Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation
Abstract
1. Introduction
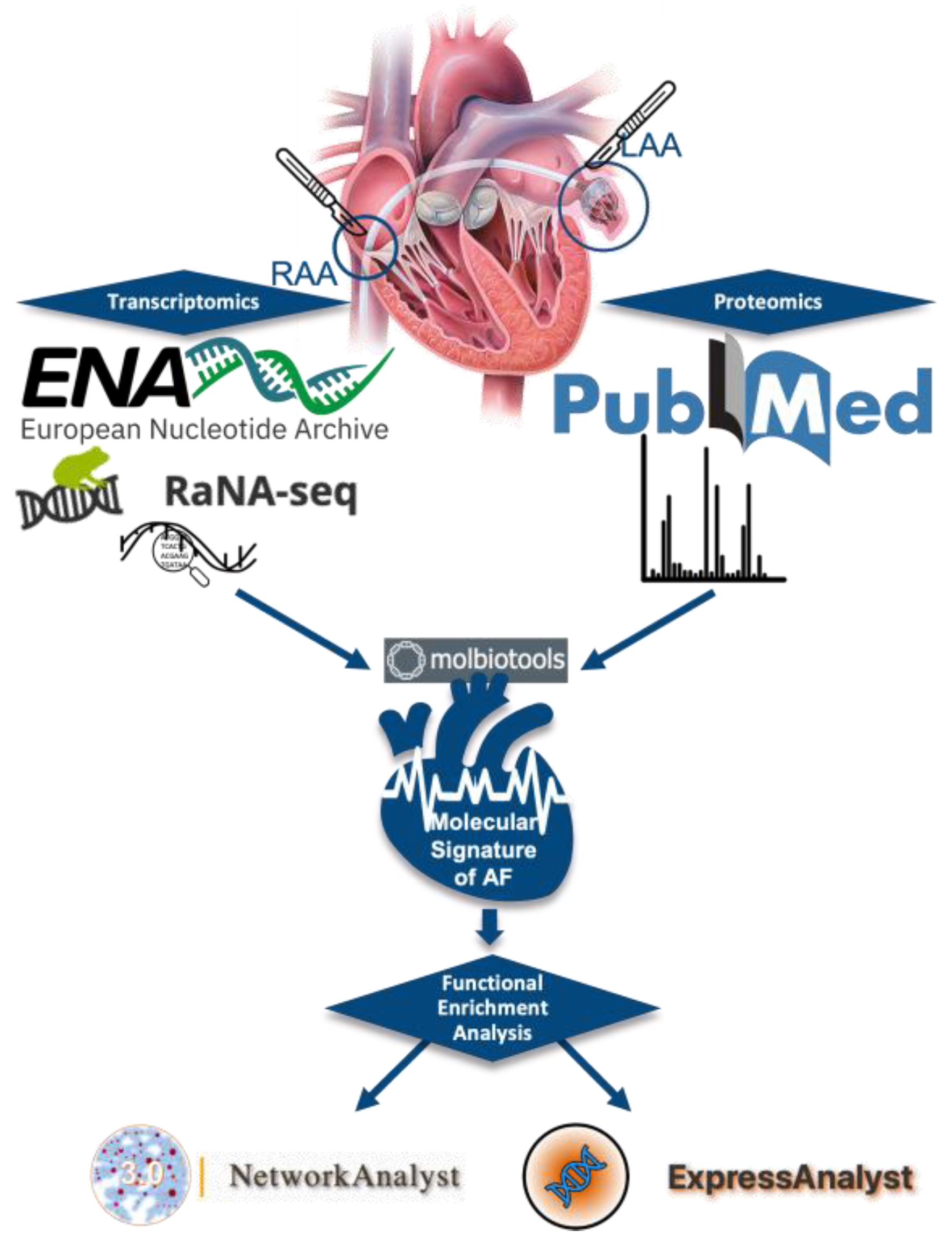
2. Materials and Methods
2.1. Data Source
2.1.1. RNA-SEQ Datasets Retrieval
2.1.2. Proteomics Data Retrieval
2.2. Data Processing
2.2.1. Identification of Differential Expression Genes
2.2.2. Selection of Differential Expression Proteins
2.2.3. Defining the AF Molecular Background
2.2.4. Functional Enrichment Analysis
2.2.5. Pathway Enrichment Analysis
3. Results
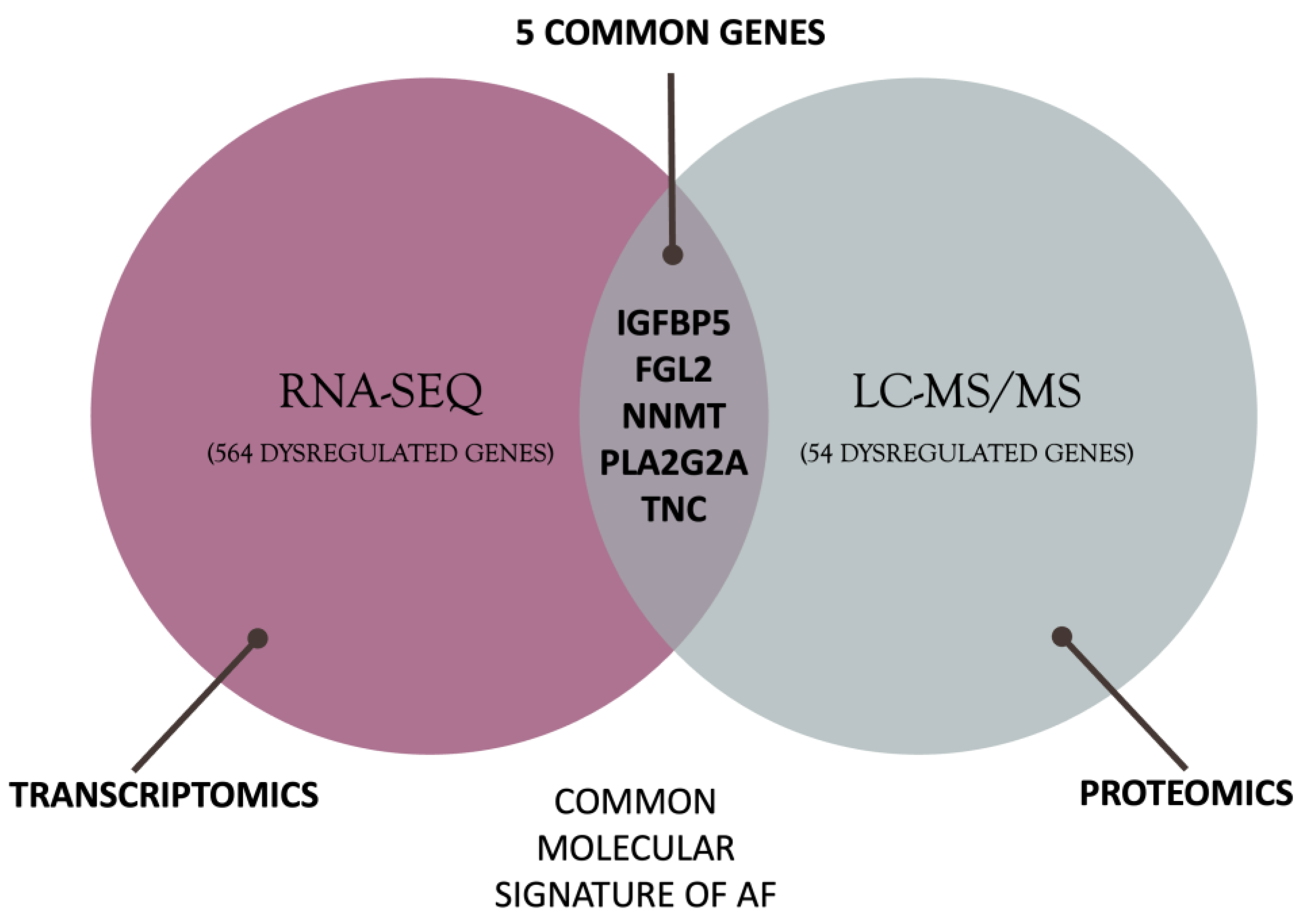
3.1. Differential Gene Expression Analysis
3.2. Differential Protein Expression Analysis
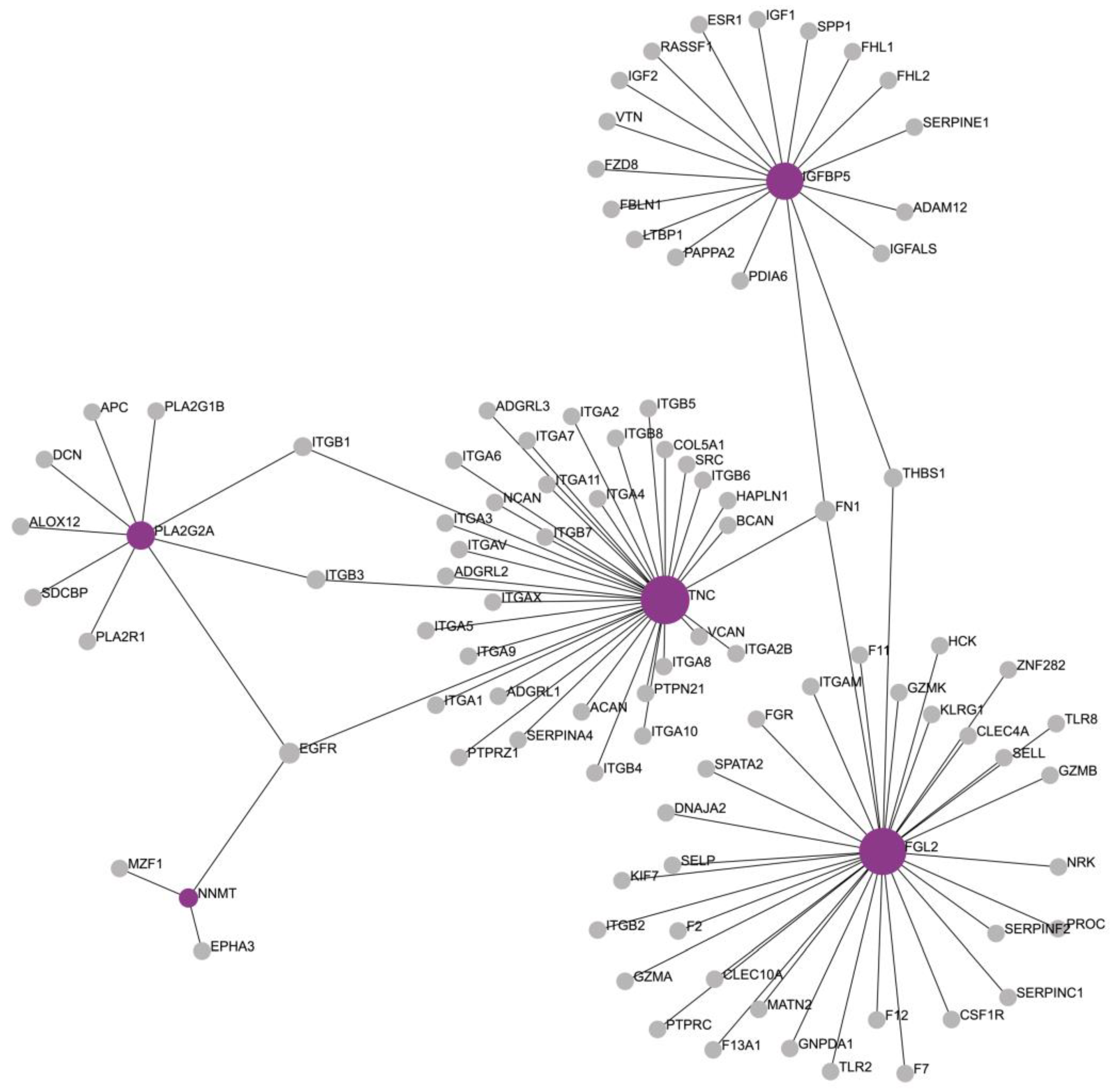
3.3. Defining the Molecular Background of AF
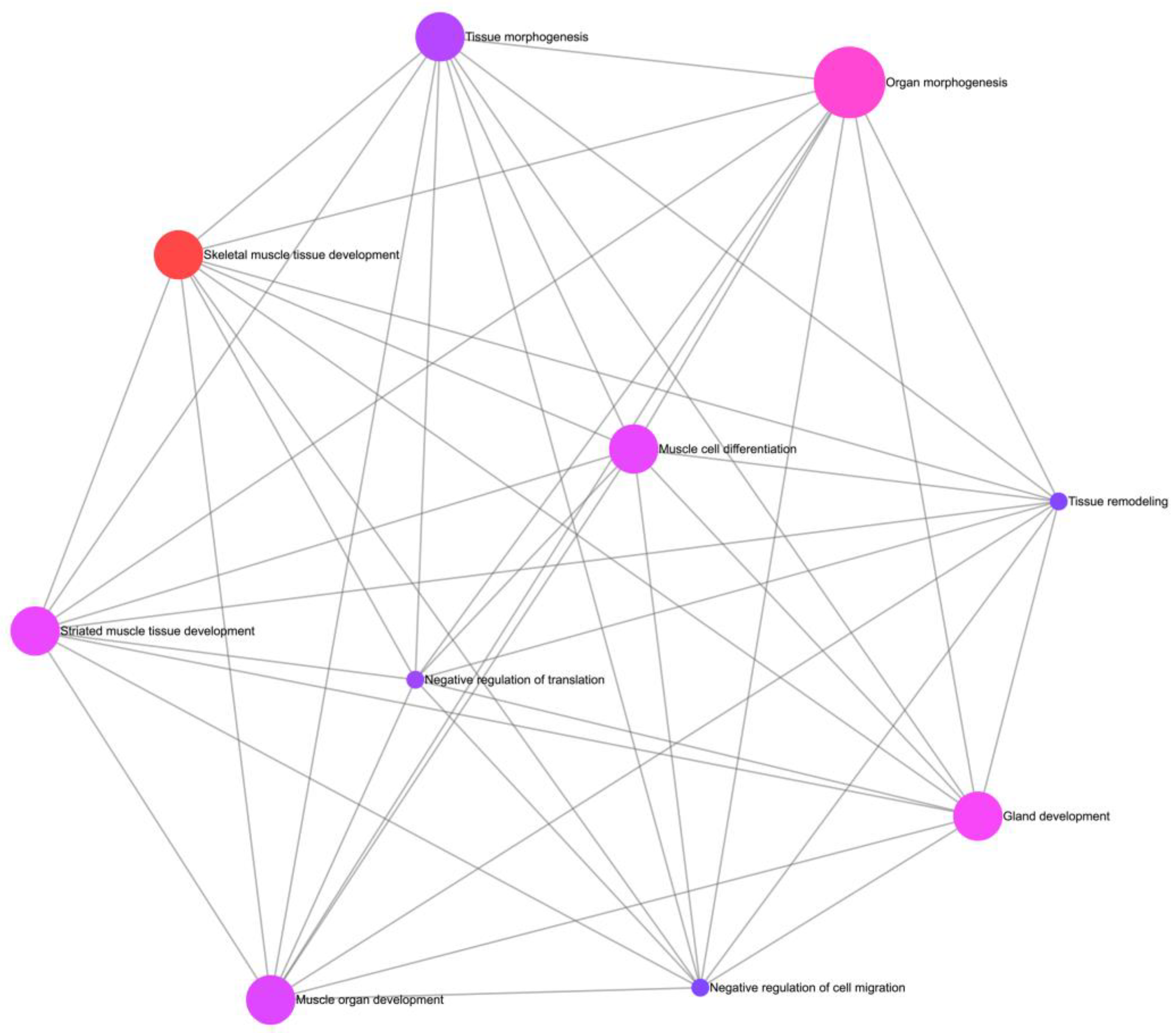
3.4. Functional Enrichment Analysis
3.5. Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lip, G.Y.; Kakar, P.; Watson, T. Atrial fibrillation--the growing epidemic. Heart 2007, 93, 542–543. [Google Scholar] [CrossRef]
- Chugh, S.S.; Blackshear, J.L.; Shen, W.K.; Hammill, S.C.; Gersh, B.J. Epidemiology and natural history of atrial fibrillation: Clinical implications. J. Am. Coll. Cardiol. 2001, 37, 371–378. [Google Scholar] [CrossRef]
- Roger, V.L.; Go, A.S.; Lloyd-Jones, D.M.; Benjamin, E.J.; Berry, J.D.; Borden, W.B.; Bravata, D.M.; Dai, S.; Ford, E.S.; Fox, C.S.; et al. Heart Disease and Stroke Statistics—2012 Update. Circulation 2012, 125, e2–e220. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Cervellin, G. Global epidemiology of atrial fibrillation: An increasing epidemic and public health challenge. Int. J. Stroke 2021, 16, 217–221. [Google Scholar] [CrossRef]
- Hart, R.G.; Halperin, J.L. Atrial Fibrillation and Stroke. Stroke 2001, 32, 803–808. [Google Scholar] [CrossRef]
- Lau, D.H.; Nattel, S.; Kalman, J.M.; Sanders, P. Modifiable Risk Factors and Atrial Fibrillation. Circulation 2017, 136, 583–596. [Google Scholar] [CrossRef]
- Nattel, S. New ideas about atrial fibrillation 50 years on. Nature 2002, 415, 219–226. [Google Scholar] [CrossRef]
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.-H.; McAnulty, J.H., Jr.; Zheng, Z.-J.; et al. Worldwide epidemiology of atrial fibrillation: A Global Burden of Disease 2010 Study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef]
- Jalife, J. Mechanisms of persistent atrial fibrillation. Curr. Opin. Cardiol. 2014, 29, 20–27. [Google Scholar]
- Forleo, G.B.; Tondo, C. Atrial fibrillation: Cure or treat? Ther. Adv. Cardiovasc. Dis. 2009, 3, 187–196. [Google Scholar] [CrossRef]
- Bertaglia, E.; Tondo, C.; De Simone, A.; Zoppo, F.; Mantica, M.; Turco, P.; Iuliano, A.; Forleo, G.; La Rocca, V.; Stabile, G. Does catheter ablation cure atrial fibrillation? Single-procedure outcome of drug-refractory atrial fibrillation ablation: A 6-year multicentre experience. Europace 2010, 12, 181–187. [Google Scholar] [CrossRef]
- Nesheiwat, Z.; Goyal, A.; Jagtap, M. Atrial Fibrillation. In StatPearls; StatPearls Publishing Copyright © 2023; StatPearls Publishing LLC: Treasure Island, FL, USA, 2023. [Google Scholar]
- Chai-Adisaksopha, C.; Hillis, C.; Isayama, T.; Lim, W.; Iorio, A.; Crowther, M. Mortality outcomes in patients receiving direct oral anticoagulants: A systematic review and meta-analysis of randomized controlled trials. J. Thromb. Haemost. 2015, 13, 2012–2020. [Google Scholar] [CrossRef]
- Boos, C.J.; Carlsson, J.; More, R.S. Rate or rhythm control in persistent atrial fibrillation? QJM Int. J. Med. 2003, 96, 881–892. [Google Scholar] [CrossRef][Green Version]
- Meyer, M.; Lustgarten, D. Beta-blockers in atrial fibrillation—Trying to make sense of unsettling results. Europace 2023, 25, 260–262. [Google Scholar] [CrossRef]
- Wijffels, M.C.E.F.; Kirchhof, C.J.H.J.; Dorland, R.; Allessie, M.A. Atrial Fibrillation Begets Atrial Fibrillation. Circulation 1995, 92, 1954–1968. [Google Scholar] [CrossRef]
- Nattel, S.; Harada, M. Atrial Remodeling and Atrial Fibrillation: Recent Advances and Translational Perspectives. J. Am. Coll. Cardiol. 2014, 63, 2335–2345. [Google Scholar] [CrossRef]
- Walters, T.E.; Nisbet, A.; Morris, G.M.; Tan, G.; Mearns, M.; Teo, E.; Lewis, N.; Ng, A.; Gould, P.; Lee, G.; et al. Progression of atrial remodeling in patients with high-burden atrial fibrillation: Implications for early ablative intervention. Heart Rhythm 2016, 13, 331–339. [Google Scholar] [CrossRef]
- Khomtchouk, B.B.; Tran, D.-T.; Vand, K.A.; Might, M.; Gozani, O.; Assimes, T.L. Cardioinformatics: The nexus of bioinformatics and precision cardiology. Brief. Bioinform. 2020, 21, 2031–2051. [Google Scholar] [CrossRef]
- Xiao, S.; Zhou, Y.; Liu, Q.; Zhang, T.; Pan, D. Identification of Pivotal MicroRNAs and Target Genes Associated with Persistent Atrial Fibrillation Based on Bioinformatics Analysis. Comput. Math. Methods Med. 2021, 2021, 6680211. [Google Scholar] [CrossRef]
- Li, J.; Wu, Z.; Zheng, D.; Sun, Y.; Wang, S.; Yan, Y. Bioinformatics analysis of the regulatory lncRNA-miRNA-mRNA network and drug prediction in patients with hypertrophic cardiomyopathy. Mol. Med. Rep. 2019, 20, 549–558. [Google Scholar] [CrossRef]
- Botto, G.L.; Luzi, M.; Sagone, A. Atrial fibrillation: The remodelling phenomenon. Eur. Heart J. Suppl. 2003, 5, H1–H7. [Google Scholar] [CrossRef]
- Bennett, P.B.; Guthrie, H.R. Trends in ion channel drug discovery: Advances in screening technologies. Trends Biotechnol. 2003, 21, 563–569. [Google Scholar] [CrossRef]
- Goette, A.; Lendeckel, U.; Klein, H.U. Signal transduction systems and atrial fibrillation. Cardiovasc. Res. 2002, 54, 247–258. [Google Scholar] [CrossRef]
- Barth, A.S.; Merk, S.; Arnoldi, E.; Zwermann, L.; Kloos, P.; Gebauer, M.; Steinmeyer, K.; Bleich, M.; Kääb, S.; Hinterseer, M.; et al. Reprogramming of the Human Atrial Transcriptome in Permanent Atrial Fibrillation. Circ. Res. 2005, 96, 1022–1029. [Google Scholar] [CrossRef]
- Nattel, S.; Burstein, B.; Dobrev, D. Atrial Remodeling and Atrial Fibrillation. Circ. Arrhythmia Electrophysiol. 2008, 1, 62–73. [Google Scholar] [CrossRef]
- Harrison, P.W.; Ahamed, A.; Aslam, R.; Alako, B.T.F.; Burgin, J.; Buso, N.; Courtot, M.; Fan, J.; Gupta, D.; Haseeb, M.; et al. The European Nucleotide Archive in 2020. Nucleic Acids Res. 2021, 49, D82–D85. [Google Scholar] [CrossRef]
- Thomas, A.M.; Cabrera, C.P.; Finlay, M.; Lall, K.; Nobles, M.; Schilling, R.J.; Wood, K.; Mein, C.A.; Barnes, M.R.; Munroe, P.B.; et al. Differentially expressed genes for atrial fibrillation identified by RNA sequencing from paired human left and right atrial appendages. Physiol. Genom. 2019, 51, 323–332. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, J.; Shao, Y. Integrative analysis reveals essential mRNA, long non-coding RNA (lncRNA), and circular RNA (circRNA) in paroxysmal and persistent atrial fibrillation patients. Anatol. J. Cardiol. 2021, 25, 414–428. [Google Scholar] [CrossRef]
- Zhu, X.; Tang, X.; Chong, H.; Cao, H.; Fan, F.; Pan, J.; Wang, D.; Zhou, Q. Expression Profiles of Circular RNA in Human Atrial Fibrillation with Valvular Heart Diseases. Front. Cardiovasc. Med. 2020, 7, 597932. [Google Scholar] [CrossRef]
- Darkow, E.; Nguyen, T.T.; Stolina, M.; Kari, F.A.; Schmidt, C.; Wiedmann, F.; Baczkó, I.; Kohl, P.; Rajamani, S.; Ravens, U.; et al. Small Conductance Ca2+ -Activated K+ (SK) Channel mRNA Expression in Human Atrial and Ventricular Tissue: Comparison Between Donor, Atrial Fibrillation and Heart Failure Tissue. Front. Physiol. 2021, 12, 650964. [Google Scholar] [CrossRef]
- Liu, Y.; Bai, F.; Tang, Z.; Liu, N.; Liu, Q. Integrative transcriptomic, proteomic, and machine learning approach to identifying feature genes of atrial fibrillation using atrial samples from patients with valvular heart disease. BMC Cardiovasc. Disord. 2021, 21, 52. [Google Scholar] [CrossRef]
- Prieto, C.; Barrios, D. RaNA-Seq: Interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics 2020, 36, 1955–1956. [Google Scholar] [CrossRef]
- Tools, M.B. Molecular Biology Tools. Available online: https://molbiotools.com (accessed on 1 March 2023).
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Yu, J.; Li, J.; Shen, J.; Du, F.; Wu, X.; Li, M.; Chen, Y.; Cho, C.H.; Li, X.; Xiao, Z.; et al. The role of Fibrinogen-like proteins in Cancer. Int. J. Biol. Sci. 2021, 17, 1079–1087. [Google Scholar] [CrossRef]
- Fan, C.; Wang, J.; Mao, C.; Li, W.; Liu, K.; Wang, Z. The FGL2 prothrombinase contributes to the pathological process of experimental pulmonary hypertension. J. Appl. Physiol. (1985) 2019, 127, 1677–1687. [Google Scholar] [CrossRef]
- Fan, C.; Chen, H.; Liu, K.; Wang, Z. Fibrinogen-like protein 2 contributes to normal murine cardiomyocyte maturation and heart development. Exp. Physiol. 2021, 106, 1559–1571. [Google Scholar] [CrossRef]
- Hozayen, S.M.; Zychowski, D.; Benson, S.; Lutsey, P.L.; Haslbauer, J.; Tzankov, A.; Kaltenborn, Z.; Usher, M.; Shah, S.; Tignanelli, C.J.; et al. Outpatient and inpatient anticoagulation therapy and the risk for hospital admission and death among COVID-19 patients. eClinicalMedicine 2021, 41, 101139. [Google Scholar] [CrossRef]
- Zheng, Z.; Yu, L.; Wu, Y.; Wu, H. FGL2 knockdown improves heart function through regulation of TLR9 signaling in the experimental autoimmune myocarditis rats. Immunol. Res. 2018, 66, 52–58. [Google Scholar] [CrossRef]
- Cerveró, J.; Segura, V.; Macías, A.; Gavira, J.J.; Montes, R.; Hermida, J. Atrial fibrillation in pigs induces left atrial endocardial transcriptional remodelling. Thromb. Haemost. 2012, 108, 742–749. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, J.E.; Kang, Y.J. Insulin Like Growth Factor Binding Protein-5 Regulates Excessive Vascular Smooth Muscle Cell Proliferation in Spontaneously Hypertensive Rats via ERK 1/2 Phosphorylation. Korean J. Physiol. Pharmacol. 2013, 17, 157–162. [Google Scholar] [CrossRef]
- James, P.L.; Jones, S.B.; Busby, W.H.; Clemmons, D.R.; Rotwein, P. A highly conserved insulin-like growth factor-binding protein (IGFBP-5) is expressed during myoblast differentiation. J. Biol. Chem. 1993, 268, 22305–22312. [Google Scholar] [CrossRef]
- Hoffmann, S.; Schmitteckert, S.; Raedecke, K.; Rheinert, D.; Diebold, S.; Roeth, R.; Weiss, B.; Granzow, M.; Niesler, B.; Griesbeck, A.; et al. Network-driven discovery yields new insight into Shox2-dependent cardiac rhythm control. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2021, 1864, 194702. [Google Scholar] [CrossRef]
- Wass, S.Y.; Offerman, E.J.; Sun, H.; Hsu, J.; Rennison, J.H.; Cantlay, C.C.; McHale, M.L.; Gillinov, A.M.; Moravec, C.; Smith, J.D.; et al. Identifying functional genes and pathways towards a unifying model for atrial fibrillation. medRxiv 2021. [Google Scholar] [CrossRef]
- Linscheid, N.; Poulsen, P.C.; Pedersen, I.D.; Gregers, E.; Svendsen, J.H.; Olesen, M.S.; Olsen, J.V.; Delmar, M.; Lundby, A. Quantitative Proteomics of Human Heart Samples Collected In Vivo Reveal the Remodeled Protein Landscape of Dilated Left Atrium without Atrial Fibrillation. Mol. Cell Proteom. 2020, 19, 1132–1144. [Google Scholar] [CrossRef]
- Chen, C.-L.; Lin, J.-L.; Lai, L.-P.; Pan, C.-H.; Huang, S.K.S.; Lin, C.-S. Altered expression of FHL1, CARP, TSC-22 and P311 provide insights into complex transcriptional regulation in pacing-induced atrial fibrillation. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2007, 1772, 317–329. [Google Scholar] [CrossRef]
- Chilukoti, R.K.; Giese, A.; Malenke, W.; Homuth, G.; Bukowska, A.; Goette, A.; Felix, S.B.; Kanaan, J.; Wollert, H.G.; Evert, K.; et al. Atrial fibrillation and rapid acute pacing regulate adipocyte/adipositas-related gene expression in the atria. Int. J. Cardiol. 2015, 187, 604–613. [Google Scholar] [CrossRef]
- Nguyen, X.-X.; Sanderson, M.; Helke, K.; Feghali-Bostwick, C. Phenotypic Characterization of Transgenic Mice Expressing Human IGFBP-5. Int. J. Mol. Sci. 2021, 22, 335. [Google Scholar] [CrossRef]
- Lu, S.; Ke, S.; Wang, C.; Xu, Y.; Li, Z.; Song, K.; Bai, M.; Zhou, M.; Yu, H.; Yin, B.; et al. NNMT promotes the progression of intrahepatic cholangiocarcinoma by regulating aerobic glycolysis via the EGFR-STAT3 axis. Oncogenesis 2022, 11, 39. [Google Scholar] [CrossRef]
- Rehan, M.; Deskin, B.; Kurundkar, A.R.; Yadav, S.; Matsunaga, Y.; Manges, J.; Smith, N.; Dsouza, K.G.; Burow, M.E.; Thannickal, V.J. Nicotinamide N-methyltransferase mediates lipofibroblast-myofibroblast transition and apoptosis resistance. J. Biol. Chem. 2023, 299, 105027. [Google Scholar] [CrossRef]
- Liu, M.; He, A.; Chu, J.; Chen, C.; Zhang, S.; He, Y.; Tao, W.; Lu, M.; Hua, M.; Ju, W.; et al. Serum N(1)-methylnicotinamide is Associated with Left Ventricular Systolic Dysfunction in Chinese. Sci. Rep. 2018, 8, 8581. [Google Scholar] [CrossRef]
- Song, Z.; Zhong, X.; Li, M.; Gao, P.; Ning, Z.; Sun, Z.; Song, X. 1-MNA Ameliorates High Fat Diet-Induced Heart Injury by Upregulating Nrf2 Expression and Inhibiting NF-κB in vivo and in vitro. Front. Cardiovasc. Med. 2021, 8, 721814. [Google Scholar] [CrossRef]
- Lemaitre, R.N.; Bartz, T.M.; King, I.B.; Brody, J.A.; McKnight, B.; Sotoodehnia, N.; Rea, T.D.; Johnson, C.O.; Mozaffarian, D.; Hesselson, S.; et al. Circulating n-3 fatty acids and trans-fatty acids, PLA2G2A gene variation and sudden cardiac arrest. J. Nutr. Sci. 2016, 5, e12. [Google Scholar] [CrossRef][Green Version]
- Boilard, E.; Lai, Y.; Larabee, K.; Balestrieri, B.; Ghomashchi, F.; Fujioka, D.; Gobezie, R.; Coblyn, J.S.; Weinblatt, M.E.; Massarotti, E.M.; et al. A novel anti-inflammatory role for secretory phospholipase A2 in immune complex-mediated arthritis. EMBO Mol. Med. 2010, 2, 172–187. [Google Scholar] [CrossRef]
- Mallat, Z.; Lambeau, G.; Tedgui, A. Lipoprotein-Associated and Secreted Phospholipases A2 in Cardiovascular Disease. Circulation 2010, 122, 2183–2200. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, C.; Luo, J.; Liang, J. Dysfunctional Network and Mutation Genes of Hypertrophic Cardiomyopathy. J. Healthc. Eng. 2022, 2022, 8680178. [Google Scholar] [CrossRef]
- Newman, M.S.; Nguyen, T.; Watson, M.J.; Hull, R.W.; Yu, H.G. Transcriptome profiling reveals novel BMI- and sex-specific gene expression signatures for human cardiac hypertrophy. Physiol. Genom. 2017, 49, 355–367. [Google Scholar] [CrossRef]
- Park Woo, J.; Jeong, D.; Oh Jae, G. Tenascin-C in Cardiac Hypertrophy and Fibrosis. J. Am. Coll. Cardiol. 2017, 70, 1616–1617. [Google Scholar] [CrossRef]
- Shimojo, N.; Hashizume, R.; Kanayama, K.; Hara, M.; Suzuki, Y.; Nishioka, T.; Hiroe, M.; Yoshida, T.; Imanaka-Yoshida, K. Tenascin-C may accelerate cardiac fibrosis by activating macrophages via the integrin αVβ3/nuclear factor-κB/interleukin-6 axis. Hypertension 2015, 66, 757–766. [Google Scholar] [CrossRef]
- Imanaka-Yoshida, K.; Tawara, I.; Yoshida, T. Tenascin-C in cardiac disease: A sophisticated controller of inflammation, repair, and fibrosis. Am. J. Physiol.-Cell Physiol. 2020, 319, C781–C796. [Google Scholar] [CrossRef]
- Imanaka-Yoshida, K. Tenascin-C in Heart Diseases-The Role of Inflammation. Int. J. Mol. Sci. 2021, 22, 5828. [Google Scholar] [CrossRef]
- Golledge, J.; Clancy, P.; Maguire, J.; Lincz, L.; Koblar, S. The role of tenascin C in cardiovascular disease. Cardiovasc. Res. 2011, 92, 19–28. [Google Scholar] [CrossRef]
- Song, L.; Wang, L.; Li, F.; Yukht, A.; Qin, M.; Ruther, H.; Yang, M.; Chaux, A.; Shah, P.K.; Sharifi, B.G. Bone Marrow-Derived Tenascin-C Attenuates Cardiac Hypertrophy by Controlling Inflammation. J. Am. Coll. Cardiol. 2017, 70, 1601–1615. [Google Scholar] [CrossRef]
- Podesser, B.K.; Kreibich, M.; Dzilic, E.; Santer, D.; Förster, L.; Trojanek, S.; Abraham, D.; Krššák, M.; Klein, K.U.; Tretter, E.V.; et al. Tenascin-C promotes chronic pressure overload-induced cardiac dysfunction, hypertrophy and myocardial fibrosis. J. Hypertens. 2018, 36, 847–856. [Google Scholar] [CrossRef]
- Perera-Gonzalez, M.; Kiss, A.; Kaiser, P.; Holzweber, M.; Nagel, F.; Watzinger, S.; Acar, E.; Szabo, P.L.; Gonçalves, I.F.; Weber, L.; et al. The Role of Tenascin C in Cardiac Reverse Remodeling Following Banding-Debanding of the Ascending Aorta. Int. J. Mol. Sci. 2021, 22, 2023. [Google Scholar] [CrossRef]
- Bachir, A.I.; Horwitz, A.R.; Nelson, W.J.; Bianchini, J.M. Actin-Based Adhesion Modules Mediate Cell Interactions with the Extracellular Matrix and Neighboring Cells. Cold Spring Harb. Perspect. Biol. 2017, 9, a023234. [Google Scholar] [CrossRef]
- Yang, S.; Plotnikov, S.V. Mechanosensitive Regulation of Fibrosis. Cells 2021, 10, 994. [Google Scholar] [CrossRef]
- Sandbo, N.; Dulin, N. Actin cytoskeleton in myofibroblast differentiation: Ultrastructure defining form and driving function. Transl. Res. 2011, 158, 181–196. [Google Scholar] [CrossRef]
- Abreu-Blanco, M.T.; Watts, J.J.; Verboon, J.M.; Parkhurst, S.M. Cytoskeleton responses in wound repair. Cell. Mol. Life Sci. 2012, 69, 2469–2483. [Google Scholar] [CrossRef]
- Ohashi, K.; Fujiwara, S.; Mizuno, K. Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J. Biochem. 2017, 161, 245–254. [Google Scholar] [CrossRef]
- Meagher, P.B.; Lee, X.A.; Lee, J.; Visram, A.; Friedberg, M.K.; Connelly, K.A. Cardiac Fibrosis: Key Role of Integrins in Cardiac Homeostasis and Remodeling. Cells 2021, 10, 770. [Google Scholar] [CrossRef]
- Portokallidou, K.; Dovrolis, N.; Ragia, G.; Atzemian, N.; Kolios, G.; Manolopoulos, V.G. Multi-omics integration to identify the genetic expression and protein signature of dilated and ischemic cardiomyopathy. Front. Cardiovasc. Med. 2023, 10, 1115623. [Google Scholar] [CrossRef]
- Kim, M.C.; Kim, C.; Wood, L.; Neal, D.; Kamm, R.D.; Asada, H.H. Integrating focal adhesion dynamics, cytoskeleton remodeling, and actin motor activity for predicting cell migration on 3D curved surfaces of the extracellular matrix. Integr. Biol. 2012, 4, 1386–1397. [Google Scholar] [CrossRef]
- Samarel, A.M. Focal adhesion signaling in heart failure. Pflug. Arch. 2014, 466, 1101–1111. [Google Scholar] [CrossRef][Green Version]
- Buckley, B.J.R.; Harrison, S.L.; Gupta, D.; Fazio-Eynullayeva, E.; Underhill, P.; Lip, G.Y.H. Atrial Fibrillation in Patients with Cardiomyopathy: Prevalence and Clinical Outcomes From Real-World Data. J. Am. Heart Assoc. 2021, 10, e021970. [Google Scholar] [CrossRef]
- Yeung, C.; Enriquez, A.; Suarez-Fuster, L.; Baranchuk, A. Atrial fibrillation in patients with inherited cardiomyopathies. Europace 2019, 21, 22–32. [Google Scholar] [CrossRef]
- Nuzzi, V.; Cannatà, A.; Manca, P.; Castrichini, M.; Barbati, G.; Aleksova, A.; Fabris, E.; Zecchin, M.; Merlo, M.; Boriani, G.; et al. Atrial fibrillation in dilated cardiomyopathy: Outcome prediction from an observational registry. Int. J. Cardiol. 2021, 323, 140–147. [Google Scholar] [CrossRef]
- Ezeani, M.; Prabhu, S. Pathophysiology and therapeutic relevance of PI3K(p110α) protein in atrial fibrillation: A non-interventional molecular therapy strategy. Pharmacol. Res. 2021, 165, 105415. [Google Scholar] [CrossRef]
- Walkowski, B.; Kleibert, M.; Majka, M.; Wojciechowska, M. Insight into the Role of the PI3K/Akt Pathway in Ischemic Injury and Post-Infarct Left Ventricular Remodeling in Normal and Diabetic Heart. Cells 2022, 11, 1553. [Google Scholar] [CrossRef]
- Ezeani, M.; Elom, S. Necessity to evaluate PI3K/Akt signalling pathway in proarrhythmia. Open Heart 2017, 4, e000596. [Google Scholar] [CrossRef]
- McMullen, J.R.; Boey, E.J.; Ooi, J.Y.; Seymour, J.F.; Keating, M.J.; Tam, C.S. Ibrutinib increases the risk of atrial fibrillation, potentially through inhibition of cardiac PI3K-Akt signaling. Blood 2014, 124, 3829–3830. [Google Scholar] [CrossRef]
- Su, F.; Zhao, L.; Zhang, S.; Wang, J.; Chen, N.; Gong, Q.; Tang, J.; Wang, H.; Yao, J.; Wang, Q.; et al. Cardioprotection by PI3K-mediated signaling is required for anti-arrhythmia and myocardial repair in response to ischemic preconditioning in infarcted pig hearts. Lab. Investig. 2015, 95, 860–871. [Google Scholar] [CrossRef][Green Version]
- Kornej, J.; Büttner, P.; Hammer, E.; Engelmann, B.; Dinov, B.; Sommer, P.; Husser, D.; Hindricks, G.; Völker, U.; Bollmann, A. Circulating proteomic patterns in AF related left atrial remodeling indicate involvement of coagulation and complement cascade. PLoS ONE 2018, 13, e0198461. [Google Scholar] [CrossRef]
- Ren, J.; Tsilafakis, K.; Chen, L.; Lekkos, K.; Kostavasili, I.; Varela, A.; Cokkinos, D.V.; Davos, C.H.; Sun, X.; Song, J.; et al. Crosstalk between coagulation and complement activation promotes cardiac dysfunction in arrhythmogenic right ventricular cardiomyopathy. Theranostics 2021, 11, 5939–5954. [Google Scholar] [CrossRef]
- Liu, B.; Shi, X.; Ding, K.; Lv, M.; Qian, Y.; Zhu, S.; Guo, C.; Zhang, Y. The Joint Analysis of Multi-Omics Data Revealed the Methylation-Expression Regulations in Atrial Fibrillation. Front. Bioeng. Biotechnol. 2020, 8, 197. [Google Scholar] [CrossRef]
- Wang, B.; Lunetta, K.L.; Dupuis, J.; Lubitz, S.A.; Trinquart, L.; Yao, L.; Ellinor, P.T.; Benjamin, E.J.; Lin, H. Integrative Omics Approach to Identifying Genes Associated with Atrial Fibrillation. Circ. Res. 2020, 126, 350–360. [Google Scholar] [CrossRef]
- Liu, L.; Huang, J.; Wei, B.; Mo, J.; Wei, Q.; Chen, C.; Yan, W.; Huang, X.; He, F.; Qin, L.; et al. Multiomics Analysis of Genetics and Epigenetics Reveals Pathogenesis and Therapeutic Targets for Atrial Fibrillation. BioMed Res. Int. 2021, 2021, 6644827. [Google Scholar] [CrossRef]
- Assum, I.; Krause, J.; Scheinhardt, M.O.; Müller, C.; Hammer, E.; Börschel, C.S.; Völker, U.; Conradi, L.; Geelhoed, B.; Zeller, T.; et al. Tissue-specific multi-omics analysis of atrial fibrillation. Nat. Commun. 2022, 13, 441. [Google Scholar] [CrossRef]

| Dataset | Samples | Age (Years) | Sex M/F | Country | Tissue Type | Platform | Reference |
|---|---|---|---|---|---|---|---|
| PRJNA526687 | 5 Permanent AF | 73.6 ± 5.12 | 5/0 | United Kingdom | Paired LAA and RAA | GPL18573 | [28] |
| 5 SR | 62.4 ± 6.87 | 5/0 | |||||
| PRJNA531935 | 3 Persistent AF | 57 (51–64) | 2/1 | China | LAA | GPL20795 | [29] |
| 3 SR | 39 (38–42) | 2/1 | |||||
| PRJNA667522 | 10 Persistent AF | 53 ± 8 | 5/5 | China | LAA | GPL16791 | [30] |
| 10 SR | 55 ± 8 | 6/4 | |||||
| PRJEB42485 | 6 Sustained AF | 67.5 ± 1.7 | 6/0 | Germany | RAA | GPL20301 | [31] |
| 7 SR | 55.8 ± 8.3 | 4/3 |
| Samples | Age (Years) | Sex M/F | Country | Tissue Type | Platform | Reference |
|---|---|---|---|---|---|---|
| 9 Persistent AF | 55.5 ± 9.0 | 4/5 | China | LAA | Agilent 300Extend Q/ExactiveTM Plus (Thermo) | [32] |
| 9 SR | 50.5 ± 6.5 | 5/4 |
| KEGG Pathways | Total | Expected | Hits | p-Value | FDR |
|---|---|---|---|---|---|
| Extracellular matrix (ECM)–receptor interaction | 82 | 0.784 | 25 | 1.84 × 10−32 | 5.85 × 10−30 |
| Focal adhesion | 199 | 1.9 | 28 | 3.2 × 10−26 | 5.08 × 10−24 |
| Regulation of actin cytoskeleton | 214 | 2.05 | 28 | 2.59 × 10−25 | 2.75 × 10−23 |
| Hypertrophic cardiomyopathy (HCM) | 85 | 0.813 | 21 | 4.95 × 10−25 | 3.48 × 10−23 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 72 | 0.689 | 20 | 5.47 × 10−25 | 3.48 × 10−23 |
| Dilated cardiomyopathy | 91 | 0.87 | 21 | 2.42 × 10−24 | 1.28 × 10−22 |
| PI3K-Akt signaling pathway | 354 | 3.39 | 30 | 1.46 × 10−21 | 6.63 × 10−20 |
| Complement and coagulation cascades | 79 | 0.756 | 13 | 3.00 × 10−13 | 1.19 × 10−11 |
| Proteoglycans in cancer | 201 | 1.92 | 17 | 3.55 × 10−12 | 1.25 × 10−10 |
| Cell adhesion molecules (CAMs) | 146 | 1.4 | 14 | 6.7 × 10−11 | 2.13 × 10−9 |
| Hematopoietic cell lineage | 97 | 0.928 | 10 | 2.22 × 10−8 | 6.42 × 10−7 |
| Phagosome | 152 | 1.45 | 11 | 1.69 × 10−7 | 4.48 × 10−6 |
| Pathways in cancer | 530 | 5.07 | 17 | 7.3 × 10−6 | 0.000179 |
| Rap1 signaling pathway | 206 | 1.97 | 10 | 2.34 × 10−5 | 0.000531 |
| Small cell lung cancer | 93 | 0.889 | 7 | 2.7 × 10−5 | 0.000572 |
| Bladder cancer | 41 | 0.392 | 4 | 0.000597 | 0.0119 |
| Leishmaniasis | 74 | 0.708 | 5 | 0.000673 | 0.0126 |
| Platelet activation | 124 | 1.19 | 6 | 0.00114 | 0.0197 |
| Malaria | 49 | 0.469 | 4 | 0.00118 | 0.0197 |
| Bacterial invasion of epithelial cells | 74 | 0.708 | 4 | 0.00536 | 0.0851 |
| Pertussis | 76 | 0.727 | 4 | 0.00589 | 0.0891 |
| Ras signaling pathway | 232 | 2.22 | 7 | 0.0065 | 0.094 |
| Tuberculosis | 179 | 1.71 | 6 | 0.00711 | 0.0983 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atzemian, N.; Dovrolis, N.; Ragia, G.; Portokallidou, K.; Kolios, G.; Manolopoulos, V.G. Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation. Biomedicines 2023, 11, 2632. https://doi.org/10.3390/biomedicines11102632
Atzemian N, Dovrolis N, Ragia G, Portokallidou K, Kolios G, Manolopoulos VG. Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation. Biomedicines. 2023; 11(10):2632. https://doi.org/10.3390/biomedicines11102632
Chicago/Turabian StyleAtzemian, Natalia, Nikolas Dovrolis, Georgia Ragia, Konstantina Portokallidou, George Kolios, and Vangelis G. Manolopoulos. 2023. "Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation" Biomedicines 11, no. 10: 2632. https://doi.org/10.3390/biomedicines11102632
APA StyleAtzemian, N., Dovrolis, N., Ragia, G., Portokallidou, K., Kolios, G., & Manolopoulos, V. G. (2023). Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation. Biomedicines, 11(10), 2632. https://doi.org/10.3390/biomedicines11102632







