Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nucleic Acids Template for Amplification
2.2. Clinical Samples and RNA Extraction
2.3. Oligonucleotide Primers
2.4. Reverse Transcription and PCR
2.5. RT-HDA Reactions
2.6. RT-LAMP Reactions
2.7. Lateral Flow Assay
2.8. Determination of the Detection Limit and the Minimal Reaction Time
2.9. Determination of Specificity, Sensitivity, Accuracy, and Reproducibility
3. Results
3.1. Verification of Oligonucleotide Primers Using PCR
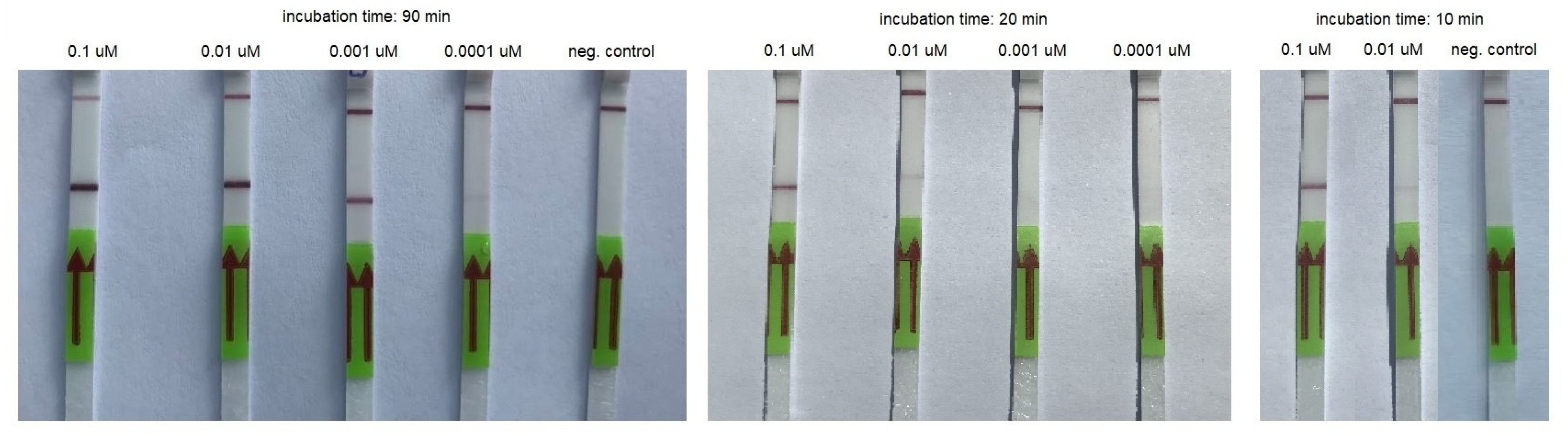
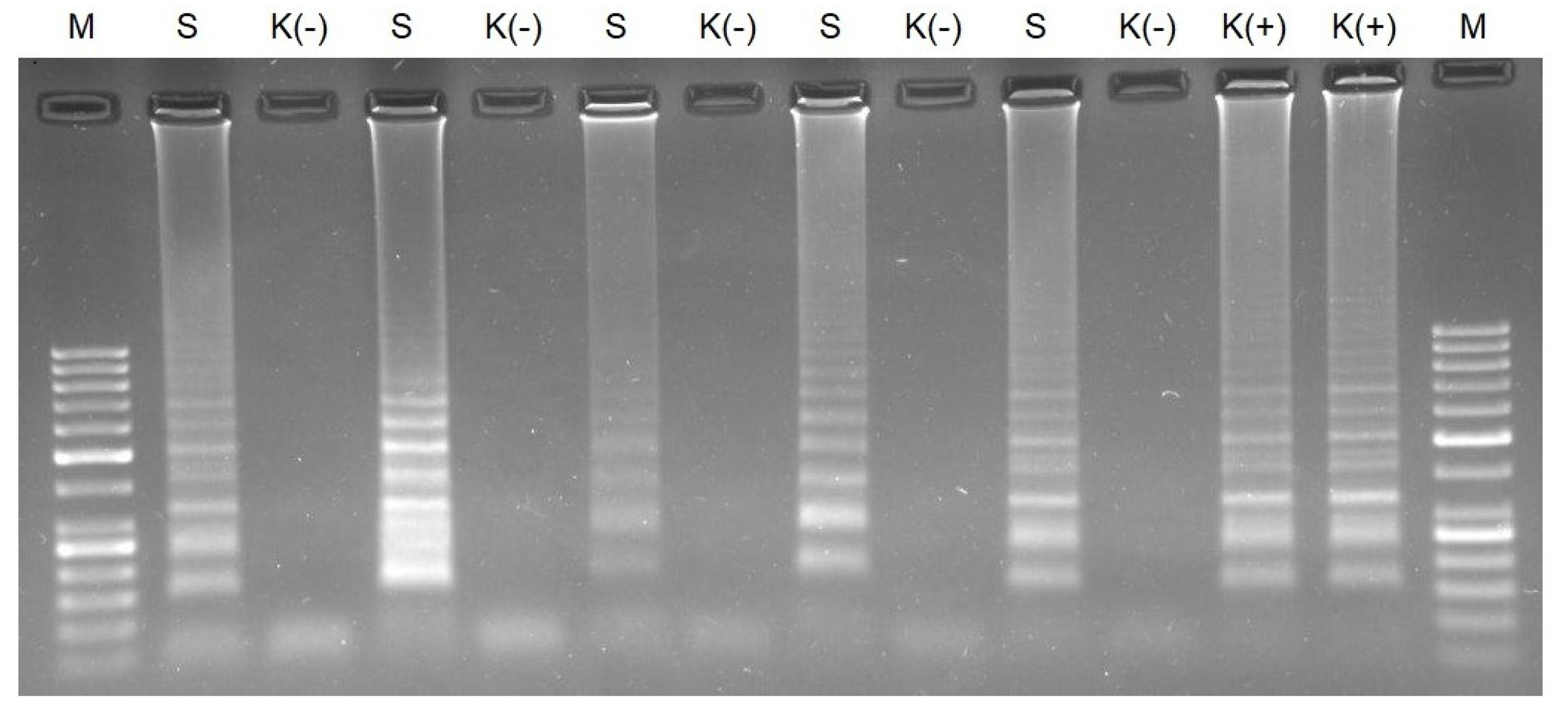
3.2. RT-HDA
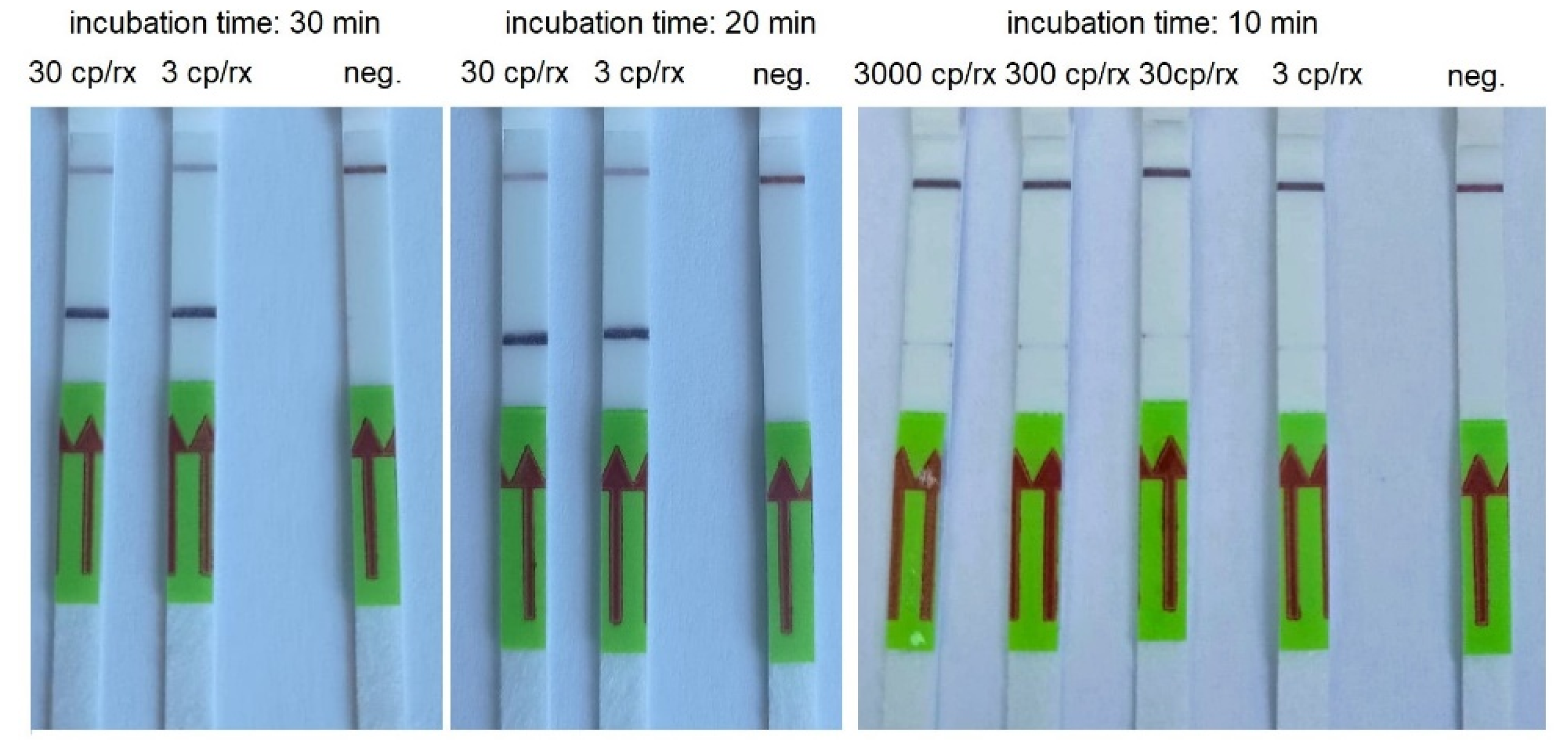
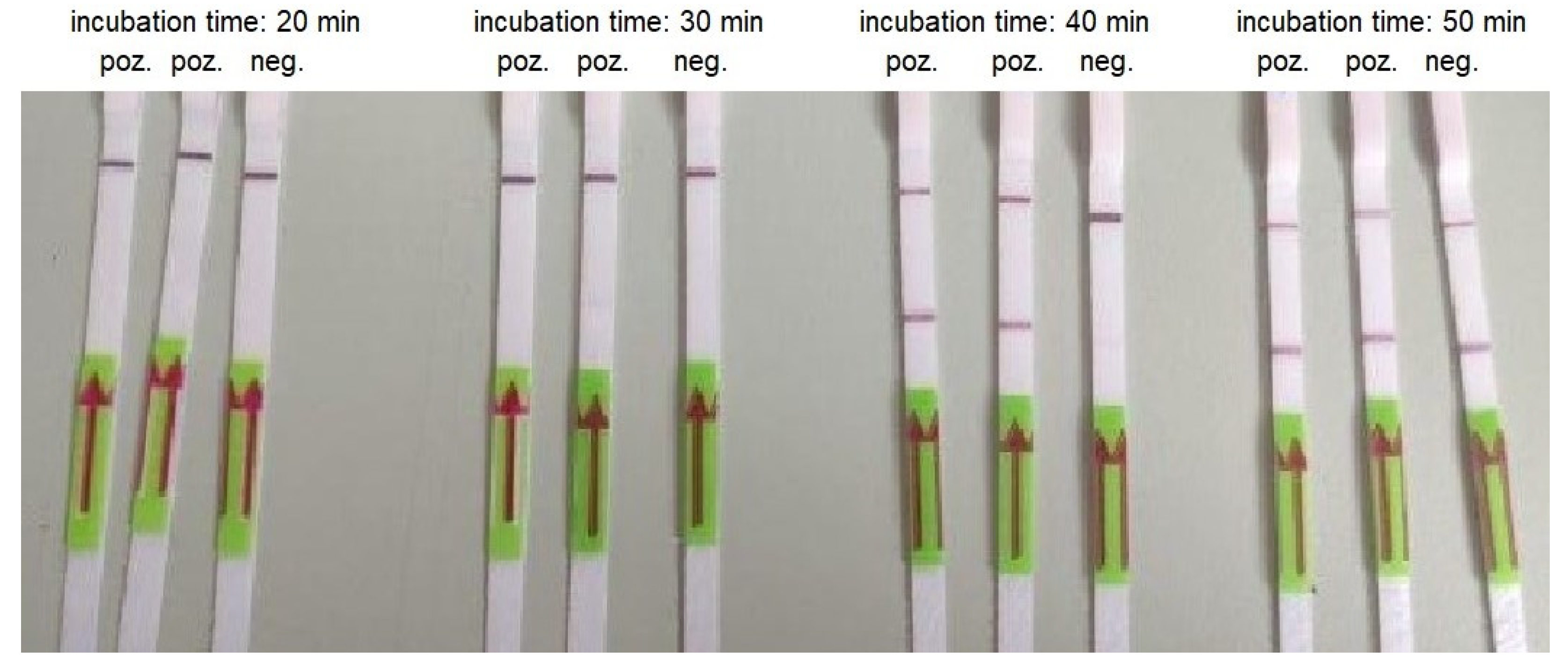
3.3. RT-LAMP
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abid, R.; Shahzad, M.K.; Sulaman, S.M.; Faheem, M.; Naeem, M.; Khan, R.; Khalil, A.A.K.; Haider, A.; Ahmad, B.; Gul, R.; et al. Therapeutic significance of nano- and biosensor technology in combating SARS-CoV-2: A review. Appl. Nanosci. 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chai, Y.; Hu, Z.; Xu, Z.; Li, M.; Chen, X.; Yang, C.; Liu, J. Recent Progress on Rapid Lateral Flow Assay-Based Early Diagnosis of COVID-19. Front. Bioeng. Biotechnol. 2022, 10, 866368. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hozumi, Y.; Yin, C.; Wei, G.W. Mutations on COVID-19 diagnostic targets. Genomics 2020, 112, 5204–5213. [Google Scholar] [CrossRef]
- Motley, M.P.; Bennett-Guerrero, E.; Fries, B.C.; Spitzer, E.D. Review of Viral Testing (Polymerase Chain Reaction) and Antibody/Serology Testing for Severe Acute Respiratory Syndrome-Coronavirus-2 for the Intensivist. Crit. Care Explor. 2020, 2, e0154. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Diagnostic Testing for SARS-CoV-2. Available online: https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2 (accessed on 18 September 2022).
- Zambry, N.S.; Obande, G.A.; Khalid, M.F.; Bustami, Y.; Hamzah, H.H.; Awang, M.S.; Aziah, I.; Manaf, A.A. Utilizing Electrochemical-Based Sensing Approaches for the Detection of SARS-CoV-2 in Clinical Samples: A Review. Biosensors 2022, 12, 473. [Google Scholar] [CrossRef] [PubMed]
- Lino, A.; Cardoso, M.A.; Gonçalves, H.M.R.; Martins-Lopes, P. SARS-CoV-2 Detection Methods. Chemosensors 2022, 10, 221. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- World Health Organisation. Protocols for 2019-Novel Coronavirus (2019-nCoV) Real-Time PCR. Available online: https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2%26download=true (accessed on 18 September 2022).
- Zang, Y.; Ren, G.; Buss, J.; Barry, A.J.; Patton, G.C.; Tanner, N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 2020, 69, 178–185. [Google Scholar] [CrossRef]
- Islam, M.M.; Koirala, D. Toward a next-generation diagnostic tool: A review on emerging isothermal nucleic acid amplification techniques for the detection of SARS-CoV-2 and other infectious viruses. Anal. Chim. Acta 2021, 1209, 339338. [Google Scholar] [CrossRef]
- Kundrod, K.A.; Natoli, M.E.; Chang, M.M.; Smith, C.A.; Paul, S.; Ogoe, D.; Goh, C.; Santhanaraj, A.; Price, A.; Eldin, K.W.; et al. Sample-to-answer, extraction-free, real-time RT-LAMP test for SARS-CoV-2 in nasopharyngeal, nasal, and saliva samples: Implications and use for surveillance testing. PLoS ONE 2020, 17, e0264130. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef] [PubMed]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, S.; Hao, X.; Dong, X.; Mao, L.; Pelechano, V.; Chen, W.H.; Yin, X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem. 2020, 66, 975–977. [Google Scholar] [CrossRef]
- Zasada, A.A.; Zacharczuk, K.; Formińska, K.; Wiatrzyk, A.; Ziółkowski, R.; Malinowska, E. Isothermal DNA amplification combined with lateral flow dipsticks for detection of biothreat agents. Anal. Biochem. 2018, 560, 60–66. [Google Scholar] [CrossRef]
- Barbieri, D.; Venturoli, S.; Rosl, F.; Rincon-Orozco, B. Detection of high-risk human papillomavirus type 16 and 18 using isothermal helicase-dependent amplification. Diagn. Microbiol. Infect. Dis. 2014, 79, 178–182. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Comparison of isothermal helicase-dependent amplification and PCR for the detection of Mycobacterium tuberculosis by an electrochemical genomagnetic assay. Anal. Bioanal. Chem. 2016, 408, 8603–8610. [Google Scholar] [CrossRef]
- Li, Y.; Kumar, N.; Gopalakrishnan, A.; Ginocchio, C.; Manji, R.; Bythrow, M.; Lemieux, B.; Kong, H. Detection and species identification of malaria parasites by isothermal tHDA amplification directly from human blood without sample preparation. J. Mol. Diagn. 2013, 15, 634–641. [Google Scholar] [CrossRef]
- Tang, W.; Chow, W.H.; Li, Y.; Kong, H.; Tang, Y.W.; Lemieux, B. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J. Infect. Dis. 2010, 201 (Suppl. S1), S46–S51. [Google Scholar] [CrossRef]
- Tong, Y.; Lemieux, B.; Kong, H. Multiple strategies to improve sensitivity, speed and robustness of isothermal nucleic acid amplification for rapid pathogen detection. BMC Biotechnol. 2011, 11, 50. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Do, J.; Mahalanabis, M.; Fan, A.; Zhao, L.; Jepeal, L.; Singh, S.K.; Klapperich, C.M. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS ONE 2013, 8, e60059. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, D.; Doseeva, V.; Rothmann, T.; Wolff, J.; Nazarenko, I. Evaluation of Chlamydia trachomatis and Neisseria gonorrhoeae detection in urine, endocervical, and vaginal specimens by a multiplexed isothermal thermophilic helicase-dependent amplification (tHDA) assay. J. Clin. Microbiol. 2011, 49, 4121–4125. [Google Scholar] [CrossRef] [PubMed]
- Zasada, A.A.; Wiatrzyk, A.; Czajka, U.; Brodzik, K.; Formińska, K.; Mosiej, E.; Prygiel, M.; Krysztopa-Grzybowska, K.; Wdowiak, K. Application of loop-mediated isothermal amplification combined with colorimetric and lateral flow dipstick visualization as the potential point-of-care testing for Corynebacterium diphtheriae. BMC Infect. Dis. 2020, 20, 308. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Helicase-depeldent isothermal amplification: A novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018, 410, 679–693. [Google Scholar] [CrossRef]
- Kolm, C.; Martzy, R.; Fuhrer, M.; Mach, R.L.; Krska, R.; Baumgartner, S.; Farnleither, A.H.; Reischer, G.H. Detection of a microbial source tracking marker by isothermal helicase-dependent amplification and a nucleic acid lateral-flow strip test. Sci. Rep. 2019, 9, 393. [Google Scholar] [CrossRef]
- Lopez-Rincon, A.; Tonda, A.; Mendoza-Maldonato, L.; Mulders, D.G.J.; Molenkamp, R.; Perez-Romero, C.; Claassen, E.; Garssen, J.; Kraneveld, A.D. Classification and specific primer design for accurate detection of SARS-CoV-2 using deep learning. Sci. Rep. 2021, 11, 947. [Google Scholar] [CrossRef]
- Mahalanabis, M.; Do, J.; Almuayad, H.; Zhang, J.Y.; Klapperich, C.M. Erratum to: An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed. Microdevices 2011, 13, 599–602. [Google Scholar] [CrossRef]
- Sharma, K.; Sharma, M.; Batra, N.; Sharma, A.; Dhillon, M.S. Diagnostic potential of multi-targeted LAMP (loop-mediated isothermal amplification) for osteoarticular tuberculosis. J. Orthop. Res. 2017, 35, 361–365. [Google Scholar] [CrossRef]
- Perchetti, G.A.; Nalla, A.K.; Huang, M.L.; Jerome, K.R.; Greninger, A.L. Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR. J. Clin. Virol. 2020, 129, 104499. [Google Scholar] [CrossRef]
- Kiatpathomchai, W.; Jaroenram, W.; Arunrut, N.; Jitrapakdee, S.; Flegel, T.W. Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods 2008, 153, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Jaroenram, W.; Kiatpathomchai, W.; Flegel, T.W. Rapid and sensitive detection of white spot syndrome virus by loop-mediated isothermal amplification combined with a lateral flow dipstick. Mol. Cell. Probes. 2009, 23, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ince, B.; Sezgintürk, M.K. Lateral flow assays for viruses diagnosis: Up-to-date technology and future prospects. TrAC Trends Anal. Chem. 2022, 157, 116725. [Google Scholar] [CrossRef] [PubMed]
- Santiago, I. Trend and innovations in biosensors for COVID-19 mass testing. ChemBioChem 2020, 21, 2880–2889. [Google Scholar] [CrossRef] [PubMed]
- Rastawicki, W.; Gierczyński, R.; Juszczyk, G.; Mitura, K.; Henry, B.M. Evaluation of PCL rapid point of care antigen test for detection of SARS-CoV-2 in nasopharyngeal swabs. J. Med. Virol. 2021, 93, 1920–1922. [Google Scholar] [CrossRef] [PubMed]
- Rastawicki, W.; Rokosz-Chudziak, N. Characteristics and assessment of the usefulness of serological tests in the diagnostic of infections caused by coronavirus SARS-CoV-2 on the basis of available manufacturer’s data and literature review. Prz. Epidemiol. 2020, 74, 49–68. [Google Scholar] [CrossRef]
- Zasada, A.A.; Zacharczuk, K.; Woźnica, K.; Główka, M.; Ziółkowski, R.; Malinowska, E. The influence of a swab type on the results of point-of-care tests. AMB Express 2020, 10, 46. [Google Scholar] [CrossRef]

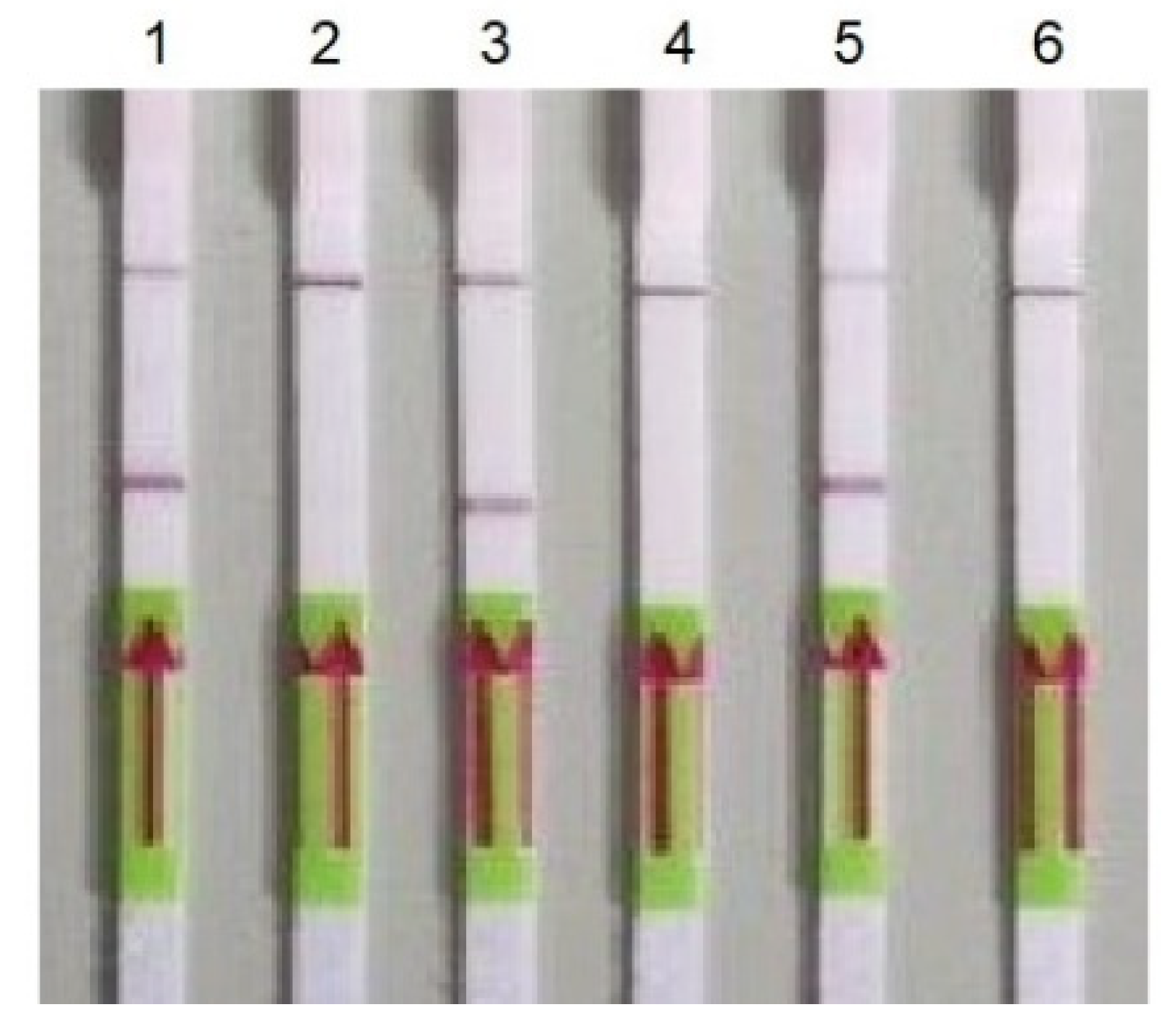
| Target | Oligonucleotide Sequence | Size |
|---|---|---|
| Gene E | ACAGGTACGTTAATAGTTAATAGCGTACTTCTTTTTCTTGCTTTCGTGGTATTCTTGCTAGTTACACTAGCCATCCTTACTGCGCTTCGATTGTGTGCGTACTGCTGCAATAT | 113 bp |
| Gene RdRP | GGTAACTGGTATGATTTCGGTGATTTCATACAAACCACGCCAGGTAGTGGAGTTCCTGTTGTAGATTCTTATT ATTCATTGTTAATGCCTATATTAACCTTGACCAG | 107 bp |
| Gene N | TTACAAACATTGGCCGCAAATTGCACAATTTGCCCCCAGTTCTTCGGAATGTCGCGC | 57 bp |
| Isothermal Amplification Assay | Primer Name | Primer Sequence | Target Gene of SARS-CoV-2 | Reference |
|---|---|---|---|---|
| RT-HDA | E_Sarbeco_F | ACAGGTACGTTAATAGTTAATAGCGT | Gene E | [8] |
| E_Sarbeco_R | ATATTGCAGCAGTACGCACACA | |||
| nCoV_IP4-14059Fw | GGTAACTGGTATGATTTCG | Gene RdRP | [9] | |
| nCoV_IP4-14146Rv | CTGGTCAAGGTTAATATAGG | |||
| 2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | Gene N | [9] | |
| 2019-nCoV_N2-R | GCGCGACATTCCGAAGAA | |||
| Gen_N-105F | TCATCACGTAGTCGCAACAG | Gene N | This study | |
| Gen_N-105R | CAAAGCAAGAGCAGCATCAC | |||
| Gen_N-90F | GGCAGTAACCAGAATGGAGAA | Gene N | This study | |
| Gen_N-90R | GGTGAACCAAGACGCAGTAT | |||
| Gen_E-107F | ATTCGTTTCGGAAGAGACAGG | Gene E | This study | |
| Gen_E-107R | ATCGAAGCGCAGTAAGGATG | |||
| RT-LAMP | E1-LF | CGCTATTAACTATTAACG | Gene E | [10] |
| E1-LB | GCGCTTCGATTGTGTGCGT | |||
| E1-F3 | TGAGTACGAACTTATGTACTCAT | |||
| E1-B3 | TTCAGATTTTTAACACGAGAGT | |||
| E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTTTTTCGTTTCGGAAGAGACAG | |||
| E1-BIP | TTGCTAGTTACACTAGCCATCCTTACTTTTGTTTTACAAG ACTCACGT | |||
| N2-LF | GGGGGCAAATTGTGCAATTTG | Gene N | [10] | |
| N2-LB | CTTCGGGAACGTGGTTGACC | |||
| N2-F3 | ACCAGGAACTAATCAGACAAG | |||
| N2-B3 | GACTTGATCTTTGAAATTTGGATCT | |||
| N2-FIP | TTCCGAAGAACGCTGAAGCGTTTTAACTGATTACAAACATTGGCC | |||
| N2-BIP | CGCATTGGCATGGAAGTCACAATTTTTTTGATGGCACCTGTGTA |
| Concentration of the Synthetic Template (µM) | Incubation Time (Min) | ||||||
|---|---|---|---|---|---|---|---|
| 90 | 75 | 60 | 45 | 30 | 20 | 10 | |
| 0.2 | + | + | + | + | + | + | + |
| 0.02 | + | + | + | + | + | (+) * | (+) |
| 0.002 | + | + | + | + | - | - | - |
| 0.0002 | (+) | (+) | - | - | - | - | - |
| Negative control | - | - | - | - | - | - | - |
| Concentration of the Synthetic Template (Copies/Reaction) | Incubation Time (Min) | ||||
|---|---|---|---|---|---|
| 60 | 45 | 30 | 20 | 10 | |
| 30000 | + | + | + | + | + |
| 3000 | + | + | + | + | (+)* |
| 300 | + | + | + | + | (+)* |
| 30 | + | + | + | + | (+)* |
| 3 | + | + | + | + | (+)* |
| Negative control | - | - | - | - | - |
| Concentration of the Template (Copies/Reaction) | Incubation Time (Min) | ||||
|---|---|---|---|---|---|
| 60 | 50 | 40 | 30 | 20 | |
| 30000 | + | + | + | + | + |
| 3000 | + | + | + | + | - |
| 300 | + | + | + | - | - |
| 30 | + | + | (+) * | - | - |
| 3 | - | - | - | - | - |
| Negative control | + | + | - | - | - |
| Type of the Test | LOD | Sensitivity | Specificity | Time |
|---|---|---|---|---|
| Antibody -targeting COVID-19 tests | 100 pf/mL–1.2 mg/mL | 44%–100% | 78%–100% | 5 min–60 min |
| Nucleic acid -targeting SARS-CoV-2 tests | 0.02 copies/reactions–2300 copies/reactions | 82%–100% | 82%–100% | <10 min–90 min |
| Antigen–targeting SARS-CoV-2 tests | 0.016 fg/mL–2.2 ng/mL | 69%–93% | 93%–100% | 5 min–20 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zasada, A.A.; Mosiej, E.; Prygiel, M.; Polak, M.; Wdowiak, K.; Formińska, K.; Ziółkowski, R.; Żukowski, K.; Marchlewicz, K.; Nowiński, A.; et al. Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay. Biomedicines 2022, 10, 2329. https://doi.org/10.3390/biomedicines10092329
Zasada AA, Mosiej E, Prygiel M, Polak M, Wdowiak K, Formińska K, Ziółkowski R, Żukowski K, Marchlewicz K, Nowiński A, et al. Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay. Biomedicines. 2022; 10(9):2329. https://doi.org/10.3390/biomedicines10092329
Chicago/Turabian StyleZasada, Aleksandra Anna, Ewa Mosiej, Marta Prygiel, Maciej Polak, Karol Wdowiak, Kamila Formińska, Robert Ziółkowski, Kamil Żukowski, Kasper Marchlewicz, Adam Nowiński, and et al. 2022. "Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay" Biomedicines 10, no. 9: 2329. https://doi.org/10.3390/biomedicines10092329
APA StyleZasada, A. A., Mosiej, E., Prygiel, M., Polak, M., Wdowiak, K., Formińska, K., Ziółkowski, R., Żukowski, K., Marchlewicz, K., Nowiński, A., Nowińska, J., Rastawicki, W., & Malinowska, E. (2022). Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay. Biomedicines, 10(9), 2329. https://doi.org/10.3390/biomedicines10092329








