Bioinformatical Design and Performance Evaluation of a Nucleocapsid- and an RBD-Based Particle Enhanced Turbidimetric Immunoassay (PETIA) to Quantify the Wild Type and Variants of Concern-Derived Immunoreactivity of SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatical Characterization and Comparison of the Coronavirus Proteins and Structural Predictions
- -
- Symbol comparison table: blosum62
- -
- Gap weight: 12
- -
- Gap length weight: 4
- -
- Consensus levels: high = 90%; low = 50%
2.2. PETIA Production
- -
- Reaction buffer (R1) = 90 µL
- -
- Sample = 10 µL
- -
- Latex bead reagent (R2) = 30 µL
- -
- Wavelength = 658 nm
- -
- Reading time = ca. 600–350 s
- -
- Result calculation = Δ [Abs600sec − Abs350sec]
2.3. Performance Comparison of S-RBD and N-Based PETIA
2.4. Quantification of Variant Cross-Reactivity
3. Results
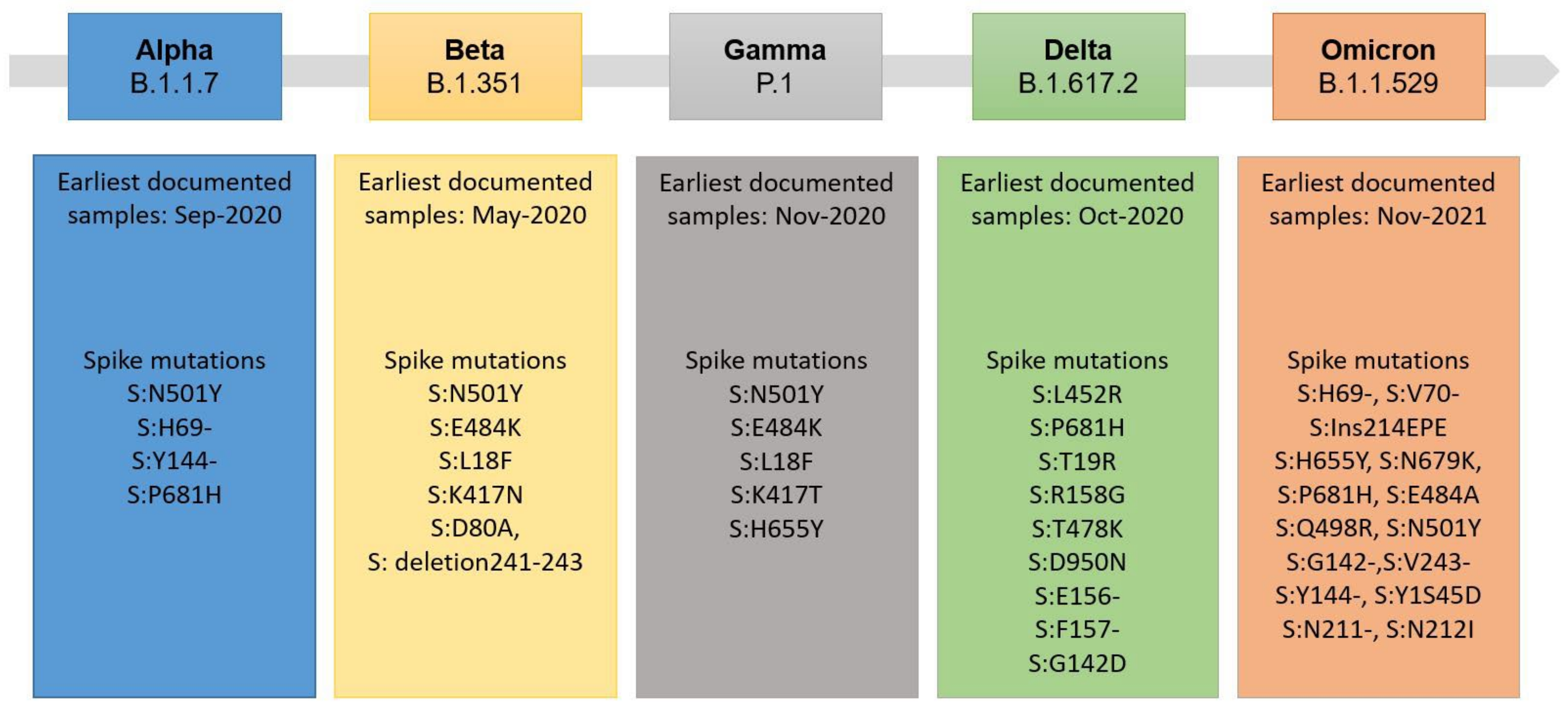
3.1. Bioinformatical Comparison of Coronavirus Proteins
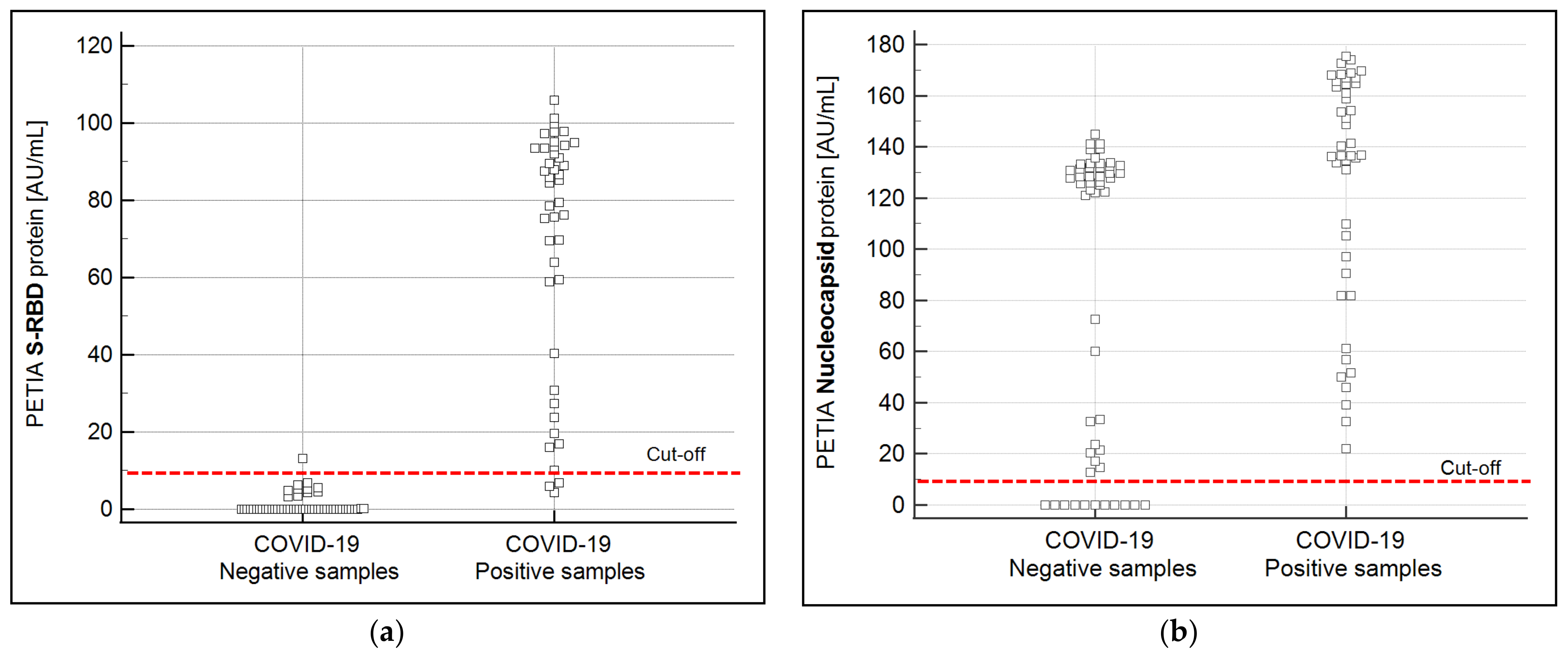
3.2. Production and Performance Comparison of S-RBD vs. N-Based PETIA
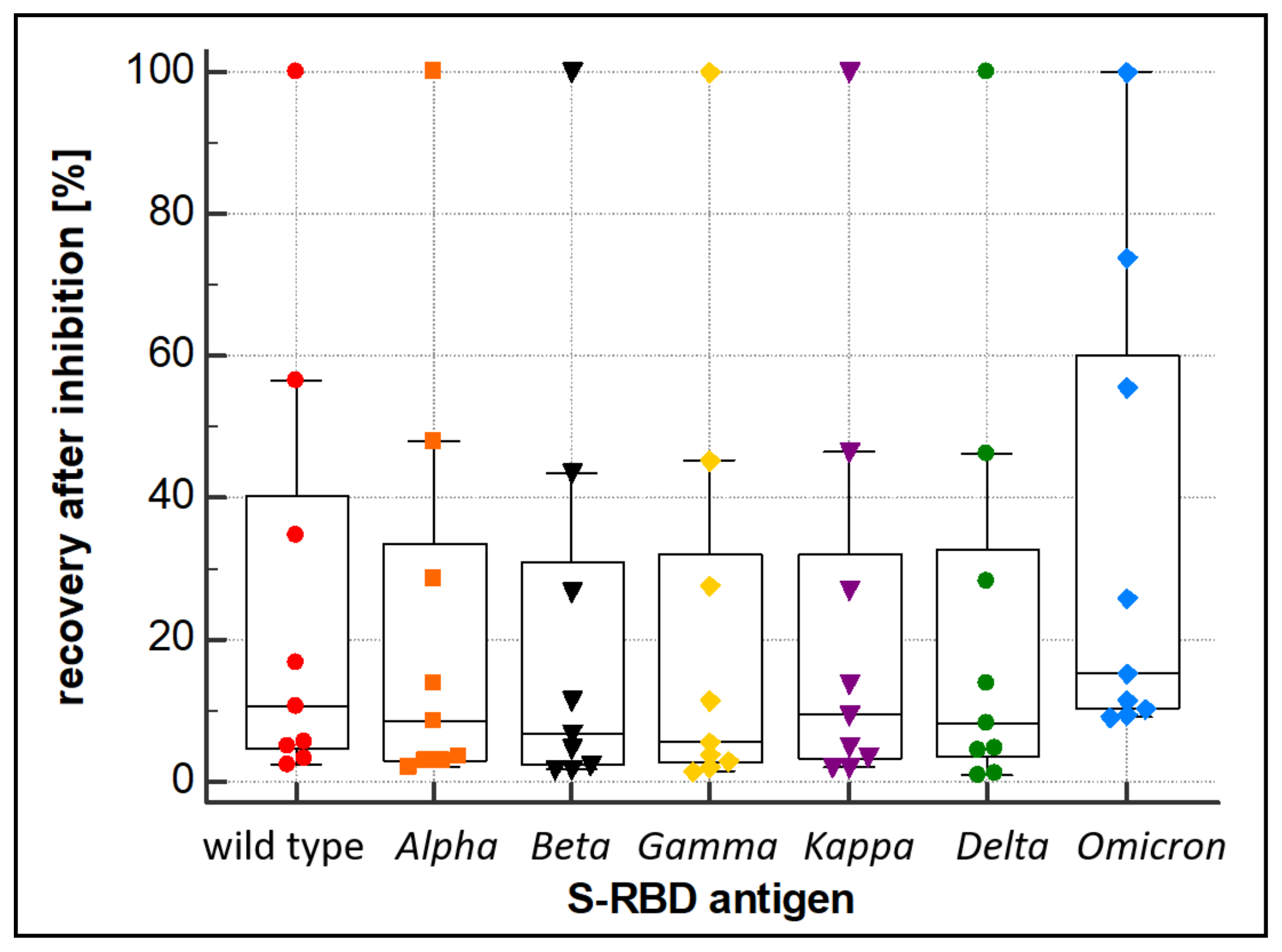
3.3. Evaluation of Variant Cross-Reactivity
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 22 January 2021).
- Stasi, C.; Fallani, S.; Voller, F.; Silvestri, C. Treatment for COVID-19: An Overview. Eur. J. Pharm. 2020, 889, 173644. [Google Scholar] [CrossRef]
- Machhi, J.; Herskovitz, J.; Senan, A.M.; Dutta, D.; Nath, B.; Oleynikov, M.D.; Blomberg, W.R.; Meigs, D.D.; Hasan, M.; Patel, M.; et al. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharm. 2020, 15, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A Comparative Overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Xiaojie, S.; Yu, L.; lei, Y.; Guang, Y.; Min, Q. Neutralizing Antibodies Targeting SARS-CoV-2 Spike Protein. Stem Cell Res. 2021, 50, 102125. [Google Scholar] [CrossRef]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C.; Rawlings, S.; Smith, D.; Das, S.; Strich, J.R.; Chertow, D.S.; Davey, R.T.; Cohen, J.I. Detection of Nucleocapsid Antibody to SARS-CoV-2 Is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Van Elslande, J.; Oyaert, M.; Lorent, N.; Vande Weygaerde, Y.; Van Pottelbergh, G.; Godderis, L.; Van Ranst, M.; André, E.; Padalko, E.; Lagrou, K.; et al. Lower Persistence of Anti-Nucleocapsid Compared to Anti-Spike Antibodies up to One Year after SARS-CoV-2 Infection. Diagn. Microbiol. Infect. Dis. 2022, 103, 115659. [Google Scholar] [CrossRef]
- Favresse, J.; Gillot, C.; Di Chiaro, L.; Eucher, C.; Elsen, M.; Van Eeckhoudt, S.; David, C.; Morimont, L.; Dogné, J.-M.; Douxfils, J. Neutralizing Antibodies in COVID-19 Patients and Vaccine Recipients after Two Doses of BNT162b2. Viruses 2021, 13, 1364. [Google Scholar] [CrossRef] [PubMed]
- Musicò, A.; Frigerio, R.; Mussida, A.; Barzon, L.; Sinigaglia, A.; Riccetti, S.; Gobbi, F.; Piubelli, C.; Bergamaschi, G.; Chiari, M.; et al. SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region. Vaccines 2021, 9, 35. [Google Scholar] [CrossRef]
- Klausberger, M.; Duerkop, M.; Haslacher, H.; Wozniak-Knopp, G.; Cserjan-Puschmann, M.; Perkmann, T.; Lingg, N.; Aguilar, P.P.; Laurent, E.; De Vos, J.; et al. A Comprehensive Antigen Production and Characterisation Study for Easy-to-Implement, Specific and Quantitative SARS-CoV-2 Serotests. EBioMedicine 2021, 67, 103348. [Google Scholar] [CrossRef]
- Mittal, A.; Manjunath, K.; Ranjan, R.K.; Kaushik, S.; Kumar, S.; Verma, V. COVID-19 Pandemic: Insights into Structure, Function, and HACE2 Receptor Recognition by SARS-CoV-2. PLOS Pathog. 2020, 16, e1008762. [Google Scholar] [CrossRef]
- Lloyd, S.B.; Kent, S.J.; Winnall, W.R. The High Cost of Fidelity. AIDS Res. Hum. Retrovir. 2014, 30, 8–16. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 6 October 2022).
- Emma Hodcroft. CoVariants. Available online: https://covariants.org/ (accessed on 7 October 2022).
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. MRNA Vaccine-Elicited Antibodies to SARS-CoV-2 and Circulating Variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Lorenzo-Redondo, R.; Ozer, E.A.; Hultquist, J.F. Covid-19: Is Omicron Less Lethal than Delta? BMJ 2022, 378, o1806. [Google Scholar] [CrossRef]
- Kleanthous, H.; Silverman, J.M.; Makar, K.W.; Yoon, I.-K.; Jackson, N.; Vaughn, D.W. Scientific Rationale for Developing Potent RBD-Based Vaccines Targeting COVID-19. npj Vaccines 2021, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Wei, H.; Li, Q.; Liu, L.; Li, B. Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis. Front. Mol. Biosci. 2021, 8, 682405. [Google Scholar] [CrossRef] [PubMed]
- Möhlendick, B.; Čiučiulkaitė, I.; Elsner, C.; Anastasiou, O.E.; Trilling, M.; Wagner, B.; Zwanziger, D.; Jöckel, K.-H.; Dittmer, U.; Siffert, W. Individuals With Weaker Antibody Responses After Booster Immunization Are Prone to Omicron Breakthrough Infections. Front. Immunol. 2022, 13, 907343. [Google Scholar] [CrossRef]
- EMA. Adapted Vaccine Targeting BA.4 and BA.5 Omicron Variants Original SARS-CoV-2 Recommended for Approval. European Medicines Agency. Available online: https://www.ema.europa.eu/en/news/adapted-vaccine-targeting-ba4-ba5-omicron-variants-original-sars-cov-2-recommended-approval (accessed on 7 October 2022).
- Zeng, B.; Gao, L.; Zhou, Q.; Yu, K.; Sun, F. Effectiveness of COVID-19 Vaccines against SARS-CoV-2 Variants of Concern: A Systematic Review and Meta-Analysis. BMC Med. 2022, 20, 200. [Google Scholar] [CrossRef]
- Regev-Yochay, G.; Gonen, T.; Gilboa, M.; Mandelboim, M.; Indenbaum, V.; Amit, S.; Meltzer, L.; Asraf, K.; Cohen, C.; Fluss, R.; et al. 4th Dose COVID MRNA Vaccines’ Immunogenicity & Efficacy Against Omicron VOC. medRxiv 2022. [Google Scholar] [CrossRef]
- Jaago, M.; Rähni, A.; Pupina, N.; Pihlak, A.; Sadam, H.; Tuvikene, J.; Avarlaid, A.; Planken, A.; Planken, M.; Haring, L.; et al. Differential Patterns of Cross-Reactive Antibody Response against SARS-CoV-2 Spike Protein Detected for Chronically Ill and Healthy COVID-19 Naïve Individuals. Sci. Rep. 2022, 12, 16817. [Google Scholar] [CrossRef]
- Rodgers, M.A.; Olivo, A.; Harris, B.J.; Lark, C.; Luo, X.; Berg, M.G.; Meyer, T.V.; Mohaimani, A.; Orf, G.S.; Goldstein, Y.; et al. Detection of SARS-CoV-2 Variants by Abbott Molecular, Antigen, and Serological Tests. J. Clin. Virol. 2022, 147, 105080. [Google Scholar] [CrossRef]
- Johns Hopkins Center for Health Security. Variants of Concern (VOCs) and Testing. COVID-19 Testing Toolkit. Available online: https://www.centerforhealthsecurity.org/covid-19TestingToolkit/testing-basics/VOCs.html (accessed on 7 October 2022).
- Lippi, G.; Plebani, M. The Critical Role of Laboratory Medicine during Coronavirus Disease 2019 (COVID-19) and Other Viral Outbreaks. Clin. Chem. Lab. Med. 2020, 58, 1063–1069. [Google Scholar] [CrossRef]
- Adeli, K. Critical Role of Laboratory Medicine in the Global Response to the COVID-19 Pandemic. Clin. Chem. Lab. Med. 2020, 58, 1019–1020. [Google Scholar] [CrossRef]
- 360Dx. Coronavirus Test Tracker: Commercially Available COVID-19 Diagnostic Tests. Available online: https://www.360dx.com/coronavirus-test-tracker-launched-covid-19-tests (accessed on 7 October 2022).
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-QPCR), Direct RT-QPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a Novel Antigen-Based Rapid Detection Test for the Diagnosis of SARS-CoV-2 in Respiratory Samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Evaluation of Rapid Antigen Test for Detection of SARS-CoV-2 Virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef] [PubMed]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, Point-of-Care Antigen and Molecular-Based Tests for Diagnosis of SARS-CoV-2 Infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. [Google Scholar] [CrossRef]
- Plebani, M.; Padoan, A.; Fedeli, U.; Schievano, E.; Vecchiato, E.; Lippi, G.; Lo Cascio, G.; Porru, S.; Palù, G. SARS-CoV-2 Serosurvey in Health Care Workers of the Veneto Region. Clin. Chem. Lab. Med. 2020, 58, 2107–2111. [Google Scholar] [CrossRef] [PubMed]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.-P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic Accuracy of Serological Tests for Covid-19: Systematic Review and Meta-Analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Herzberg, J.; Fischer, B.; Lindenkamp, C.; Becher, H.; Becker, A.-K.; Honarpisheh, H.; Guraya, S.Y.; Strate, T.; Knabbe, C. Persistence of Immune Response in Health Care Workers After Two Doses BNT162b2 in a Longitudinal Observational Study. Front. Immunol. 2022, 13, 701. [Google Scholar] [CrossRef]
- Brehm, J.; Spaeth, A.; Dreßler, L.; Masetto, T.; Dannenberg, R.; Peter, C.; Grimmler, M. SARS-CoV-2 Antibody Progression and Neutralizing Potential in Mild Symptomatic COVID-19 Patients—A Comparative Long Term Post-Infection Study. Front. Immunol. 2022, 13, 915338. [Google Scholar] [CrossRef]
- Weidner, L.; Gänsdorfer, S.; Unterweger, S.; Weseslindtner, L.; Drexler, C.; Farcet, M.; Witt, V.; Schistal, E.; Schlenke, P.; Kreil, T.R.; et al. Quantification of SARS-CoV-2 Antibodies with Eight Commercially Available Immunoassays. J. Clin. Virol. 2020, 129, 104540. [Google Scholar] [CrossRef] [PubMed]
- Roche. Cobas e 601 Module. Available online: https://diagnostics.roche.com/global/en/products/instruments/cobas-e-601.html (accessed on 10 March 2021).
- Spaeth, A.; Masetto, T.; Brehm, J.; Wey, L.; Kochem, C.; Brehm, M.; Peter, C.; Grimmler, M. Characterization of the Diagnostic Performance of a Novel COVID-19 PETIA in Comparison to Four Routine N-, S- and RBD-Antigen Based Immunoassays. Diagnostics 2021, 11, 1332. [Google Scholar] [CrossRef]
- Lippi, G.; Adeli, K.; Plebani, M. Commercial Immunoassays for Detection of Anti-SARS-CoV-2 Spike and RBD Antibodies: Urgent Call for Validation against New and Highly Mutated Variants. Clin. Chem. Lab. Med. 2022, 60, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple Sequence Alignment with Hierarchical Clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Borque, L.; Bellod, L.; Rus, A.; Seco, M.L.; Galisteo-González, F. Development and Validation of an Automated and Ultrasensitive Immunoturbidimetric Assay for C-Reactive Protein. Clin. Chem. 2000, 46, 1839–1842. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, M.P.S.; Martín-Molina, A.; Ramos, J.; Rus, A.; Borque, L.; Forcada, J.; Galisteo-González, F. Amino, Chloromethyl and Acetal-Functionalized Latex Particles for Immunoassays: A Comparative Study. J. Immunol. Methods 2004, 287, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181. [Google Scholar] [CrossRef]
- Sheahan, T.; Deming, D.; Donaldson, E.; Pickles, R.; Baric, R. Resurrection of an “Extinct” SARS-CoV Isolate GD03 from LATE 2003. Nidoviruses 2006, 581, 547–550. [Google Scholar] [CrossRef]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The Receptor Binding Domain of the Viral Spike Protein Is an Immunodominant and Highly Specific Target of Antibodies in SARS-CoV-2 Patients. Sci. Immunol. 2020, 5, eabc8413. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Hernández-Carralero, E.; Paz-Cabrera, M.C.; Cabrera, E.; Hernández-Reyes, Y.; Hernández-Fernaud, J.R.; Gillespie, D.A.; Salido, E.; Hernández-Porto, M.; Freire, R. The Nucleocapsid Protein Triggers the Main Humoral Immune Response in COVID-19 Patients. Biochem. Biophys. Res. Commun. 2021, 543, 45–49. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- World Medical Association. WMA Declaration of Helsinki—Ethical Principles for Medical Research Involving Human Subjects. Available online: https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/ (accessed on 11 November 2022).



| Virus | Identity of Related Viral Proteins to SARS-CoV-2 (%) (Score) | ||
|---|---|---|---|
| S-RBD SARS-CoV-2 | S Protein SARS-CoV-2 | N Protein SARS-CoV-2 | |
| SARS-CoV-1 * | 73.1% (896) | 76.0% (5119) | 90.5% (1993) |
| MERS-CoV | 24.7% (86) | 34.1% (1249) | 50.9% (855) |
| HCoV-OC43 | 27.3% (130) | 37.3% (1108) | 42.0% (397) |
| HCoV-HKU1 | 24.5% (112) | 35.8% (1040) | 36.7% (424) |
| HCoV-NL63 | - * (42) | 33.3% (706) | 32.1% (260) |
| HCoV-229E | - * (35) | 34.4% (754) | 26.4% (153) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wey, L.; Masetto, T.; Spaeth, A.; Brehm, J.; Kochem, C.; Reinhart, M.; Müller, H.; Kempin, U.; Lorenz, F.; Peter, C.; et al. Bioinformatical Design and Performance Evaluation of a Nucleocapsid- and an RBD-Based Particle Enhanced Turbidimetric Immunoassay (PETIA) to Quantify the Wild Type and Variants of Concern-Derived Immunoreactivity of SARS-CoV-2. Biomedicines 2023, 11, 160. https://doi.org/10.3390/biomedicines11010160
Wey L, Masetto T, Spaeth A, Brehm J, Kochem C, Reinhart M, Müller H, Kempin U, Lorenz F, Peter C, et al. Bioinformatical Design and Performance Evaluation of a Nucleocapsid- and an RBD-Based Particle Enhanced Turbidimetric Immunoassay (PETIA) to Quantify the Wild Type and Variants of Concern-Derived Immunoreactivity of SARS-CoV-2. Biomedicines. 2023; 11(1):160. https://doi.org/10.3390/biomedicines11010160
Chicago/Turabian StyleWey, Leoni, Thomas Masetto, Alexander Spaeth, Jessica Brehm, Christian Kochem, Marco Reinhart, Holger Müller, Uwe Kempin, Franziska Lorenz, Christoph Peter, and et al. 2023. "Bioinformatical Design and Performance Evaluation of a Nucleocapsid- and an RBD-Based Particle Enhanced Turbidimetric Immunoassay (PETIA) to Quantify the Wild Type and Variants of Concern-Derived Immunoreactivity of SARS-CoV-2" Biomedicines 11, no. 1: 160. https://doi.org/10.3390/biomedicines11010160
APA StyleWey, L., Masetto, T., Spaeth, A., Brehm, J., Kochem, C., Reinhart, M., Müller, H., Kempin, U., Lorenz, F., Peter, C., & Grimmler, M. (2023). Bioinformatical Design and Performance Evaluation of a Nucleocapsid- and an RBD-Based Particle Enhanced Turbidimetric Immunoassay (PETIA) to Quantify the Wild Type and Variants of Concern-Derived Immunoreactivity of SARS-CoV-2. Biomedicines, 11(1), 160. https://doi.org/10.3390/biomedicines11010160












