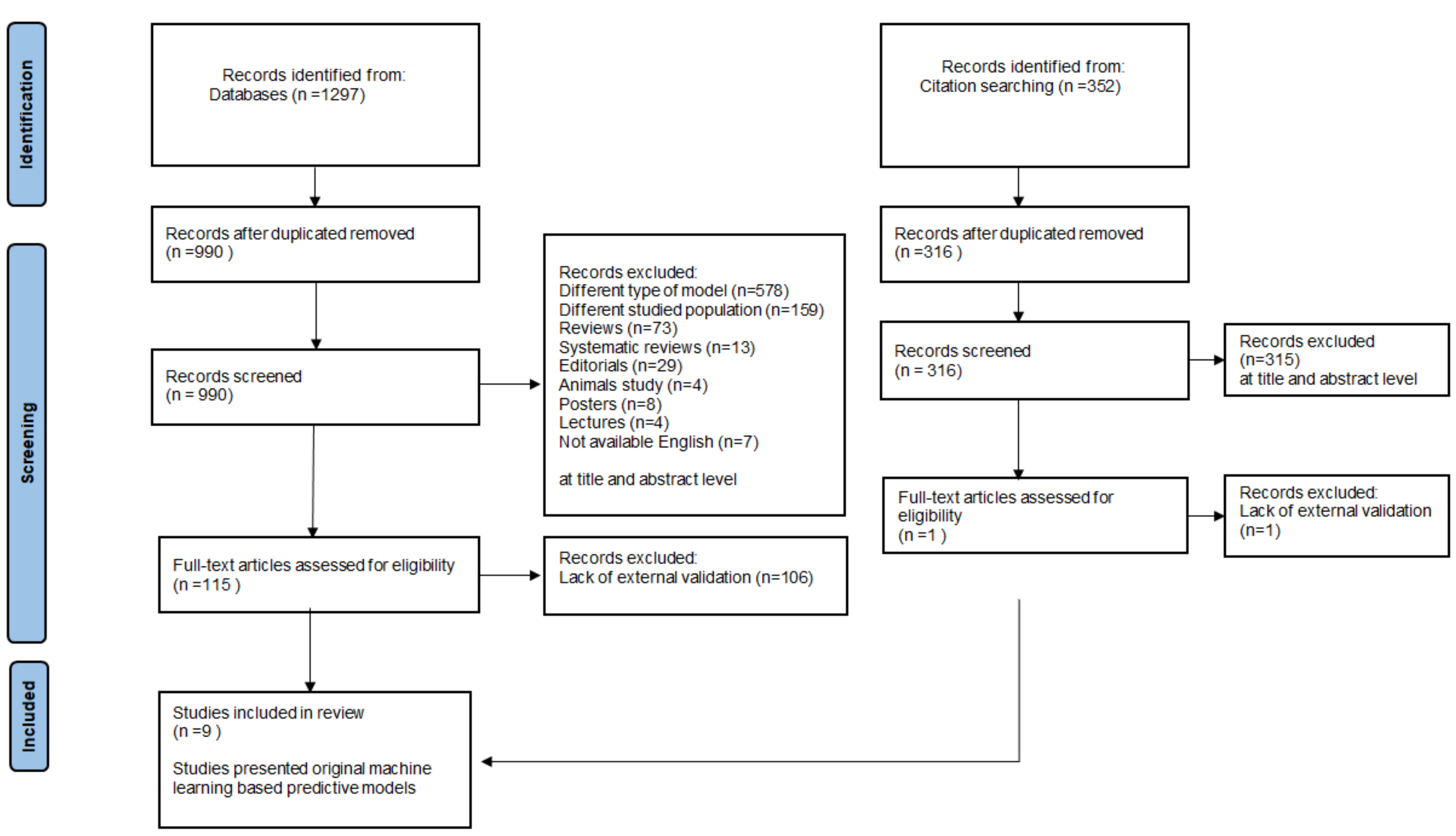
3.1. The Review Process
3.2. Comparison of the Predictive Value
3.3. Relevant Studies
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savarese, G.; Becher, P.M.; Lund, L.H.; Seferovic, P.; Rosano, G.M.C.; Coats, A.J.S. Global burden of heart failure: A comprehensive and updated review of epidemiology. Cardiovasc. Res. 2022, cvac013. [Google Scholar] [CrossRef] [PubMed]
- Ambrosy, A.P.; Fonarow, G.C.; Butler, J.; Chioncel, O.; Greene, S.J.; Vaduganathan, M.; Nodari, S.; Lam, C.S.P.; Sato, N.; Shah, A.N.; et al. The global health and economic burden of hospitalizations for heart failure: Lessons learned from hospitalized heart failure registries. J. Am. Coll. Cardiol. 2014, 63, 1123–1133. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, M.A.; Olesen, K.K.W.; Koul, S.; Gale, C.P.; Rylance, R.; Jernberg, T.; Baron, T.; Spaak, J.; James, S.; Lindahl, B.; et al. Development and validation of an artificial neural network algorithm to predict mortality and admission to hospital for heart failure after myocardial infarction: A nationwide population-based study. Lancet Digit. Health 2022, 4, e37–e45. [Google Scholar] [CrossRef]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef]
- Ahmad, T.; Lund, L.H.; Rao, P.; Ghosh, R.; Warier, P.; Vaccaro, B.; Dahlström, U.; O’Connor, C.M.; Felker, G.M.; Desai, N.R. Machine Learning Methods Improve Prognostication, Identify Clinically Distinct Phenotypes, and Detect Heterogeneity in Response to Therapy in a Large Cohort of Heart Failure Patients. J. Am. Heart Assoc. 2018, 7, e008081. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Walsh, C.G.; Ribeiro, J.D.; Franklin, J.C. Predicting suicide attempts in adolescents with longitudinal clinical data and machine learning. J. Child Psychol. Psychiatry 2018, 59, 1261–1270. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q.; et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: A 5-year multicentre prospective registry analysis. Eur. Heart J. 2016, 38, 500–507. [Google Scholar] [CrossRef] [Green Version]
- Henneges, C.; Morbach, C.; Sahiti, F.; Scholz, N.; Frantz, S.; Ertl, G.; Angermann, C.E.; Störk, S. Sex-Specific Bimodal Clustering ofLeft Ventricular Ejection Fraction in Patients with Acute Heart Failure. ESC Heart Fail. 2021, 9, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.M.; Reed, B.P.; DiSomma, S.; Nanayakkara, P.; Moyer, M.; Millis, S.; Levy, P. Presenting Phenotypes of Acute HeartFailure Patients in the ED: Identification and Implications. Am. J. Emerg. Med. 2017, 35, 536–542. [Google Scholar] [CrossRef]
- Ahmad, T.; Desai, N.; Wilson, F.; Schulte, P.; Dunning, A.; Jacoby, D.; Allen, L.; Fiuzat, M.; Rogers, J.; Felker, G.M.; et al. ClinicalImplications of Cluster Analysis-Based Classification of Acute Decompensated Heart Failure and Correlation with BedsideHemodynamic Profiles. PLoS ONE 2016, 11, e0145881. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.; Błaziak, M.; Jura, M.; Iwanek, G.; Zdanowicz, A.; Guzik, M.; Borkowski, A.; Gajewski, P.; Biegus, J.; Siennicka, A.; et al. Novel Phenotyping for Acute Heart Failure-Unsupervised Machine Learning-Based Approach. Biomedicines 2022, 10, 1514. [Google Scholar] [CrossRef]
- Asyali, M.H. Discrimination power of long-term heart rate variability measures. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat No03CH37439), Cancun, Mexico, 17–21 September 2003; Volume 1, pp. 200–203. [Google Scholar]
- Melillo, P.; Fusco, R.; Sansone, M.; Bracale, M.; Pecchia, L. Discrimination power of long-term heart rate variability measures for chronic heart failure detection. Med. Biol. Eng. Comput. 2011, 49, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, L.; Wang, Q.; Zhou, G.; Wang, Y.; Jiang, Q. A new approach to detect congestive heart failure using short-term heart rate variability measures. PLoS ONE 2014, 9, e93399. [Google Scholar] [CrossRef]
- Chen, W.; Liu, G.; Su, S.; Jiang, Q.; Nguyen, H. A CHF detection method based on deep learning with RR intervals. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 3369–3372. [Google Scholar]
- Chen, W.; Zheng, L.; Li, K.; Wang, Q.; Liu, G.; Jiang, Q. A Novel and Effective Method for Congestive Heart Failure Detection and Quantification Using Dynamic Heart Rate Variability Measurement. PLoS ONE 2016, 11, e0165304. [Google Scholar] [CrossRef]
- Cikes, M.; Sanchez-Martinez, S.; Claggett, B.; Duchateau, N.; Piella, G.; Butakoff, C.; Pouleur, A.C.; Knappe, D.; Biering-Sørensen, T.; Kutyifa, V.; et al. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur. J. Heart Fail. 2019, 21, 74–85. [Google Scholar] [CrossRef]
- Feeny, A.K.; Rickard, J.; Patel, D.; Toro, S.; Trulock, K.M.; Park, C.J.; LaBarbera, M.A.; Varma, N.; Niebauer, M.J.; Sinha, S.; et al. Machine Learning Prediction of Response to Cardiac Resynchronization Therapy. Circ. Arrhythmia Electrophysiol. 2019, 12, e007316. [Google Scholar] [CrossRef]
- Schmitz, B.; De Maria, R.; Gatsios, D.; Chrysanthakopoulou, T.; Landolina, M.; Gasparini, M.; Campolo, J.; Parolini, M.; Sanzo, A.; Galimberti, P.; et al. Identification of genetic markers for treatment success in heart failure patients: Insight from cardiac resynchronization therapy. Circ. Cardiovasc. Genet. 2014, 7, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Peressutti, D.; Sinclair, M.; Bai, W.; Jackson, T.; Ruijsink, J.; Nordsletten, D.; Asner, L.; Hadjicharalambous, M.; Rinaldi, C.A.; Rueckert, D.; et al. A framework for combining a motion atlas with non-motion information to learn clinically useful biomarkers: Application to cardiac resynchronisation therapy response prediction. Med. Image Anal. 2017, 35, 669–684. [Google Scholar] [CrossRef] [PubMed]
- Karanasiou, G.S.; Tripoliti, E.E.; Papadopoulos, T.G.; Kalatzis, F.G.; Goletsis, Y.; Naka, K.K.; Bechlioulis, A.; Errachid, A.; Fotiadis, D.I. Predicting adherence of patients with HF through machine learning techniques. Healthc. Technol. Lett. 2016, 3, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Dini, F.L.; Ballo, P.; Badano, L.; Barbier, P.; Chella, P.; Conti, U.; De Tommasi, S.M.; Galderisi, M.; Ghio, S.; Magagnini, E.; et al. Validation of an echo-Doppler decision model to predict left ventricular filling pressure in patients with heart failure independently of ejection fraction. Eur. J. Echocardiogr. 2010, 11, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Graven, L.J.; Higgins, M.K.; Reilly, C.M.; Dunbar, S.B. Heart Failure Symptoms Profile Associated with Depressive Symptoms. Clin. Nurs. Res. 2018, 29, 73–83. [Google Scholar] [CrossRef]
- Lagu, T.; Pekow, P.S.; Shieh, M.-S.; Stefan, M.; Pack, Q.R.; Kashef, M.A.; Atreya, A.R.; Valania, G.; Slawsky, M.T.; Lindenauer, P.K. Validation and Comparison of Seven Mortality Prediction Models for Hospitalized Patients With Acute Decompensated Heart Failure. Circ. Heart Fail. 2016, 9, e002912. [Google Scholar] [CrossRef]
- Pocock, S.J.; Ariti, C.A.; Mcmurray, J.; Maggioni, A.P.; Køber, L.; Squire, I.B.; Swedberg, K.; Dobson, J.; Poppe, K.K.; Whalley, G.; et al. Predicting survival in heart failure: A risk score based on 39 372 patients from 30 studies. Eur. Heart J. 2012, 34, 1404–1413. [Google Scholar] [CrossRef]
- Mortazavi, B.J.; Downing, N.S.; Bucholz, E.M.; Dharmarajan, K.; Manhapra, A.; Li, S.-X.; Negahban, S.N.; Krumholz, H.M. Analysis of Machine Learning Techniques for Heart Failure Readmissions. Circ. Cardiovasc. Qual. Outcomes 2016, 9, 629–640. [Google Scholar] [CrossRef]
- Luo, W.; Phung, Q.-D.; Tran, T.; Gupta, S.; Rana, S.; Karmakar, C.; Shilton, A.; Yearwood, J.L.; Dimitrova, N.; Ho, T.B.; et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J. Med Internet Res. 2016, 18, e323. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.F.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Kmet, L.M.; Lee, R.C.; Cook, L.S. Standard quality assessment criteria for Evaluating Primary Research Papers from a Variety of Fields. HTA Initiat. 2004, 13, 4. [Google Scholar]
- Luo, C.; Zhu, Y.; Zhu, Z.; Li, R.; Chen, G.; Wang, Z. A machine learning-based risk stratification tool for in-hospital mortality of intensive care unit patients with heart failure. J. Transl. Med. 2022, 20, 1–9. [Google Scholar] [CrossRef]
- Adler, E.D.; Voors, A.A.; Klein, L.; Macheret, F.; Braun, O.O.; Urey, M.A.; Zhu, W.; Sama, I.; Tadel, M.; Campagnari, C.; et al. Improving risk prediction in heart failure using machine learning. Eur. J. Heart Fail. 2019, 22, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.-M.; Kim, K.-H.; Jeon, K.-H.; Lee, S.E.; Lee, H.-Y.; Cho, H.-J.; Choi, J.O.; Jeon, E.-S.; Kim, M.-S.; Kim, J.-J.; et al. Artificial intelligence algorithm for predicting mortality of patients with acute heart failure. PLoS ONE 2019, 14, e0219302. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Cerna, A.E.U.; Good, C.W.; Sauers, N.M.; Schneider, G.; Hartzel, D.N.; Leader, J.B.; Kirchner, H.L.; Hu, Y.; Riviello, D.M.; et al. A Machine Learning Approach to Management of Heart Failure Populations. JACC Heart Fail. 2020, 8, 578–587. [Google Scholar] [CrossRef]
- Chirinos, J.A.; Orlenko, A.; Zhao, L.; Basso, M.D.; Cvijic, M.E.; Li, Z.; Spires, T.E.; Yarde, M.; Wang, Z.; Seiffert, D.A.; et al. Multiple Plasma Biomarkers for Risk Stratification in Patients With Heart Failure and Preserved Ejection Fraction. J. Am. Coll. Cardiol. 2020, 75, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.M.; Kim, K.H.; Jeon, K.H.; Park, J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiography 2019, 36, 213–218. [Google Scholar] [CrossRef]
- Mahajan, S.M.; Ghani, R. Using ensemble machine learning methods for predicting risk of readmission for heart failure. Stud. Health Technol. Inform. 2019, 264, 243–247. [Google Scholar]
- Mahajan, S.M.; Ghani, R. Combining structured and unstructured data for predicting risk of readmission for heart failure patients. Stud. Health Technol. Inform. 2019, 264, 238–242. [Google Scholar]
- Kakarmath, S.; Golas, S.; Felsted, J.; Kvedar, J.; Jethwani, K.; Agboola, S. Validating a Machine Learning Algorithm to Predict 30-Day Re-Admissions in Patients With Heart Failure: Protocol for a Prospective Cohort Study. JMIR Res. Protoc. 2018, 7, e176. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 1–510. [Google Scholar] [CrossRef]
- Understanding AUC—ROC Curve|by Sarang Narkhede|Towards Data Science. Available online: https://towardsdatascience.com/understanding-auc-roc-curve-68b2303cc9c5 (accessed on 24 July 2022).
- Januzzi, J.L., Jr.; Sakhuja, R.; O’Donoghue, M.; Baggish, A.L.; Anwaruddin, S.; Chae, C.U.; Cameron, R.; Krauser, D.G.; Tung, R.; Camargo, C.A., Jr.; et al. Utility of Amino-Terminal Pro–Brain Natriuretic Peptide Testing for Prediction of 1-Year Mortality in Patients With Dyspnea Treated in the Emergency Department. Arch. Intern. Med. 2006, 166, 315–320. [Google Scholar] [CrossRef] [PubMed]
- McKie, P.M.; Cataliotti, A.; Lahr, B.D.; Martin, F.L.; Redfield, M.M.; Bailey, K.R.; Rodeheffer, R.J.; Burnett, J.C. The Prognostic Value of N-Terminal Pro–B-Type Natriuretic Peptide for Death and Cardiovascular Events in Healthy Normal and Stage A/B Heart Failure Subjects. J. Am. Coll. Cardiol. 2010, 55, 2140–2147. [Google Scholar] [CrossRef] [Green Version]
- Royston, P.; Altman, D.G. External validation of a Cox prognostic model: Principles and methods. BMC Med. Res. Methodol. 2013, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Siontis, G.C.M.; Tzoulaki, I.; Castaldi, P.J.; Ioannidis, J.P.A. External validation of new risk prediction models is infrequent and reveals worse prognostic discrimination. J. Clin. Epidemiol. 2015, 68, 25–34. [Google Scholar] [PubMed]
- Lemeshow, S.; Klar, J.; Teres, D.; Avrunin, J.S.; Gehlbach, S.H.; Rapoport, J.; Rué, M. Mortality probability models for patients in the intensive care unit for 48 or 72 hours: A prospective, multicenter study. Crit. Care Med. 1994, 22, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Adrie, C.; Francais, A.; Alvarez-Gonzalez, A.; Mounier, R.; Azoulay, E.; Zahar, J.-R.; Clec’H, C.; Goldgran-Toledano, D.; Hammer, L.; Descorps-Declere, A.; et al. Model for predicting short-term mortality of severe sepsis. Crit. Care 2009, 13, R72. [Google Scholar] [CrossRef]
- Agbor, V.N.; Essouma, M.; Ntusi, N.A.; Nyaga, U.F.; Bigna, J.J.; Noubiap, J.J. Heart failure in sub-Saharan Africa: A contemporaneous systematic review and meta-analysis. Int. J. Cardiol. 2018, 257, 207–215. [Google Scholar] [CrossRef]
- Bahrami, H.; Kronmal, R.; Bluemke, D.A.; Olson, J.; Shea, S.; Liu, K.; Burke, G.L.; Lima, J.A. Differences in the incidence of congestive heart failure by ethnicity: The multi-ethnic study of atherosclerosis. Arch. Intern. Med. 2008, 168, 2138–2145. [Google Scholar] [CrossRef]
- Bazoukis, G.; Stavrakis, S.; Zhou, J.; Bollepalli, S.C.; Tse, G.; Zhang, Q.; Singh, J.P.; Armoundas, A.A. Machine learning versus conventional clinical methods in guiding management of heart failure patients—A systematic review. Heart Fail. Rev. 2020, 26, 23–34. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable ai for trees. Nat. Mach. Intell. 2020, 2, 2522–5839. [Google Scholar] [CrossRef]
- Duchnowski, P.; Hryniewiecki, T.; Koźma, M.; Mariusz, K.; Piotr, S. High-sensitivity troponin T is a prognostic marker of hemodynamic instability in patients undergoing valve surgery. Biomark Med. 2018, 12, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Duchnowski, P.; Hryniewiecki, T.; Kuśmierczyk, M.; Szymański, P. Postoperative high-sensitivity troponin T as a predictor of sudden cardiac arrest in patients undergoing cardiac surgery. Cardiol. J. 2019, 26, 777–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacciatore, F.; Abete, P.; Mazzella, F.; Furgi, G.; Nicolino, A.; Longobardi, G.; Testa, G.; Langellotto, A.; Infante, T.; Napoli, C.; et al. Six-minute walking test but not ejection fraction predicts mortality in elderly patients undergoing cardiac rehabilitation following coronary artery bypass grafting. Eur. J. Prev. Cardiol. 2011, 19, 1401–1409. [Google Scholar] [CrossRef]
- Briongos-Figuero, S.; Estévez, A.; Pérez, M.L.; Martínez-Ferrer, J.B.; García, E.; Viñolas, X.; Arenal, Á.; Alzueta, J.; Muñoz-Aguilera, R. Prognostic role of NYHA class in heart failure patients undergoing primary prevention ICD therapy. ESC Heart Fail. 2019, 7, 280–284. [Google Scholar] [CrossRef]
- Al-Tamimi, M.A.A.; Gillani, S.W.; Abd Alhakam, M.E.; Sam, K.G. Factors Associated with Hospital Readmission of Heart Failure Patients. Front. Pharmacol. 2021, 12, 732760. [Google Scholar] [PubMed]
- Baert, A.; Clays, E.; Bolliger, L.; De Smedt, D.; Lustrek, M.; Vodopija, A.; Bohanec, M.; Puddu, P.E.; Ciancarelli, M.C.; Schiariti, M.; et al. A Personal Decision Support System for Heart Failure Management (HeartMan): Study protocol of the HeartMan randomized controlled trial. BMC Cardiovasc. Disord. 2018, 18, 186. [Google Scholar] [CrossRef]
- Sengupta, P.P.; Shrestha, S.; Kagiyama, N.; Hamirani, Y.; Kulkarni, H.; Yanamala, N.; Bing, R.; Chin, C.W.L.; Pawade, T.A.; Messika-Zeitoun, D.; et al. A Machine-Learning Framework to Identify Distinct Phenotypes of Aortic Stenosis Severity. JACC Cardiovasc. Imaging 2021, 14, 1707–1720. [Google Scholar] [CrossRef]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef]
- Ghazi, L.; Ahmad, T.; Wilson, F.P. A Clinical Framework for Evaluating Machine Learning Studies. JACC Heart Fail. 2022, in press. [Google Scholar] [CrossRef]

| No. | Variables | MAGGIC | GWTG-HF |
|---|---|---|---|
| 1. | Age | X | X |
| 2. | COPD history | X | X |
| 3. | Systolic blood pressure | X | X |
| 4. | Gender | X | |
| 5. | Diabetes | X | |
| 6. | Heart failure diagnosed within the last 18 months | X | |
| 7. | Current smoker | X | |
| 8. | NYHA class | X | |
| 9. | Receives beta blockers | X | |
| 10. | Receives ACEI/ARB | X | |
| 11. | BMI | X | |
| 12. | Creatinine | X | |
| 13. | Ejection fraction | X | |
| 14. | BUN | X | |
| 15. | Sodium | X | |
| 16. | Black race | X | |
| 17. | Heart rate | X |
| No | Author | Year | Data Source | Population | Setting | Validation Size | Number of Variables | Outcome | Quality |
|---|---|---|---|---|---|---|---|---|---|
| 1. | C. Luo et al. [33] | 2022 | MIMIC-III database | 5676 | inpatient | 1349 | 24 | in-hospital mortality in intensive care unit | 87% |
| 2. | E. Adler et al. [34] | 2020 | UCSD database | 5822 | inpatient and outpatient | 1512 + 888 | 8 | general mortality | 80% |
| 3. | J. Kwon et al. [35] | 2019 | KorAHF | 2165 | inpatient | 4759 | 23 | in-hospital, 12-month and 36-month mortality | 87% |
| 4. | L. Jing et al. [36] | 2020 | Geisinger EHR | 26,971 | inpatient and outpatient | 548 | 209 | 1-year all-cause mortality | 73% |
| 5. | J. Chirinos [37] | 2020 | TOPCAT | 379 | inpatient and outpatient | 156 | 12 | composite endpoints of death or heart failure–related hospitalization | 87% |
| 6. | J. Kwon et al. [38] | 2019 | register | 20,651 | inpatient | 1560 | 65 | in-hospital mortality | 87% |
| 7. | S. Mahajan [39] | 2019 | register | 27,714 | inpatient | 8531 | Not reported | 30-day readmission | 66% |
| 8. | S. Mahajan [40] | 2019 | register | 1279 | inpatient | 340 | Not reported | 30-day readmission | 66% |
| 9. | S. Kakarmath et al. [41] | The protocol for the study | |||||||
| No. | Author | Algorithm | AUC for ML in IV | AUC for MAGGIC in IV | AUC for GWTG-HF in IV | AUC for ML in EV | AUC for MAGGIC in EV | AUC for GWTG-HF in EV |
|---|---|---|---|---|---|---|---|---|
| 1. | C. Luo et al. [33] | XGBoost | 0.831 | - | 0.667 | 0.809 | - | - |
| 2. | E. Adler et al. [34] | boosted decision tree | 0.88 | - | 0.74 | 0.81–0.84 | - | 0.758 |
| 3. | J. Kwon et al. [35] | deep neural network | - | - | - | 0.88—in-hospital mortality 0.782—12-month mortality 0.813—36-month mortality | 0.718—12-month mortality 0.729—36-month mortality | 0.728—in-hospital mortality |
| 4. | L. Jing et al. [36] | XGBoost | 0.77 | - | - | 0.78 | - | - |
| 5. | J. Chirinos [37] | created by the tree-based pipeline optimizer platform | 0.743 (C-index) | 0.621 (C-Index) | - | 0.717 (C-index) | 0.622 (C-index) | - |
| 6. | J. Kwon et al. [38] | deep neural network | 0.912—(HD) | - | - | 0.913 (HF) 0.898 (HD) 0.958 (CAD) | 0.806 (HF) | 0.783 (HF) |
| 7. | S. Mahajan [39] | ensemble ML | - | - | - | 0.6987 | - | - |
| 8. | S. Mahajan [40] | created by NLP process | - | - | - | 0.6494 | - | - |
| 9. | S. Kakarmath et al. [41] | The protocol for the study | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Błaziak, M.; Urban, S.; Wietrzyk, W.; Jura, M.; Iwanek, G.; Stańczykiewicz, B.; Kuliczkowski, W.; Zymliński, R.; Pondel, M.; Berka, P.; et al. An Artificial Intelligence Approach to Guiding the Management of Heart Failure Patients Using Predictive Models: A Systematic Review. Biomedicines 2022, 10, 2188. https://doi.org/10.3390/biomedicines10092188
Błaziak M, Urban S, Wietrzyk W, Jura M, Iwanek G, Stańczykiewicz B, Kuliczkowski W, Zymliński R, Pondel M, Berka P, et al. An Artificial Intelligence Approach to Guiding the Management of Heart Failure Patients Using Predictive Models: A Systematic Review. Biomedicines. 2022; 10(9):2188. https://doi.org/10.3390/biomedicines10092188
Chicago/Turabian StyleBłaziak, Mikołaj, Szymon Urban, Weronika Wietrzyk, Maksym Jura, Gracjan Iwanek, Bartłomiej Stańczykiewicz, Wiktor Kuliczkowski, Robert Zymliński, Maciej Pondel, Petr Berka, and et al. 2022. "An Artificial Intelligence Approach to Guiding the Management of Heart Failure Patients Using Predictive Models: A Systematic Review" Biomedicines 10, no. 9: 2188. https://doi.org/10.3390/biomedicines10092188
APA StyleBłaziak, M., Urban, S., Wietrzyk, W., Jura, M., Iwanek, G., Stańczykiewicz, B., Kuliczkowski, W., Zymliński, R., Pondel, M., Berka, P., Danel, D., Biegus, J., & Siennicka, A. (2022). An Artificial Intelligence Approach to Guiding the Management of Heart Failure Patients Using Predictive Models: A Systematic Review. Biomedicines, 10(9), 2188. https://doi.org/10.3390/biomedicines10092188






