Deep Learning for Bone Mineral Density and T-Score Prediction from Chest X-rays: A Multicenter Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Registration and Patient Data Collection
2.2. Data Preparation
2.3. Splitting the Dataset
2.4. Image Preprocessing and Machine Learning
2.5. Statistical Analysis
2.5.1. Regression of BMD
2.5.2. Classification of T-Score
3. Results
3.1. Patient Characteristics
3.2. Predictive Performance of Deep Learning Model
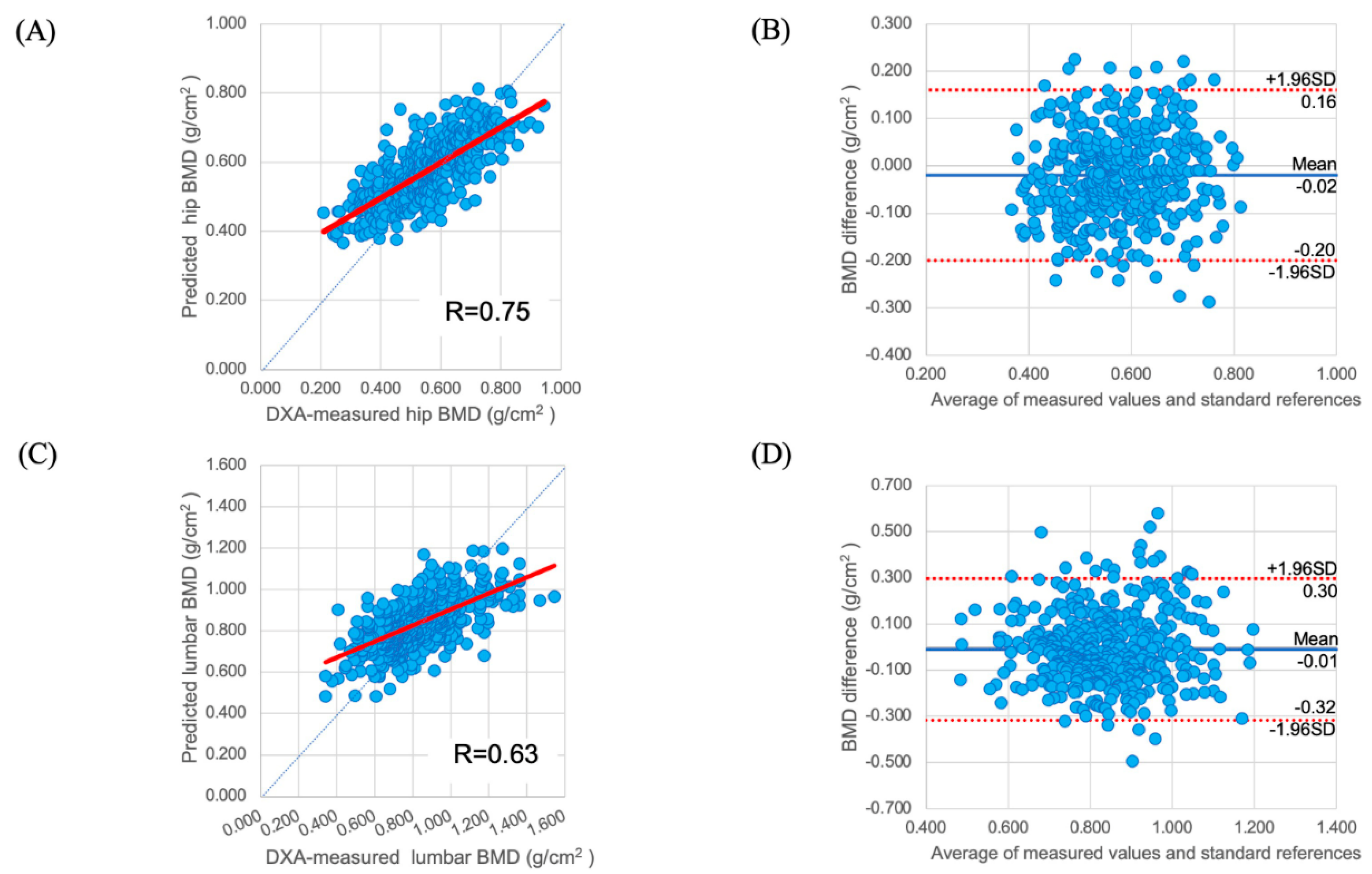
3.2.1. Regression of BMD
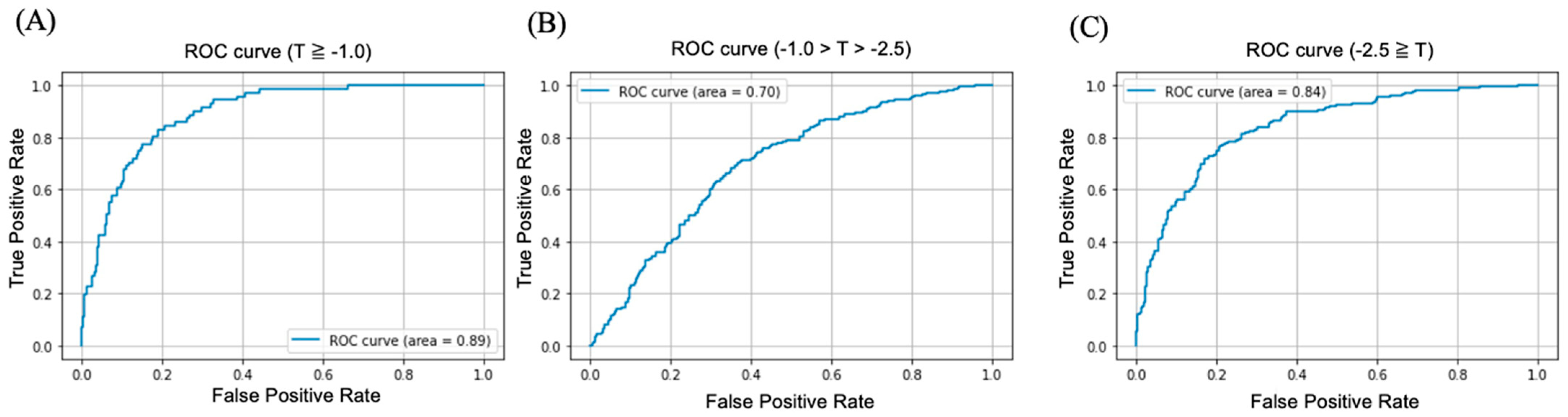
3.2.2. Classification of T-Score
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sözen, T.; Özışık, L.; Başaran, N.Ç. Overview and management of osteoporosis. Eur. J. Rheumatol. 2017, 4, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yoshida, H. Low bone mineral density at the femoral neck is a predictor of increased mortality in elderly Japanese women. Osteoporos. Int. 2010, 21, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Ensrud, K.E.; Thompson, D.E.; Cauley, J.A.; Nevitt, M.C.; Kado, D.M.; Hochberg, M.C.; Santora, A.C., II; Black, D.M. Prevalent vertebral deformities predict mortality and hospitalization in older women with low bone mass. Fracture Intervention Trial Research Group. J. Am. Geriatr. Soc. 2000, 48, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.D.; Center, J.R.; Eisman, J.A.; Nguyen, T.V. Bone loss, weight loss, and weight fluctuation predict the mortality risk in elderly men and women. J. Bone Miner Res. 2007, 22, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.J. Fracture Liaison Services: UK Experience. Osteoporos. Int. 2011, 22 (Suppl. S3), 487–494. [Google Scholar] [CrossRef]
- US Preventive Services Task Force. Clinical guideline: Screening for osteoporosis: U.S. preventive services task force recommendation statement. Encycl. Ann. Intern. Med. 2011, 154, 356–364. [Google Scholar] [CrossRef]
- Mueller, D.; Gandjour, A. Cost-effectiveness of using clinical risk factors with and without DXA for osteoporosis screening in postmenopausal women. Value Health 2009, 12, 1106–1117. [Google Scholar] [CrossRef]
- Sim, M.F.V.; Stone, M.; Johansen, A.; Evans, W. Cost effectiveness analysis of BMD referral for DXA using ultrasound as a selective pre-screen in a group of women with low trauma colle fractures. Technol. Health Care 2000, 8, 277–284. [Google Scholar] [CrossRef]
- Orimo, H.; Nakamura, T.; Hosoi, T.; Iki, M.; Uenishi, K.; Endo, N.; Ohta, H.; Shiraki, M.; Sugimoto, T.; Suzuki, T.; et al. Japanese 2011 guidelines for prevention and treatment of osteoporosis—executive summary. Arch. Osteoporos. 2012, 7, 3–20. [Google Scholar] [CrossRef]
- Sedlak, C.A.; Doheny, M.O.; Jones, S.L. Osteoporosis education programs: Changing knowledge and behaviors. Public Health Nurs. 2000, 17, 398–402. [Google Scholar] [CrossRef]
- Sato, M.; Vietri, J.; Flynn, J.A.; Fujiwara, S. Bone fractures and feeling at risk for osteoporosis among women in Japan: Patient characteristics and outcomes in the National Health and Wellness Survey. Arch. Osteoporos. 2014, 9, 199. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A. Triage screening for osteoporosis in dental clinics using panoramic radiographs. Oral Dis. 2010, 16, 316–327. [Google Scholar] [CrossRef]
- Kumar, D.A.; Anburajan, M. The role of hip and chest radiographs in osteoporotic evaluation among the South Indian women population: A comparative scenario with DXA. J. Endocrinol. Investig. 2014, 37, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhou, X.; Fujita, H.; Kubo, K.Y. Age-related changes in trabecular and cortical bone microstructure. Int. J. Endocrinol. 2013, 2013, 213234. [Google Scholar] [CrossRef] [PubMed]
- Holcombe, S.A.; Hwang, E.; Derstine, B.A.; Wang, S.C. Measurement of rib cortical bone thickness and cross-section using CT. Med. Image Anal. Elsevier B.V. 2018, 49, 27–34. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on image classification. Proc. IEEE Int. Conf. Comput. Vis. 2015, 2015, 1026–1034. [Google Scholar]
- Smets, J.; Shevroja, E.; Hügle, T.; Leslie, W.D.; Hans, D. Machine learning solutions for osteoporosis-a review. J. Bone Miner Res. 2021, 36, 833–851. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Chae, D.S.; Park, S.J.; Yoon, J. A novel approach for evaluating bone mineral density of hips based on a Sobel gradient-based map of radiographs utilizing a convolutional neural network. Comput. Biol. Med. Elsevier Ltd. 2021, 132, 104298. [Google Scholar] [CrossRef]
- Hsieh, C.I.; Zheng, K.; Lin, C.; Mei, L.; Lu, L.; Li, W.; Chen, F.-P.; Wang, Y.; Zhou, X.; Wang, F.; et al. Automated bone mineral density prediction and fracture risk assessment using plain radiographs via deep learning. Nat. Commun. 2021, 12, 5472. [Google Scholar] [CrossRef]
- Yamamoto, N.; Sukegawa, S.; Kitamura, A.; Goto, R.; Noda, T.; Nakano, K.; Takabatake, K.; Kawai, H.; Nagatsuka, H.; Kawasaki, K.; et al. Deep learning for osteoporosis classification using hip radiographs and patient clinical covariates. Biomolecules 2020, 10, 1534. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, K.; Ning, Z.; Wang, K.; Dong, Y.; Liu, X.; Wang, J.; Zhu, C.; Yu, Q.; Duan, Y.; et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone 2020, 140, 115561. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Yamamoto, K.; Matsuzawa, H.; Kobayashi, T. Development of a fast screening method for osteoporosis using chest X-ray images and machine learning. Can. J. Biomed. Res. Technol. 2020, 3, 3–9. [Google Scholar]
- Jang, M.; Kim, M.; Bae, S.J.; Lee, S.H.; Koh, J.M.; Kim, N. Opportunistic osteoporosis screening using chest radiographs with deep learning: Development and external validation with a cohort dataset. J. Bone Miner Res. 2022, 37, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Yamamoto, N.; Sukegawa, S.; Yamashita, K.; Manabe, M.; Nakano, K.; Takabatake, K.; Kawai, H.; Ozaki, T.; Kawasaki, K.; Nagatsuka, H.; et al. Effects of patient clinical variables on osteoporosis classification using hip X-rays in deep learning analysis. Medicina 2021, 57, 846. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): TRIPOD statement. BMC Med. 2015, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Dimai, H.P. Use of dual-energy X-ray absorptiometry (DXA) for diagnosis and fracture risk assessment; WHO-criteria, T- and Z-scores, and reference databases. Bone Elsevier Inc. 2017, 104, 39–43. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern. Recognit. 2016, 2016, 770–778. [Google Scholar]
- Mukaka, M.M. Statistics corner: A guide to the appropriate use of correlation coefficients in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Liu, G.; Peacock, M.; Eilam, O.; Dorulla, G.; Braunstein, E.; Johnston, C.C. Effect of osteoarthritis in the lumbar spine and hip on bone mineral density and osteoporosis diagnosis in elderly men and women. Osteoporos. Int. 1997, 7, 564–569. [Google Scholar] [CrossRef]
- Siris, E.S.; Chen, Y.T.; Abbott, T.A.; Barrett-Connor, E.; Miller, P.D.; Wehren, L.E.; Berger, M.L. Bone mineral density thresholds for pharmacological interventions to prevent fractures. Arch. Intern. Med. 2004, 164, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Kanis, J.A.; Cooper, C.; Rizzoli, R.; Reginster, J.Y. Scientific Advisory Board of the European Society for Clinical and Economic Aspects of Osteoporosis (ESCEO) and the Committees of Scientific Advisors and National Societies of the International Osteoporosis Foundation (IOF). Correction to: European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos. Int. 2020, 31, 209. [Google Scholar] [PubMed]
- Camacho, P.M.; Petak, S.M.; Binkley, N.; Diab, D.L.; Eldeiry, L.S.; Farooki, A.; Harris, S.T.; Hurley, D.L.; Kelly, J.; Lewiecki, M.; et al. American Association of Clinical Endocrinologists/American College of Endocrinology Clinical Practice Guidelines for The Diagnosis and Treatment of Postmenopausal Osteoporosis-2020 Update. Endocr. Pract. 2020, 26 (Suppl. S1), 1–46. [Google Scholar] [CrossRef]
- Cadarette, S.M.; Jaglal, S.B.; Murray, T.M.; McIsaac, W.J.; Joseph, L.; Brown, J.P. Evaluation of decision rules for referring women for bone densitometry by dual-energy X-ray absorptiometry. J. Am. Med. Assoc. 2001, 286, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Information Service. Available online: https://ganjoho.jp/reg_stat/index.html (accessed on 16 May 2022).
- Yoshimura, N.; Muraki, S.; Oka, H.; Mabuchi, A.; En-Yo, Y.; Yoshida, M.; Saika, A.; Yoshida, H.; Suzuki, T.; Yamamoto, S.; et al. Prevalence of knee osteoarthritis, lumbar spondylosis, and osteoporosis in Japanese men and women: The research on osteoarthritis/osteoporosis against disability study. J. Bone Miner Metab. 2009, 27, 620–628. [Google Scholar] [CrossRef]
- Cummings, S.R.; Black, D.M.; Thompson, D.E.; Applegate, W.B.; Barrett-Connor, E.; Musliner, T.A.; Palermo, L.; Prineas, R.; Rubin, S.M.; Scott, J.C.; et al. Effect of alendronate on risk of fracture in women with low bone density but without vertebral fractures: Results from the Fracture Intervention. Trial. J. Am. Med. Assoc. 1998, 280, 2077–2082. [Google Scholar] [CrossRef]
- Kim, D.W.; Jang, H.Y.; Kim, K.W.; Shin, Y.; Park, S.H. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: Results from recently published papers. Korean J. Radiol. 2019, 20, 405–410. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Cruz, A.S.; Lins, H.C.; Medeiros, R.V.A.; Filho, J.M.F.; da Silva, S.G. Artificial intelligence on the identification of risk groups for osteoporosis, a general review. Biomed. Eng. Online 2018, 17, 12. [Google Scholar] [CrossRef]
- Hirano, K.; Imagama, S.; Hasegawa, Y.; Ito, Z.; Muramoto, A.; Ishiguro, N. The influence of locomotive syndrome on health-related quality of life in a community-living population. Mod. Rheumatol. 2013, 23, 939–944. [Google Scholar] [CrossRef]
- Haugeberg, G.; Uhlig, T.; Falch, J.A.; Halse, J.I.; Kvien, T.K. Bone mineral density and frequency of osteoporosis in female patients with rheumatoid arthritis: Results from 394 patients in the Oslo County rheumatoid arthritis register. Arthritis Rheum. 2000, 43, 522–530. [Google Scholar] [CrossRef]

| Training Dataset | Validation Dataset | Test Dataset | Overall | ||
|---|---|---|---|---|---|
| Participant | 12,529 | 1790 | 3580 | 17,899 | |
| age (years), mean ± SD | 71.94 ± 10.05 | 71.24 ± 10.94 | 71.54 ± 11.27 | 71.57 ± 10.75 | |
| Sex | Female (%) | 10,544 (84.16%) | 1508 (84.25%) | 3008 (84.02%) | 15,060 (84.14%) |
| Male (%) | 1985 (15.84%) | 282 (15.75%) | 572 (15.98%) | 2839 (15.86%) | |
| BMD (g/cm2), mean ± SD | Lumbar | 0.88 ± 0.19 | 0.89 ± 0.21 | 0.88 ± 0.20 | 0.88 ± 0.20 |
| Hip | 0.58 ± 0.12 | 0.59 ± 0.15 | 0.58 ± 0.13 | 0.58 ± 0.13 | |
| T-score mean ± SD | Lumbar | −1.51 ± 1.56 | −1.53 ± 1.68 | −1.51 ± 1.60 | −1.52 ± 1.61 |
| Hip | −2.145 ± 1.17 | −2.15 ± 1.40 | −2.16 ± 1.10 | −2.15 ± 1.22 | |
| T-score categories, n (%) | Normal | 2204 (17.59%) | 317 (17.71%) | 631 (17.63%) | 3152 (17.61%) |
| Osteopenia | 7287 (58.16%) | 1038 (57.99%) | 2079 (58.07%) | 10,404 (58.13%) | |
| Osteoporosis | 3038 (24.25%) | 435 (24.30%) | 870 (24.30%) | 4343 (24.26%) |
| AUC (95% CI) | Accuracy (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | |
|---|---|---|---|---|
| T-score ≥ −1.0 | 0.89 (0.86–0.91) | 74.89 (71.21–77.45) | 90.14 (87.35–92.41) | 72.24 (68.32–75.80) |
| −1.0 > T-score > −2.5 | 0.70 (0.68–0.72) | 66.06 (63.65–68.39) | 71.28 (69.01–73.53) | 62.35 (59.94–64.77) |
| −2.5 ≥ T-score | 0.84 (0.82–0.86) | 76.47 (75.52–79.90) | 81.25 (74.94–79.36) | 73.68 (76.32–80.65) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, Y.; Yamamoto, N.; Inagaki, N.; Iesaki, Y.; Asamoto, T.; Suzuki, T.; Takahara, S. Deep Learning for Bone Mineral Density and T-Score Prediction from Chest X-rays: A Multicenter Study. Biomedicines 2022, 10, 2323. https://doi.org/10.3390/biomedicines10092323
Sato Y, Yamamoto N, Inagaki N, Iesaki Y, Asamoto T, Suzuki T, Takahara S. Deep Learning for Bone Mineral Density and T-Score Prediction from Chest X-rays: A Multicenter Study. Biomedicines. 2022; 10(9):2323. https://doi.org/10.3390/biomedicines10092323
Chicago/Turabian StyleSato, Yoichi, Norio Yamamoto, Naoya Inagaki, Yusuke Iesaki, Takamune Asamoto, Tomohiro Suzuki, and Shunsuke Takahara. 2022. "Deep Learning for Bone Mineral Density and T-Score Prediction from Chest X-rays: A Multicenter Study" Biomedicines 10, no. 9: 2323. https://doi.org/10.3390/biomedicines10092323
APA StyleSato, Y., Yamamoto, N., Inagaki, N., Iesaki, Y., Asamoto, T., Suzuki, T., & Takahara, S. (2022). Deep Learning for Bone Mineral Density and T-Score Prediction from Chest X-rays: A Multicenter Study. Biomedicines, 10(9), 2323. https://doi.org/10.3390/biomedicines10092323





