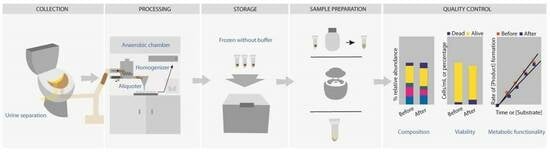
From-Toilet-to-Freezer: A Review on Requirements for an Automatic Protocol to Collect and Store Human Fecal Samples for Research Purposes
Abstract
:1. Introduction
2. Differences Amongst Reported Human Fecal Microbiota Protocols
2.1. Criteria for Human Fecal Material Donors
2.2. Urine–Feces Separation
2.3. Collection of the Fecal Sample
2.4. Anaerobic Conditions
2.5. Buffer or No Buffer in Fecal Material Processing
2.6. Temperature and Time for Processing and Storage of Fecal Material
2.7. Parameters to Evaluate Stability upon Storage with Respect to Composition, Viability and Metabolic Functionality
3. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Haller, D. The Gut Microbiome in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Koleva, P.T.; Kim, J.S.; Scott, J.A.; Kozyrskyj, A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 265–277. [Google Scholar] [CrossRef]
- Scotti, E.; Boué, S.; Sasso, G.L.; Zanetti, F.; Belcastro, V.; Poussin, C.; Sierro, N.; Battey, J.; Gimalac, A.; Ivanov, N.V. Exploring the microbiome in health and disease: Implications for toxicology. Toxicol. Res. Appl. 2017, 1, 2397847317741884. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Shah, Y.A.; Hussain, M.; Rabail, R.; Socol, C.T.; Hassoun, A.; Pateiro, M.; Lorenzo, J.M.; Rusu, A.V. Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 2022, 13, 999001. [Google Scholar] [CrossRef] [PubMed]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Machiels, K.; Sabino, J.; Vandermosten, L.; Joossens, M.; Arijs, I.; de Bruyn, M.; Eeckhaut, V.; Van Assche, G.; Ferrante, M.; Verhaegen, J. Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut 2017, 66, 79–88. [Google Scholar] [CrossRef]
- Zou, J.; Xiao, Z.; Wu, Y.; Yang, J.; Cui, N. Noninvasive fecal testing for colorectal cancer. Clin. Chim. Acta 2022, 524, 123–131. [Google Scholar] [CrossRef]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Yan, S.; Huang, J.; Chen, Z.; Jiang, Z.; Li, X.; Chen, Z. Metabolomics in gut microbiota: Applications and challenges. Science Bulletin 2016, 61, 1151–1153. [Google Scholar] [CrossRef]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 2019, 76, 473–493. [Google Scholar] [CrossRef]
- Mullish, B.H.; Pechlivanis, A.; Barker, G.F.; Thursz, M.R.; Marchesi, J.R.; McDonald, J.A. Functional microbiomics: Evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods 2018, 149, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; Gahan, C.G. Bile acid modifications at the microbe-host interface: Potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef]
- Hara, E. Relationship between obesity, gut microbiome and hepatocellular carcinoma development. Dig. Dis. 2015, 33, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Wewalka, M.; Patti, M.-E.; Barbato, C.; Houten, S.M.; Goldfine, A.B. Fasting serum taurine-conjugated bile acids are elevated in type 2 diabetes and do not change with intensification of insulin. J. Clin. Endocrinol. Metab. 2014, 99, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Prinz, P.; Hofmann, T.; Ahnis, A.; Elbelt, U.; Goebel-Stengel, M.; Klapp, B.F.; Rose, M.; Stengel, A. Plasma bile acids show a positive correlation with body mass index and are negatively associated with cognitive restraint of eating in obese patients. Front. Neurosci. 2015, 9, 199. [Google Scholar] [CrossRef]
- Behr, C.; Sperber, S.; Jiang, X.; Strauss, V.; Kamp, H.; Walk, T.; Herold, M.; Beekmann, K.; Rietjens, I.M.C.M.; Van Ravenzwaay, B. Microbiome-related metabolite changes in gut tissue, cecum content and feces of rats treated with antibiotics. Toxicol. Appl. Pharmacol. 2018, 355, 198–210. [Google Scholar] [CrossRef]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Giuffrè, M.; Campigotto, M.; Campisciano, G.; Comar, M.; Crocè, L.S. A story of liver and gut microbes: How does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol.-Gastrointest. Liver Physiol. 2020, 318, G889–G906. [Google Scholar] [CrossRef]
- Chiou, K.L.; Bergey, C.M. Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces. Sci. Rep. 2018, 8, 1975. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Molino, S.; Navajas-Porras, B.; Valverde-Moya, Á.J.; Hinojosa-Nogueira, D.; López-Maldonado, A.; Pastoriza, S.; Rufián-Henares, J.Á. An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality. Nat. Protoc. 2021, 16, 3186–3209. [Google Scholar] [CrossRef]
- Wu, W.-K.; Chen, C.-C.; Panyod, S.; Chen, R.-A.; Wu, M.-S.; Sheen, L.-Y.; Chang, S.-C. Optimization of fecal sample processing for microbiome study—The journey from bathroom to bench. J. Formos. Med. Assoc. 2019, 118, 545–555. [Google Scholar] [CrossRef]
- Santiago, A.; Panda, S.; Mengels, G.; Martinez, X.; Azpiroz, F.; Dore, J.; Guarner, F.; Manichanh, C. Processing faecal samples: A step forward for standards in microbial community analysis. BMC Microbiol. 2014, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.-E. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Kassam, Z.; Dubois, N.; Ramakrishna, B.; Ling, K.; Qazi, T.; Smith, M.; Kelly, C.R.; Fischer, M.; Allegretti, J.R.; Budree, S. Donor screening for fecal microbiota transplantation. N. Engl. J. Med. 2019, 381, 2070–2072. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zaman, A.; Ramakrishna, B.; Olesen, S.W. Stool banking for fecal microbiota transplantation: Methods and operations at a large stool bank. Front. Cell. Infect. Microbiol. 2021, 11, 622949. [Google Scholar] [CrossRef]
- van Dongen, K.C.; Belzer, C.; Bakker, W.; Rietjens, I.M.; Beekmann, K. Inter-and Intraindividual Differences in the Capacity of the Human Intestinal Microbiome in Fecal Slurries to Metabolize Fructoselysine and Carboxymethyllysine. J. Agric. Food Chem. 2022, 70, 11759–11768. [Google Scholar] [CrossRef]
- Wang, Q.; Spenkelink, B.; Boonpawa, R.; Rietjens, I.M. Use of physiologically based pharmacokinetic modeling to predict human gut microbial conversion of daidzein to S-Equol. J. Agric. Food Chem. 2021, 70, 343–352. [Google Scholar] [CrossRef]
- Wei, J.; Dai, W.; Pan, X.; Zhong, Y.; Xu, N.; Ye, P.; Wang, J.; Li, J.; Yang, F.; Luo, J. Identifying the Novel Gut Microbial Metabolite Contributing to Metabolic Syndrome in Children Based on Integrative Analyses of Microbiome-Metabolome Signatures. Microbiol. Spectr. 2023, 11, e03771-22. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; Pérez-Martínez, P.; Andrés-Lacueva, C.; Cardona, F.; Tinahones, F.; Queipo-Ortuño, M.I. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. 2016, 7, 1775–1787. [Google Scholar] [CrossRef]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004, 70, 3575–3581. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.; Wang, A.; Su, W.; Rozenbloom, S.R.; Taibi, A.; Comelli, E.M.; Wolever, T.M. Age, dietary fiber, breath methane, and fecal short chain fatty acids are interrelated in Archaea-positive humans. J. Nutr. 2013, 143, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.; Ching, J.Y.; Chan, V.C.; Lam, T.Y.; Shum, J.P.; Luk, A.K.; Wong, S.S.; Ng, S.C.; Ng, S.S.; Wu, J.C. Diagnostic accuracy of a qualitative fecal immunochemical test varies with location of neoplasia but not number of specimens. Clin. Gastroenterol. Hepatol. 2015, 13, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Levi, Z.; Rozen, P.; Hazazi, R.; Vilkin, A.; Waked, A.; Maoz, E.; Birkenfeld, S.; Leshno, M.; Niv, Y. A quantitative immunochemical fecal occult blood test for colorectal neoplasia. Ann. Intern. Med. 2007, 146, 244–255. [Google Scholar] [CrossRef]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal. Chim. Acta 2018, 1030, 1–24. [Google Scholar] [CrossRef]
- Kaisar, M.M.; Brienen, E.A.; Djuardi, Y.; Sartono, E.; Yazdanbakhsh, M.; Verweij, J.J.; Supali, T.; Van Lieshout, L. Improved diagnosis of Trichuris trichiura by using a bead-beating procedure on ethanol preserved stool samples prior to DNA isolation and the performance of multiplex real-time PCR for intestinal parasites. Parasitology 2017, 144, 965–974. [Google Scholar] [CrossRef]
- Hashemi, S.; Han, M.; Kim, T.; Kim, Y. (Eds.) Innovative toilet technologies for smart and green cities. In Proceedings of the 8th Conference International Forum on Urbanism (IFoU), Incheon, Republic of Korea, 2–24 June 2015. [Google Scholar]
- Magri, M.E.; Philippi, L.S.r.; Vinnerås, B. Inactivation of pathogens in feces by desiccation and urea treatment for application in urine-diverting dry toilets. Appl. Environ. Microbiol. 2013, 79, 2156–2163. [Google Scholar] [CrossRef]
- Vinnerås, B.; Jönsson, H. The performance and potential of faecal separation and urine diversion to recycle plant nutrients in household wastewater. Bioresour. Technol. 2002, 84, 275–282. [Google Scholar] [CrossRef]
- Zakaria, F.; Ćurko, J.; Muratbegovic, A.; Garcia, H.A.; Hooijmans, C.M.; Brdjanovic, D. Evaluation of a smart toilet in an emergency camp. Int. J. Disaster Risk Reduct. 2018, 27, 512–523. [Google Scholar] [CrossRef]
- Mbonu, C.; Kilanko, O.; Kilanko, M.; Babalola, P. Smart Toilets and Toilet Gadgets in Sustainable Smart Cities: An Overview of Personal Health Monitoring. In Advanced Manufacturing in Biological, Petroleum, and Nanotechnology Processing: Application Tools for Design, Operation, Cost Management, and Environmental Remediation; Springer: Berlin/Heidelberg, Germany, 2022; pp. 143–156. [Google Scholar]
- Eom, Y.S.; Oh, H.; Cho, J.; Kim, J. Social acceptance and willingness to pay for a smart Eco-toilet system producing a Community-based bioenergy in Korea. Sustain. Energy Technol. Assess. 2021, 47, 101400. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Q.; He, T.; Guo, X.; Dong, B.; Lee, J.; Lee, C. Artificial intelligence of toilet (AI-Toilet) for an integrated health monitoring system (IHMS) using smart triboelectric pressure sensors and image sensor. Nano Energy 2021, 90, 106517. [Google Scholar] [CrossRef]
- Ge, T.J.; Chan, C.T.; Lee, B.J.; Liao, J.C.; Park, S.-m. Smart toilets for monitoring COVID-19 surges: Passive diagnostics and public health. NPJ Digit. Med. 2022, 5, 39. [Google Scholar] [CrossRef]
- Park, S.-m.; Won, D.D.; Lee, B.J.; Escobedo, D.; Esteva, A.; Aalipour, A.; Ge, T.J.; Kim, J.H.; Suh, S.; Choi, E.H. A mountable toilet system for personalized health monitoring via the analysis of excreta. Nat. Biomed. Eng. 2020, 4, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.-H.; Lee, H.-K. User health information analysis with a urine and feces separable smart toilet system. IEEE Access 2018, 6, 78751–78765. [Google Scholar] [CrossRef]
- Shafik, A. A concept of the anatomy of the anal sphincter mechanism and the physiology of defecation. Dis. Colon Rectum 1987, 30, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, W.; Sanchez, I.; Mommaerts, K.; Frasquilho, S.; Betsou, F. An independent evaluation of the CryoXtract Instruments’ CXT350 frozen sample aliquotter using tissue and fecal biospecimens. Biopreserv. Biobank. 2016, 14, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Papanicolas, L.E.; Choo, J.M.; Wang, Y.; Leong, L.E.; Costello, S.P.; Gordon, D.L.; Wesselingh, S.L.; Rogers, G.B. Bacterial viability in faecal transplants: Which bacteria survive? EBioMedicine 2019, 41, 509–516. [Google Scholar] [CrossRef]
- Salonen, A.; Nikkilä, J.; Jalanka-Tuovinen, J.; Immonen, O.; Rajilić-Stojanović, M.; Kekkonen, R.A.; Palva, A.; de Vos, W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 2010, 81, 127–134. [Google Scholar] [CrossRef]
- Gorzelak, M.A.; Gill, S.K.; Tasnim, N.; Ahmadi-Vand, Z.; Jay, M.; Gibson, D.L. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS ONE 2015, 10, e0134802. [Google Scholar] [CrossRef]
- Wu, G.D.; Lewis, J.D.; Hoffmann, C.; Chen, Y.-Y.; Knight, R.; Bittinger, K.; Hwang, J.; Chen, J.; Berkowsky, R.; Nessel, L. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010, 10, 206. [Google Scholar] [CrossRef]
- Wesolowska-Andersen, A.; Bahl, M.I.; Carvalho, V.; Kristiansen, K.; Sicheritz-Pontén, T.; Gupta, R.; Licht, T.R. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Dong, T.; Chen, M.; He, L.; Wang, T.; Liu, X.; Chang, H.; Mao, J.-H.; Hang, B.; Snijders, A.M. Systematic analysis of impact of sampling regions and storage methods on fecal gut microbiome and metabolome profiles. Msphere 2020, 5, e00763-00719. [Google Scholar] [CrossRef] [PubMed]
- Amarenco, G. Bristol Stool Chart: Prospective and monocentric study of “stools introspection” in healthy subjects. Prog. En Urol. J. De L’association Fr. D’urologie Et De La Soc. Fr. D’urologie 2014, 24, 708–713. [Google Scholar]
- Irmscher, S.B.; Gibis, M.; Herrmann, K.; Kohlus, R.; Weiss, J. Development of a novel homogenizer using the vane pump-grinder technology for the production of meat batter. J. Food Eng. 2016, 169, 10–17. [Google Scholar] [CrossRef]
- Bellali, S.; Lagier, J.-C.; Raoult, D.; Bou Khalil, J. Among live and dead bacteria, the optimization of sample collection and processing remains essential in recovering gut microbiota components. Front. Microbiol. 2019, 10, 1606. [Google Scholar] [CrossRef]
- Lehne, G.; Müller, A.; Schwedes, J. Mechanical disintegration of sewage sludge. Water Sci. Technol. 2001, 43, 19–26. [Google Scholar] [CrossRef]
- Loesche, W.J. Oxygen sensitivity of various anaerobic bacteria. Appl. Microbiol. 1969, 18, 723–727. [Google Scholar] [CrossRef]
- Finegold, S.M. Anaerobic infections in humans: An overview. Anaerobe 1995, 1, 3–9. [Google Scholar] [CrossRef]
- Van der Waaij, L.; Mesander, G.; Limburg, P.; Van der Waaij, D. Direct flow cytometry of anaerobic bacteria in human feces. Cytom. J. Int. Soc. Anal. Cytol. 1994, 16, 270–279. [Google Scholar] [CrossRef]
- Marteau, P.; Pochart, P.; Doré, J.; Béra-Maillet, C.; Bernalier, A.; Corthier, G. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 2001, 67, 4939–4942. [Google Scholar] [CrossRef]
- Heilig, H.G.; Zoetendal, E.G.; Vaughan, E.E.; Marteau, P.; Akkermans, A.D.; de Vos, W.M. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 2002, 68, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Chen, J.; Amir, A.; Shi, J.; Abnet, C.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of collection methods for fecal samples in microbiome studies. Am. J. Epidemiol. 2017, 185, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Schwintner, C.; Affagard, H.; Joël, D. Microorganism Sampling Method, Microorganism Sampling Device and Sampling Kit Comprising Such a Sampling Device. U.S. Patent 11,013,498, 25 May 2021. [Google Scholar]
- Li, P.; Zhang, T.T.; Zhang, Y. Measuring the flushing-generated flow and aerosols in lavatory of commercial aircraft. Build. Environ. 2022, 214, 108948. [Google Scholar] [CrossRef]
- Postgate, J.; Hunter, J. On the survival of frozen bacteria. Microbiology 1961, 26, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Burz, S.D.; Abraham, A.-L.; Fonseca, F.; David, O.; Chapron, A.; Béguet-Crespel, F.; Cénard, S.; Le Roux, K.; Patrascu, O.; Levenez, F. A guide for ex vivo handling and storage of stool samples intended for fecal microbiota transplantation. Sci. Rep. 2019, 9, 8897. [Google Scholar] [CrossRef] [PubMed]
- Achá, S.; Kühn, I.; Mbazima, G.; Colque-Navarro, P.; Möllby, R. Changes of viability and composition of the Escherichia coli flora in faecal samples during long time storage. J. Microbiol. Methods 2005, 63, 229–238. [Google Scholar] [CrossRef]
- Tap, J.; Cools-Portier, S.; Pavan, S.; Druesne, A.; Öhman, L.; Törnblom, H.; Simren, M.; Derrien, M. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci. Rep. 2019, 9, 601. [Google Scholar] [CrossRef]
- Goldman, S.L.; Sanders, J.G.; Yan, W.; Denice, A.; Cornwall, M.; Ivey, K.N.; Taylor, E.N.; Gunderson, A.R.; Sheehan, M.J.; Mjungu, D. Culture-enriched community profiling improves resolution of the vertebrate gut microbiota. Mol. Ecol. Resour. 2022, 22, 122–136. [Google Scholar] [CrossRef]
- Hale, V.L.; Tan, C.L.; Knight, R.; Amato, K.R. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J. Microbiol. Methods 2015, 113, 16–26. [Google Scholar] [CrossRef]
- Conrads, G.; Abdelbary, M.M. Challenges of next-generation sequencing targeting anaerobes. Anaerobe 2019, 58, 47–52. [Google Scholar] [CrossRef]
- Tedjo, D.I.; Jonkers, D.M.; Savelkoul, P.H.; Masclee, A.A.; van Best, N.; Pierik, M.J.; Penders, J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS ONE 2015, 10, e0126685. [Google Scholar] [CrossRef]
- Choo, J.M.; Leong, L.E.; Rogers, G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015, 5, 16350. [Google Scholar] [CrossRef]
- Hisada, T.; Endoh, K.; Kuriki, K. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch. Microbiol. 2015, 197, 919–934. [Google Scholar] [CrossRef]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Klaenhammer, T.R.; Ringel, Y. Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage. PLoS ONE 2012, 7, e46953. [Google Scholar] [CrossRef] [PubMed]
- Dore, J.; Ehrlich, S.; Levenez, F.; Pelletier, E.; Alberti, A.; Bertrand, L.; Bork, P.; Costea, P.; Sunagawa, S.; Guarner, F. IHMS Sop 02 V1: Standard operating procedure for fecal samples self-collection. Int. Hum. Microbiome Stand 2015, 2, 1–13. [Google Scholar]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 2016, 1, e00021-16. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, N.A.; Walker, A.W.; Berry, S.H.; Duncan, S.H.; Farquarson, F.M.; Louis, P.; Thomson, J.M.; Consortium, U.I.G.; Satsangi, J.; Flint, H.J.; et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE 2014, 9, e88982. [Google Scholar] [CrossRef]
- Shah, N.; Tang, H.; Doak, T.G.; Ye, Y. Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics. In Pacific Symposium on Biocomputing 2011; Altman, R.B., Dunker, A.K., Hunter, L., Murray, T.A., Klein, T.E., Eds.; World Scientific: Kohala Coast, HI, USA, 2011; pp. 165–176. [Google Scholar] [CrossRef]
- Bilinski, J.; Dziurzynski, M.; Grzesiowski, P.; Podsiadly, E.; Stelmaszczyk-Emmel, A.; Dzieciatkowski, T.; Dziewit, L.; Basak, G.W. Multimodal approach to assessment of Fecal Microbiota donors based on three complementary methods. J. Clin. Med. 2020, 9, 2036. [Google Scholar] [CrossRef]
- Wilson, I.D.; Nicholson, J.K. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Vandeputte, D.; Tito, R.Y.; Vanleeuwen, R.; Falony, G.; Raes, J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 2017, 41 (Suppl. S1), S154–S167. [Google Scholar] [CrossRef] [PubMed]
- Mathay, C.; Hamot, G.; Henry, E.; Georges, L.; Bellora, C.; Lebrun, L.; de Witt, B.; Ammerlaan, W.; Buschart, A.; Wilmes, P.; et al. Method optimization for fecal sample collection and fecal DNA extraction. Biopreserv. Biobank. 2015, 13, 79–93. [Google Scholar] [CrossRef]
- Otterpohl, R. Options for alternative types of sewerage and treatment systems directed to improvement of the overall performance. Water Sci. Technol. 2002, 45, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hofstaedter, C.E.; Zhao, C.; Mattei, L.; Tanes, C.; Clarke, E.; Lauder, A.; Sherrill-Mix, S.; Chehoud, C.; Kelsen, J. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Morgan, X.C.; Segata, N.; Waldron, L.; Reyes, J.; Earl, A.M.; Giannoukos, G.; Boylan, M.R.; Ciulla, D.; Gevers, D. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA 2014, 111, E2329–E2338. [Google Scholar] [CrossRef]
- Bikov, A.; Szabo, H.; Piroska, M.; Kunos, L.; Szily, M.; Ligeti, B.; Makra, N.; Szabo, D.; Tarnoki, D.L.; Tarnoki, A.D. Gut Microbiome in Patients with Obstructive Sleep Apnoea. Appl. Sci. 2022, 12, 2007. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef]
- Ogilvie, L.A.; Jones, B.V. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: A mechanism and marker of disease? Gut 2012, 61, 1642–1643. [Google Scholar] [CrossRef] [PubMed]
- Hustoft, T.; Hausken, T.; Ystad, S.; Valeur, J.; Brokstad, K.; Hatlebakk, J.; Lied, G.A. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol. Motil. 2017, 29, e12969. [Google Scholar] [CrossRef]
- Landini, L.; Dadson, P.; Gallo, F.; Honka, M.-J.; Cena, H. Microbiota in Anorexia Nervosa–Potential for Treatment. Nutr. Res. Rev. 2022, 1–51. [Google Scholar] [CrossRef]
- Ruiz, A.; Cerdó, T.; Jáuregui, R.; Pieper, D.H.; Marcos, A.; Clemente, A.; García, F.; Margolles, A.; Ferrer, M.; Campoy, C.; et al. One-year calorie restriction impacts gut microbial composition but not its metabolic performance in obese adolescents. Environ. Microbiol. 2017, 19, 1536–1551. [Google Scholar] [CrossRef]
- Zhang, Y.-K.; Zhang, Q.; Wang, Y.-L.; Zhang, W.-Y.; Hu, H.-Q.; Wu, H.-Y.; Sheng, X.-Z.; Luo, K.-J.; Zhang, H.; Wang, M.; et al. A Comparison study of age and colorectal cancer-related gut bacteria. Front. Cell. Infect. Microbiol. 2021, 11, 606490. [Google Scholar] [CrossRef]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.-L.; et al. Gut microbiota is altered in patients with Alzheimer’s disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef]
- Ding, H.; Yi, X.; Zhang, X.; Wang, H.; Liu, H.; Mou, W.-W. Imbalance in the gut microbiota of children with autism spectrum disorders. Front. Cell. Infect. Microbiol. 2021, 11, 572752. [Google Scholar] [CrossRef]
- Li, Z.; Lu, G.; Li, Z.; Wu, B.; Luo, E.; Qiu, X.; Guo, J.; Xia, Z.; Zheng, C.; Su, Q.; et al. Altered actinobacteria and firmicutes phylum associated epitopes in patients with Parkinson’s disease. Front. Immunol. 2021, 12, 632482. [Google Scholar] [CrossRef]
- Otaka, M.; Kikuchi-Hayakawa, H.; Ogura, J.; Ishikawa, H.; Yomogida, Y.; Ota, M.; Hidese, S.; Ishida, I.; Aida, M.; Matsuda, K.; et al. Effect of Lacticaseibacillus paracasei strain Shirota on improvement in depressive symptoms, and its association with abundance of Actinobacteria in gut microbiota. Microorganisms 2021, 9, 1026. [Google Scholar] [CrossRef]
- Jia, W.; Rajani, C.; Kaddurah-Daouk, R.; Li, H. Expert insights: The potential role of the gut microbiome-bile acid-brain axis in the development and progression of Alzheimer’s disease and hepatic encephalopathy. Med. Res. Rev. 2020, 40, 1496–1507. [Google Scholar] [CrossRef]
- Valentini, F.; Evangelisti, M.; Arpinelli, M.; Di Nardo, G.; Borro, M.; Simmaco, M.; Villa, M.P. Gut microbiota composition in children with obstructive sleep apnoea syndrome: A pilot study. Sleep Med. 2020, 76, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Knox, N.C.; Forbes, J.D.; Peterson, C.-L.; Van Domselaar, G.; Bernstein, C.N. The gut microbiome in inflammatory bowel disease: Lessons learned from other immune-mediated inflammatory diseases. Off. J. Am. Coll. Gastroenterol. ACG 2019, 114, 1051–1070. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: A systematic review. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef]
- Wan, Y.; Yuan, J.; Li, J.; Li, H.; Yin, K.; Wang, F.; Li, D. Overweight and underweight status are linked to specific gut microbiota and intestinal tricarboxylic acid cycle intermediates. Clin. Nutr. 2020, 39, 3189–3198. [Google Scholar] [CrossRef]
- Xu, P.; Li, M.; Zhang, J.; Zhang, T. Correlation of intestinal microbiota with overweight and obesity in Kazakh school children. BMC Microbiol. 2012, 12, 283. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.-A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Doifode, T.; Giridharan, V.V.; Generoso, J.S.; Bhatti, G.; Collodel, A.; Schulz, P.E.; Forlenza, O.V.; Barichello, T. The impact of the microbiota-gut-brain axis on Alzheimer’s disease pathophysiology. Pharmacol. Res. 2021, 164, 105314. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Lin, C.-H.; Chen, C.-C.; Chiang, H.-L.; Liou, J.-M.; Chang, C.-M.; Lu, T.-P.; Chuang, E.Y.; Tai, Y.-C.; Cheng, C.; Lin, H.-Y.; et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflammation 2019, 16, 129. [Google Scholar] [CrossRef]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.A.; Gyamfi, A.; Cong, X.S. Altered composition of gut microbiota in depression: A systematic review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Sung, C.M.; Lin, Y.-F.; Chen, K.-F.; Ke, H.-M.; Huang, H.-Y.; Gong, Y.-N.; Tsai, W.-S.; You, J.-F.; Lu, M.J.; Cheng, H.-T.; et al. Predicting clinical outcomes of cirrhosis patients with hepatic encephalopathy from the fecal microbiome. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 301–318.e2. [Google Scholar] [CrossRef]
- James, M.M.; Pal, N.; Sharma, P.; Kumawat, M.; Shubham, S.; Verma, V.; Tiwari, R.R.; Singh, B.; Nagpal, R.; Sarma, D.K.; et al. Role of butyrogenic Firmicutes in type-2 diabetes. J. Diabetes Metab. Disord. 2022, 21, 1873–1882. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. (Eds.) The gut microbiota and inflammatory bowel disease. In Semin in Immunopathol; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Kassinen, A.; Krogius-Kurikka, L.; Mäkivuokko, H.; Rinttilä, T.; Paulin, L.; Corander, J.; Malinen, E.; Apajalahti, J.; Palva, A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007, 133, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Li, Y.; Li, R.; Du, Y.; Xu, X.; Xue, Y.; Gao, D.; Gao, T.; Sheng, Z.; Zhang, L.; Tuo, H. Features of gut microbiota in patients with idiopathic Parkinson’s disease. Zhonghua Yi Xue Za Zhi 2020, 100, 1017–1022. [Google Scholar]
- Huang, Y.; Shi, X.; Li, Z.; Shen, Y.; Shi, X.; Wang, L.; Li, G.; Yuan, Y.; Wang, J.; Zhang, Y.; et al. Possible association of Firmicutes in the gut microbiota of patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 2018, 3329–3337. [Google Scholar] [CrossRef]
- Zuo, Z.; Fan, H.; Tang, X.D.; Chen, Y.M.; Xun, L.T.; Li, Y.; Song, Z.; Zhai, H. Effect of different treatments and alcohol addiction on gut microbiota in minimal hepatic encephalopathy patients. Exp. Ther. Med. 2017, 14, 4887–4895. [Google Scholar] [CrossRef]
- Li, Q.; Xu, T.; Shao, C.; Gao, W.; Wang, M.; Dong, Y.; Wang, X.; Lu, F.; Li, D.; Tan, H.; et al. Obstructive sleep apnea is related to alterations in fecal microbiome and impaired intestinal barrier function. Sci. Rep. 2023, 13, 778. [Google Scholar] [CrossRef]
- Wei, B.; Wang, Y.; Xiang, S.; Jiang, Y.; Chen, R.; Hu, N. Alterations of gut microbiome in patients with type 2 diabetes mellitus who had undergone cholecystectomy. Am. J. Physiol.-Endocrinol. Metab. 2021, 320, E113–E121. [Google Scholar] [CrossRef]
- Sheehan, D.; Moran, C.; Shanahan, F. The microbiota in inflammatory bowel disease. J. Gastroenterol. 2015, 50, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Zhou, J.; Su, Y.; Mao, K.; Wu, J.; Zhu, C.; He, L.; Cui, Y. Gut microbiota composition and functional prediction in diarrhea-predominant irritable bowel syndrome. BMC Gastroenterol. 2021, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, M.; Xue, J.; Huang, J.; Zhuang, R.; Zhou, X.; Zhang, H.; Fu, Q.; Hao, Y. Body mass index differences in the gut microbiota are gender specific. Front. Microbiol. 2018, 9, 1250. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef]
- Bello-Medina, P.C.; Hernández-Quiroz, F.; Pérez-Morales, M.; González-Franco, D.A.; Cruz-Pauseno, G.; García-Mena, J.; Díaz-Cintra, S.; Pacheco-López, G. Spatial memory and gut microbiota alterations are already present in early adulthood in a pre-clinical transgenic model of Alzheimer’s disease. Front. Neurosci. 2021, 15, 595583. [Google Scholar] [CrossRef]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef]
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H.; et al. Gut microbiota in patients with Parkinson’s disease in southern China. Park. Relat. Disord. 2018, 53, 82–88. [Google Scholar] [CrossRef]
- Rai, R.; Saraswat, V.A.; Dhiman, R.K. Gut microbiota: Its role in hepatic encephalopathy. J. Clin. Exp. Hepatol. 2015, 5, S29–S36. [Google Scholar] [CrossRef]
- Zhang, Z.; Tian, T.; Chen, Z.; Liu, L.; Luo, T.; Dai, J. Characteristics of the gut microbiome in patients with prediabetes and type 2 diabetes. PeerJ 2021, 9, e10952. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Barandouzi, Z.A.; Lee, J.; del Carmen Rosas, M.; Chen, J.; Henderson, W.A.; Starkweather, A.R.; Cong, X.S. Associations of neurotransmitters and the gut microbiome with emotional distress in mixed type of irritable bowel syndrome. Sci. Rep. 2022, 12, 1648. [Google Scholar] [CrossRef] [PubMed]
- Huey, S.L.; Jiang, L.; Fedarko, M.W.; McDonald, D.; Martino, C.; Ali, F.; Russell, D.G.; Udipi, S.A.; Thorat, A.; Thakker, V.; et al. Nutrition and the gut microbiota in 10- to 18-month-old children living in urban slums of Mumbai, India. Msphere 2020, 5, e00731-20. [Google Scholar] [CrossRef]
- Abdullah, B.; Daud, S.; Aazmi, M.S.; Idorus, M.Y.; Mahamooth, M.I.J. Gut microbiota in pregnant Malaysian women: A comparison between trimesters, body mass index and gestational diabetes status. BMC Pregnancy Childbirth 2022, 22, 152. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, R.; Shu, R.; Yu, J.; Li, H.; Long, H.; Jin, S.; Li, S.; Hu, Q.; Yao, F.; et al. Study of the relationship between microbiome and colorectal cancer susceptibility using 16SrRNA sequencing. BioMed Res. Int. 2020, 2020, 7828392. [Google Scholar] [CrossRef]
- Franceschi, F.; Ojetti, V.; Candelli, M.; Covino, M.; Cardone, S.; Potenza, A.; Simeoni, B.; Gabrielli, M.; Sabia, L.; Gasbarrini, G.; et al. Microbes and Alzheimer’disease: Lessons from H. pylori and GUT microbiota. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 426–430. [Google Scholar]
- Plaza-Díaz, J.; Gómez-Fernández, A.; Chueca, N.; Torre-Aguilar MJdl Gil, Á.; Perez-Navero, J.L.; Flores-Rojas, K.; Martín-Borreguero, P.; Solis-Urra, P.; Ruiz-Ojeda, F.J.; Garcia, F.; et al. Autism spectrum disorder (ASD) with and without mental regression is associated with changes in the fecal microbiota. Nutrients 2019, 11, 337. [Google Scholar] [CrossRef]
- Zapała, B.; Stefura, T.; Wójcik-Pędziwiatr, M.; Kabut, R.; Bałajewicz-Nowak, M.; Milewicz, T.; Dudek, A.; Stój, A.; Rudzińska-Bar, M. Differences in the composition of gut microbiota between patients with parkinson’s disease and healthy controls: A cohort study. J. Clin. Med. 2021, 10, 5698. [Google Scholar] [CrossRef]
- Simpson, C.A.; Mu, A.; Haslam, N.; Schwartz, O.S.; Simmons, J.G. Feeling down? A systematic review of the gut microbiota in anxiety/depression and irritable bowel syndrome. J. Affect. Disord. 2020, 266, 429–446. [Google Scholar] [CrossRef]
- Galli, J.; Calò, L.; Posteraro, B.; Rossi, G.; Sterbini, F.P.; Paludetti, G.; Sanguinetti, M. Pediatric oropharyngeal microbiome: Mapping in chronic tonsillitis and tonsillar hypertrophy. Int. J. Pediatr. Otorhinolaryngol. 2020, 139, 110478. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, M.; Yang, J.; Xu, Q.; Liang, C.; Chen, B.; Zhang, J.; Yang, Y.; Wang, H.; Shang, Y.; et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 2019, 66, 485–493. [Google Scholar] [CrossRef]
- Damman, C.J.; Miller, S.I.; Surawicz, C.M.; Zisman, T.L. The microbiome and inflammatory bowel disease: Is there a therapeutic role for fecal microbiota transplantation? Off. J. Am. Coll. Gastroenterol. ACG 2012, 107, 1452–1459. [Google Scholar] [CrossRef]
- Pimentel, M.; Lembo, A. Microbiome and its role in irritable bowel syndrome. Dig. Dis. Sci. 2020, 65, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Sen Gupta, S.; Bhattacharya, T.; Yadav, D.; Barik, A.; Chowdhury, A.; Das, B.; Mande, S.S.; Nair, G.B. Gut microbiomes of Indian children of varying nutritional status. PLoS ONE 2014, 9, e95547. [Google Scholar] [CrossRef] [PubMed]
- Graessler, J.; Qin, Y.; Zhong, H.; Zhang, J.; Licinio, J.; Wong, M.-L.; Xu, A.; Chavakis, T.; Bornstein, A.B.; Ehrhart-Bornstein, M.; et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: Correlation with inflammatory and metabolic parameters. Pharm. J. 2013, 13, 514–522. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Q.; Shu, X.O.; Steinwandel, M.D.; Blot, W.J.; Zheng, W.; Long, J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer 2019, 144, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Kaiyrlykyzy, A.; Kozhakhmetov, S.; Babenko, D.; Zholdasbekova, G.; Alzhanova, D.; Olzhayev, F.; Baibulatova, A.; Kushugulova, A.R.; Askarova, S. Study of gut microbiota alterations in Alzheimer’s dementia patients from Kazakhstan. Sci. Rep. 2022, 12, 15115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zou, R.; Guo, M.; Duan, M.; Li, Q.; Zheng, H. Comparison of gut microbiota between adults with autism spectrum disorder and obese adults. PeerJ 2021, 9, e10946. [Google Scholar] [CrossRef]
- Lubomski, M.; Xu, X.; Holmes, A.J.; Muller, S.; Yang, J.Y.; Davis, R.L.; Sue, C.M. The gut microbiome in Parkinson’s disease: A longitudinal study of the impacts on disease progression and the use of device-assisted therapies. Front. Aging Neurosci. 2022, 14, 875261. [Google Scholar] [CrossRef]
- Paudel, D.; Uehara, O.; Giri, S.; Yoshida, K.; Morikawa, T.; Kitagawa, T.; Matsuoka, H.; Miura, H.; Toyofuku, A.; Kuramitsu, Y.; et al. Effect of psychological stress on the oral-gut microbiota and the potential oral-gut-brain axis. JPN Dent. Sci. Rev. 2022, 58, 365–375. [Google Scholar] [CrossRef]
- Ling, Z.; Liu, X.; Cheng, Y.; Jiang, X.; Jiang, H.; Wang, Y.; Li, L. Decreased diversity of the oral microbiota of patients with hepatitis B virus-induced chronic liver disease: A pilot project. Sci. Rep. 2015, 5, 17098. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.R.; Carroll, I.; Azcarate-Peril, M.A.; Rochette, A.D.; Heinberg, L.J.; Peat, C.; Steffen, F.; Manderino, L.M.; Mitchell, J.; Gunstad, J. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017, 38, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Salguero, M.V.; Al-Obaide, M.A.; Singh, R.; Siepmann, T.; Vasylyeva, T.L. Dysbiosis of Gram-negative gut microbiota and the associated serum lipopolysaccharide exacerbates inflammation in type 2 diabetic patients with chronic kidney disease. Exp. Ther. Med. 2019, 18, 3461–3469. [Google Scholar] [CrossRef] [PubMed]
- Salem, F.; Kindt, N.; Marchesi, J.R.; Netter, P.; Lopez, A.; Kokten, T.; Danese, S.; Jouzeau, J.-Y.; Peyrin-Biroulet, L.; Moulin, D. Gut microbiome in chronic rheumatic and inflammatory bowel diseases: Similarities and differences. United Eur. Gastroenterol. J. 2019, 7, 1008–1032. [Google Scholar] [CrossRef]
- Barandouzi, Z.A.; Lee, J.; Maas, K.; Starkweather, A.R.; Cong, X.S. Altered gut microbiota in irritable bowel syndrome and its association with food components. J. Pers. Med. 2021, 11, 35. [Google Scholar] [CrossRef]
- de Clercq, N.C.; Frissen, M.N.; Davids, M.; Groen, A.K.; Nieuwdorp, M. Weight gain after fecal microbiota transplantation in a patient with recurrent underweight following clinical recovery from anorexia nervosa. Psychother. Psychosom. 2019, 88, 58–60. [Google Scholar] [CrossRef]
- Barlow, G.M.; Yu, A.; Mathur, R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr. Clin. Pract. 2015, 30, 787–797. [Google Scholar] [CrossRef]
- Huang, R.; He, K.; Duan, X.; Xiao, J.; Wang, H.; Xiang, G. Changes of intestinal microflora in colorectal cancer patients after surgical resection and chemotherapy. Comput. Math. Methods Med. 2022, 2022, 1940846. [Google Scholar] [CrossRef]
- Askarova, S.; Umbayev, B.; Masoud, A.-R.; Kaiyrlykyzy, A.; Safarova, Y.; Tsoy, A.; Olzhayev, F.; Kushugulova, A. The links between the gut microbiome, aging, modern lifestyle and Alzheimer’s disease. Front. Cell. Infect. Microbiol. 2020, 10, 104. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Hemmings, S.M.; Malan-Muller, S.; van den Heuvel, L.L.; Demmitt, B.A.; Stanislawski, M.A.; Smith, D.G.; Bohr, A.D.; Stamper, C.E.; Hyde, E.R.; Morton, J.T.; et al. The microbiome in posttraumatic stress disorder and trauma-exposed controls: An exploratory study. Psychosom. Med. 2017, 79, 936. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, K.; Tang, S.; Lv, Y.; Wang, Q.; Wang, Z.; Luo, B.; Niu, J.; Zhu, Y.; Guo, W.; et al. Restoration of the gut microbiota is associated with a decreased risk of hepatic encephalopathy after TIPS. JHEP Rep. 2022, 4, 100448. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef] [PubMed]
- Deledda, A.; Palmas, V.; Heidrich, V.; Fosci, M.; Lombardo, M.; Cambarau, G.; Lai, A.; Melis, M.; Loi, E.; Loviselli, A.; et al. Dynamics of Gut Microbiota and Clinical Variables after Ketogenic and Mediterranean Diets in Drug-Naïve Patients with Type 2 Diabetes Mellitus and Obesity. Metabolites 2022, 12, 1092. [Google Scholar] [CrossRef]
- Chang, C.-C.; Liu, C.-Y.; Su, I.-C.; Lee, Y.-J.; Yeh, H.-J.; Chen, W.-C.; Yu, C.-J.; Kao, W.-Y.; Liu, Y.-C.; Huang, C.-J. Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 11422. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-S.; Wang, Y.-L.; Zhang, W.-Y.; Li, K.-D.; Luo, X.-F.; Cui, Y.-L. Puerarin from Pueraria lobata alleviates the symptoms of irritable bowel syndrome-diarrhea. Food Funct. 2021, 12, 2211–2224. [Google Scholar] [CrossRef]
- Han, D.-S.; Wu, W.-K.; Liu, P.-Y.; Yang, Y.-T.; Hsu, H.-C.; Kuo, C.-H.; Wu, M.-S.; Wang, T.-G. Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin. Nutr. 2022, 41, 1491–1500. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, J.; Yang, M.; Qian, C.; Liu, Y.; Qi, Y.; Feng, R.; Yang, M.; Liu, W.; Ma, J. Low Doses of Sucralose Alter Fecal Microbiota in High-Fat Diet-Induced Obese Rats. Front. Nutr. 2021, 8, 1151. [Google Scholar] [CrossRef]
- Ubeda, C.; Vázquez-Carretero, M.D.; Luque-Tirado, A.; Ríos-Reina, R.; Rubio-Sánchez, R.; Franco-Macías, E.; García-Miranda, P.; Calonge, M.L.; Peral, M.J. Fecal Volatile Organic Compounds and Microbiota Associated with the Progression of Cognitive Impairment in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 24, 707. [Google Scholar] [CrossRef]
- Li, N.; Chen, H.; Cheng, Y.; Xu, F.; Ruan, G.; Ying, S.; Tang, W.; Chen, L.; Chen, M.; Lv, L.; et al. Fecal microbiota transplantation relieves gastrointestinal and autism symptoms by improving the gut microbiota in an open-label study. Front. Cell. Infect. Microbiol. 2021, 11, 948. [Google Scholar]
- Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ishida, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Short-chain fatty acid-producing gut microbiota is decreased in Parkinson’s disease but not in rapid-eye-movement sleep behavior disorder. mSystems 2020, 5, e00797-20. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Choi, J.; Lee, H.-J. Flavonoid-rich orange juice intake and altered gut microbiome in young adults with depressive symptom: A randomized controlled study. Nutrients 2020, 12, 1815. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wan, Y.-J.Y. The role of gut microbiota in liver disease development and treatment. Liver Res. 2019, 3, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, X.; Zheng, X.; Xia, Y.; Fu, Y.; Li, X.; Qian, Y.; Zou, J.; Zhao, A.; Guan, J.; et al. Pediatric obstructive sleep apnea is associated with changes in the oral microbiome and urinary metabolomics profile: A pilot study. J. Clin. Sleep Med. 2018, 14, 1559–1567. [Google Scholar] [CrossRef]
- Bailey, M.J.; Naik, N.N.; Wild, L.E.; Patterson, W.B.; Alderete, T.L. Exposure to air pollutants and the gut microbiota: A potential link between exposure, obesity, and type 2 diabetes. Gut Microbes 2020, 11, 1188–1202. [Google Scholar] [CrossRef]
- Humbel, F.; Rieder, J.H.; Franc, Y.; Juillerat, P.; Scharl, M.; Misselwitz, B.; Schreiner, P.; Begré, S.; Rogler, G.; von Känel, R.; et al. Association of alterations in intestinal microbiota with impaired psychological function in patients with inflammatory bowel diseases in remission. Clin. Gastroenterol. Hepatol. 2020, 18, 2019–2029.e11. [Google Scholar] [CrossRef]
- Jalanka, J.; Lam, C.; Bennett, A.; Hartikainen, A.; Crispie, F.; Finnegan, L.A.; Cotter, P.D.; Spiller, R. Colonic gene expression and fecal microbiota in diarrhea-predominant irritable bowel syndrome: Increased toll-like receptor 4 but minimal inflammation and no response to mesalazine. J. Neurogastroenterol. Motil. 2021, 27, 279. [Google Scholar] [CrossRef]
- Eggers, S.; Safdar, N.; Sethi, A.K.; Suen, G.; Peppard, P.E.; Kates, A.E.; Skarlupka, J.H.; Kanarek, M.; Malecki, K.M. Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environ. Int. 2019, 133, 105122. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Sugahara, H.; Shimada, K.; Mitsuyama, E.; Kuhara, T.; Yasuoka, A.; Kondo, T.; Abe, K.; Xiao, J.-Z. Therapeutic potential of Bifidobacterium breve strain A1 for preventing cognitive impairment in Alzheimer’s disease. Sci. Rep. 2017, 7, 13510. [Google Scholar] [CrossRef]
- Williams, B.L.; Hornig, M.; Parekh, T.; Lipkin, W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012, 3, e00261-11. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.E.; Stringer, A.; Bobrovskaya, L. Rotenone induces gastrointestinal pathology and microbiota alterations in a rat model of Parkinson’s disease. Neurotoxicology 2018, 65, 174–185. [Google Scholar] [CrossRef]
- Zheng, S.; Zhu, Y.; Wu, W.; Zhang, Q.; Wang, Y.; Wang, Z.; Yang, F. A correlation study of intestinal microflora and first-episode depression in Chinese patients and healthy volunteers. Brain Behav. 2021, 11, e02036. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Ridlon, J.M.; Hylemon, P.B.; Thacker, L.R.; Heuman, D.M.; Smith, S.; Sikaroodi, M.; Gillevet, P.M. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G168–G175. [Google Scholar] [CrossRef]
- Bikov, A.; Dragonieri, S.; Csoma, B.; Mazzuca, C.; Finamore, P.; Rocchi, G.; Putignani, L.; Guarino, M.; Scarlata, S. The Role of Gut Bacteriome in Asthma, Chronic Obstructive Pulmonary Disease and Obstructive Sleep Apnoea. Microorganisms 2022, 10, 2457. [Google Scholar] [CrossRef]
- Fugmann, M.; Breier, M.; Rottenkolber, M.; Banning, F.; Ferrari, U.; Sacco, V.; Grallert, H.; Parhofer, K.G.; Seissler, J.; Clavel, T.; et al. The stool microbiota of insulin resistant women with recent gestational diabetes, a high risk group for type 2 diabetes. Sci. Rep. 2015, 5, 13212. [Google Scholar] [CrossRef]
- Alam, M.T.; Amos, G.C.; Murphy, A.R.; Murch, S.; Wellington, E.M.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef]
- Peter, J.; Fournier, C.; Durdevic, M.; Knoblich, L.; Keip, B.; Dejaco, C.; Trauner, M.; Moser, G. A microbial signature of psychological distress in irritable bowel syndrome. Psychosom. Med. 2018, 80, 698. [Google Scholar] [CrossRef]
- Aguilar, T.; Nava, G.M.; Olvera-Ramírez, A.M.; Ronquillo, D.; Camacho, M.; Zavala, G.A.; Caamaño, M.C.; Acevedo-Whitehouse, K.; Rosado, J.L.; García, O.P. Gut bacterial families are associated with body composition and metabolic risk markers in school-aged children in rural Mexico. Child. Obes. 2020, 16, 358–366. [Google Scholar] [CrossRef]
- Cho, K.Y. Lifestyle modifications result in alterations in the gut microbiota in obese children. BMC Microbiol. 2021, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Lauka, L.; Reitano, E.; Carra, M.C.; Gaiani, F.; Gavriilidis, P.; Brunetti, F.; De’angelis, G.L.; Sobhani, I.; De’angelis, N. Role of the intestinal microbiome in colorectal cancer surgery outcomes. World J. Surg. Oncol. 2019, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.; Maji, A.; Dhakan, D.B.; Saxena, R.; Mohan, B.; Anto, M.M.; Agarwal, N.; Grace, T.; Sharma, V.K. Gut microbial dysbiosis in Indian children with autism spectrum disorders. Microb. Ecol. 2018, 76, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Martinez, G.; Chin, B.; Camarillo, C.; Herrera, G.V.; Yang, B.; Sarosiek, I.; Perez, R.G. A pilot microbiota study in Parkinson’s disease patients versus control subjects, and effects of FTY720 and FTY720-mitoxy therapies in parkinsonian and multiple system atrophy mouse models. J. Park. Dis. 2020, 10, 185–192. [Google Scholar] [CrossRef]
- Zheng, P.; Yang, J.; Li, Y.; Wu, J.; Liang, W.; Yin, B.; Tan, X.; Huang, Y.; Chai, T.; Zhang, H.; et al. Gut microbial signatures can discriminate unipolar from bipolar depression. Adv. Sci. 2020, 7, 1902862. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Sanyal, A.J.; Hylemon, P.B.; Sterling, R.K.; Stravitz, R.T.; Fuchs, M.; Ridlon, J.M.; Daita, K.; Monteith, P.; et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS ONE 2013, 8, e60042. [Google Scholar] [CrossRef]
- Farré, N.; Farré, R.; Gozal, D. Sleep apnea morbidity: A consequence of microbial-immune cross-talk? Chest 2018, 154, 754–759. [Google Scholar] [CrossRef]
- Gonai, M.; Shigehisa, A.; Kigawa, I.; Kurasaki, K.; Chonan, O.; Matsuki, T.; Yoshida, Y.; Aida, M.; Hamano, K.; Terauchi, Y. Galacto-oligosaccharides ameliorate dysbiotic Bifidobacteriaceae decline in Japanese patients with type 2 diabetes. Benef. Microbes 2017, 8, 705–716. [Google Scholar] [CrossRef]
- Teofani, A.; Marafini, I.; Laudisi, F.; Pietrucci, D.; Salvatori, S.; Unida, V.; Biocca, S.; Monteleone, G.; Desideri, A. Intestinal taxa abundance and diversity in inflammatory bowel disease patients: An analysis including covariates and confounders. Nutrients 2022, 14, 260. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Yuan, Y.; Leontiadis, G.I.; Tse, F.; Surette, M.; Moayyedi, P. Gut microbiota in patients with irritable bowel syndrome—A systematic review. Gastroenterology 2019, 157, 97–108. [Google Scholar] [CrossRef]
- Nakayama, J.; Watanabe, K.; Jiang, J.; Matsuda, K.; Chao, S.-H.; Haryono, P.; La-Ongkham, O.; Sarwoko, M.-A.; Sujaya, I.N.; Zhao, L.; et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015, 5, 8397. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.; Howland, G.; West, M.; Hockey, M.; Marx, W.; Loughman, A.; O’hely, M.; Jacka, F.; Rocks, T. The effect of ultra-processed very low-energy diets on gut microbiota and metabolic outcomes in individuals with obesity: A systematic literature review. Obes. Res. Clin. Pract. 2020, 14, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Youssef, O.; Lahti, L.; Kokkola, A.; Karla, T.; Tikkanen, M.; Ehsan, H.; Carpelan-Holmström, M.; Koskensalo, S.; Böhling, T.; Rautelin, H.; et al. Stool microbiota composition differs in patients with stomach, colon, and rectal neoplasms. Dig. Dis. Sci. 2018, 63, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef]
- Xie, X.; Li, L.; Wu, X.; Hou, F.; Chen, Y.; Shi, L.; Liu, Q.; Zhu, K.; Jiang, Q.; Feng, Y.; et al. Alteration of the fecal microbiota in Chinese children with autism spectrum disorder. Autism Res. 2022, 15, 996–1007. [Google Scholar] [CrossRef]
- Shen, T.; Yue, Y.; He, T.; Huang, C.; Qu, B.; Lv, W.; Lai, H.Y. The association between the gut microbiota and Parkinson’s disease, a meta-analysis. Front. Aging Neurosci. 2021, 13, 636545. [Google Scholar] [CrossRef]
- Knuesel, T.; Mohajeri, M.H. The role of the gut microbiota in the development and progression of major depressive and bipolar disorder. Nutrients 2022, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Salzman, N.H.; Acharya, C.; Sterling, R.K.; White, M.B.; Gavis, E.A.; Fagan, A.; Hayward, M.; Holtz, M.L.; Matherly, S.; et al. Fecal microbial transplant capsules are safe in hepatic encephalopathy: A phase 1, randomized, placebo-controlled trial. Hepatology 2019, 70, 1690–1703. [Google Scholar] [CrossRef]
- Muñiz Pedrogo, D.A.; Chen, J.; Hillmann, B.; Jeraldo, P.; Al-Ghalith, G.; Taneja, V.; Davis, J.M.; Knights, D.; Nelson, H.; A Faubion, W.; et al. An increased abundance of Clostridiaceae characterizes arthritis in inflammatory bowel disease and rheumatoid arthritis: A cross-sectional study. Inflamm. Bowel Dis. 2019, 25, 902–913. [Google Scholar] [CrossRef]
- Ricci, C.; Rizzello, F.; Valerii, M.C.; Spisni, E.; Gionchetti, P.; Turroni, S.; Candela, M.; D’amico, F.; Spigarelli, R.; Bellocchio, I.; et al. Geraniol treatment for irritable bowel syndrome: A double-blind randomized clinical trial. Nutrients 2022, 14, 4208. [Google Scholar] [CrossRef]
- Hourigan, S.; Grigoryan, Z.; Kim, S.; Chirumamilla, S.; Rabizadeh, S.; Golub, J.; Saeed, S.A.; Elson, C.O.; Oliva-Hemker, M.; Sears, C.L.; et al. Decreased Diversity of the Fecal Microbiome in Pediatric Carriage of Clostridium difficile. Gastroenterology 2014, 146, S-13. [Google Scholar] [CrossRef]
- Golloso-Gubat, M.J.; Ducarmon, Q.R.; Tan, R.C.A.; Zwittink, R.D.; Kuijper, E.J.; Nacis, J.S.; Santos, N.L.C. Gut microbiota and dietary intake of normal-weight and overweight Filipino children. Microorganisms 2020, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Dominianni, C.; Shapiro, J.A.; Church, T.R.; Wu, J.; Miller, G.; Yuen, E.; Freiman, H.; Lustbader, I.; Salik, J.; et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016, 4, 69. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-C.; Chang, C.-C.; Huang, C.-W.; Nouchi, R.; Cheng, C.-H. Gut microbiota in patients with Alzheimer’s disease spectrum: A systematic review and meta-analysis. Aging 2022, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Srikantha, P.; Mohajeri, M.H. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef]
- Painold, A.; Mörkl, S.; Kashofer, K.; Halwachs, B.; Dalkner, N.; Bengesser, S.; Birner, A.; Fellendorf, F.; Platzer, M.; Queissner, R.; et al. A step ahead: Exploring the gut microbiota in inpatients with bipolar disorder during a depressive episode. Bipolar Disord. 2019, 21, 40–49. [Google Scholar] [CrossRef]
- Haraguchi, M.; Miuma, S.; Masumoto, H.; Ichikawa, T.; Kanda, Y.; Sasaki, R.; Fukushima, M.; Miyaaki, H.; Taura, N.; Nakao, K. Bacteroides in colonic mucosa-associated microbiota affects the development of minimal hepatic encephalopathy in patients with cirrhosis. Hepatol. Int. 2019, 13, 482–489. [Google Scholar] [CrossRef]
- Durgan, D.J.; Ganesh, B.P.; Cope, J.L.; Ajami, N.J.; Phillips, S.C.; Petrosino, J.F.; Hollister, E.B.; Bryan, R.M., Jr. Role of the gut microbiome in obstructive sleep apnea–induced hypertension. Hypertension 2016, 67, 469–474. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Wang, X.; Yu, X.; Hu, C.; Zhang, X. The family Coriobacteriaceae is a potential contributor to the beneficial effects of Roux-en-Y gastric bypass on type 2 diabetes. Surg. Obes. Relat. Dis. 2018, 14, 584–593. [Google Scholar] [CrossRef]
- Abecia, L.; Hoyles, L.; Khoo, C.; Frantz, N.; McCartney, A.L. Effects of a novel galactooligosaccharide on the faecal microbiota of healthy and inflammatory bowel disease cats during a randomized, double-blind, cross-over feeding study. Int. J. Probiotics Prebiotics 2010, 5, 61–68. [Google Scholar]
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Mörkl, S.; Lackner, S.; Müller, W.; Gorkiewicz, G.; Kashofer, K.; Oberascher, A.; Painold, A.; Holl, A.; Holzer, P.; Meinitzer, A.; et al. Gut microbiota and body composition in anorexia nervosa inpatients in comparison to athletes, overweight, obese, and normal weight controls. Int. J. Eat. Disord. 2017, 50, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Alderete, T.L.; Jones, R.B.; Chen, Z.; Kim, J.S.; Habre, R.; Lurmann, F.; Gilliland, F.D.; Goran, M.I. Exposure to traffic-related air pollution and the composition of the gut microbiota in overweight and obese adolescents. Environ. Res. 2018, 161, 472–478. [Google Scholar] [CrossRef]
- Wang, M.; Cao, J.; Gong, C.; Amakye, W.K.; Yao, M.; Ren, J. Exploring the microbiota-Alzheimer’s disease linkage using short-term antibiotic treatment followed by fecal microbiota transplantation. Brain Behav. Immun. 2021, 96, 227–238. [Google Scholar] [CrossRef]
- Chen, K.; Fu, Y.; Wang, Y.; Liao, L.; Xu, H.; Zhang, A.; Zhang, J.; Fan, L.; Ren, J.; Fang, B. Therapeutic effects of the in vitro cultured human gut microbiota as transplants on altering gut microbiota and improving symptoms associated with autism spectrum disorder. Microb. Ecol. 2020, 80, 475–486. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Yuan, W.-L.; Ye, M.; Yin, L.; Wang, S.-J. Changes in the intestinal microbiota of patients with Parkinson’s disease and their clinical significance. Int. J. Clin. Pharmacol. Ther. 2022, 61, 48–58. [Google Scholar] [CrossRef]
- Lin, Y.; Yan, G.; Feng, F.; Wang, M.; Long, F. Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy. Open Life Sci. 2022, 17, 139–154. [Google Scholar] [CrossRef]
- Hu, C.; Wang, P.; Yang, Y.; Li, J.; Jiao, X.; Yu, H.; Wei, Y.; Li, J.; Qin, Y. Chronic intermittent hypoxia participates in the pathogenesis of atherosclerosis and perturbs the formation of intestinal microbiota. Front. Cell. Infect. Microbiol. 2021, 11, 560201. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, Y.; Li, X.; Zhu, L.; Wang, X.; Li, L.; Sun, H.; Han, X.; Li, J. Myricetin relieves the symptoms of type 2 diabetes mice and regulates intestinal microflora. Biomed. Pharmacother. 2022, 153, 113530. [Google Scholar] [CrossRef]
- Regner, E.H.; Ohri, N.; Stahly, A.; Gerich, M.E.; Fennimore, B.P.; Ir, D.; Jubair, W.K.; Görg, C.; Siebert, J.; Robertson, C.E.; et al. Functional intraepithelial lymphocyte changes in inflammatory bowel disease and spondyloarthritis have disease specific correlations with intestinal microbiota. Arthritis Res. Ther. 2018, 20, 149. [Google Scholar] [CrossRef]
- Ng, S.C.; Lam, E.F.C.; Lam, T.T.Y.; Chan, Y.; Law, W.; Tse, P.C.H.; A Kamm, M.; Sung, J.J.Y.; Chan, F.K.L.; Wu, J.C.Y. Effect of probiotic bacteria on the intestinal microbiota in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2013, 28, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Brandwein, M.; Katz, I.; Katz, A.; Kohen, R. Beyond the gut: Skin microbiome compositional changes are associated with BMI. Hum. Microbiome J. 2019, 13, 100063. [Google Scholar] [CrossRef]
- Palomo-Buitrago, M.E.; Sabater-Masdeu, M.; Moreno-Navarrete, J.M.; Caballano-Infantes, E.; Arnoriaga-Rodríguez, M.; Coll, C.; Ramió, L.; Palomino-Schätzlein, M.; Gutiérrez-Carcedo, P.; Pérez-Brocal, V.; et al. Glutamate interactions with obesity, insulin resistance, cognition and gut microbiota composition. Acta Diabetol. 2019, 56, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Liu, R.; Chu, G.; Wang, Y.; Cui, H.; Zhang, T.; Bi, K.; Gao, P.; Song, Z.; Li, Q. A Comprehensive Analysis of Microflora and Metabolites in the Development of Ulcerative Colitis into Colorectal Cancer Based on the Lung–Gut Correlation Theory. Molecules 2022, 27, 5838. [Google Scholar] [CrossRef]
- Ling, Z.; Zhu, M.; Yan, X.; Cheng, Y.; Shao, L.; Liu, X.; Jiang, R.; Wu, S. Structural and functional dysbiosis of fecal microbiota in Chinese patients with Alzheimer’s disease. Front. Cell Dev. Biol. 2021, 8, 634069. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. NPJ Park. Dis. 2020, 6, 11. [Google Scholar] [CrossRef]
- Chen, Y.-h.; Xue, F.; Yu, S.-f.; Li, X.-s.; Liu, L.; Jia, Y.-y.; Yan, W.-j.; Tan, Q.-r.; Wang, H.-n.; Peng, Z.-w. Gut microbiota dysbiosis in depressed women: The association of symptom severity and microbiota function. J. Affect. Disord. 2021, 282, 391–400. [Google Scholar] [CrossRef]
- Yan, R.; Wang, K.; Wang, Q.; Jiang, H.; Lu, Y.; Chen, X.; Zhang, H.; Su, X.; Du, Y.; Chen, L.; et al. Probiotic Lactobacillus casei Shirota prevents acute liver injury by reshaping the gut microbiota to alleviate excessive inflammation and metabolic disorders. Microb. Biotechnol. 2022, 15, 247–261. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Mao, X.; Tao, Y.; Ran, X.; Zhao, H.; Xiong, J.; Li, L. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS ONE 2017, 12, e0172774. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, N.; Yoon, H.; Kim, Y.S.; Choi, S.I.; Park, J.H.; Lee, D.H. Compositional and functional changes in the gut microbiota in irritable bowel syndrome patients. Gut Liver 2021, 15, 253. [Google Scholar] [CrossRef]
- Lv, Y.; Qin, X.; Jia, H.; Chen, S.; Sun, W.; Wang, X. The association between gut microbiota composition and BMI in Chinese male college students, as analysed by next-generation sequencing. Br. J. Nutr. 2019, 122, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, J.; Hong, F.; Wang, S.; Jin, X.; Xue, T.; Jia, L.; Zhai, Y. Melatonin prevents obesity through modulation of gut microbiota in mice. J. Pineal Res. 2017, 62, e12399. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, C.; Liu, R.; Gao, L.; Ou, S.; Liu, L.; Peng, X. Ganoderma lucidum polysaccharide alleviating colorectal cancer by alteration of special gut bacteria and regulation of gene expression of colonic epithelial cells. J. Funct. Foods 2018, 47, 127–135. [Google Scholar] [CrossRef]
- Sheng, C.; Yang, K.; He, B.; Du, W.; Cai, Y.; Han, Y. Combination of gut microbiota and plasma amyloid-β as a potential index for identifying preclinical Alzheimer’s disease: A cross-sectional analysis from the SILCODE study. Alzheimers Res. Ther. 2022, 14, 35. [Google Scholar] [CrossRef]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef]
- Murros, K.E. Hydrogen sulfide produced by gut bacteria may induce Parkinson’s disease. Cells 2022, 11, 978. [Google Scholar] [CrossRef]
- Rhee, S.J.; Kim, H.; Lee, Y.; Lee, H.J.; Park, C.H.K.; Yang, J.; Kim, Y.-K.; Ahn, Y.M. The association between serum microbial DNA composition and symptoms of depression and anxiety in mood disorders. Sci. Rep. 2021, 11, 13987. [Google Scholar] [CrossRef]
- Szabo, H.; Piroska, M.; Hernyes, A.; Zoldi, L.; Juhasz, J.; Ligeti, B.; Makra, N.; Szabo, D.; Bikov, A.; Kunos, L.; et al. The Relationship between Atherosclerosis and Gut Microbiome in Patients with Obstructive Sleep Apnoea. Appl. Sci. 2022, 12, 11484. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The role of Enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 521-e248. [Google Scholar] [CrossRef]
- Million, M.; Diallo, A.; Raoult, D. Gut microbiota and malnutrition. Microb. Pathog. 2017, 106, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Alcoholado, L.; Ordóñez, R.; Otero, A.; Plaza-Andrade, I.; Laborda-Illanes, A.; Medina, J.A.; Ramos-Molina, B.; Gómez-Millán, J.; Queipo-Ortuño, M.I. Gut microbiota-mediated inflammation and gut permeability in patients with obesity and colorectal cancer. Int. J. Mol. Sci. 2020, 21, 6782. [Google Scholar] [CrossRef]
- Zhuang, Z.; Yang, R.; Wang, W.; Qi, L.; Huang, T. Associations between gut microbiota and Alzheimer’s disease, major depressive disorder, and schizophrenia. J. Neuroinflammation 2020, 17, 288. [Google Scholar] [CrossRef]
- Lou, M.; Cao, A.; Jin, C.; Mi, K.; Xiong, X.; Zeng, Z.; Pan, X.; Qie, J.; Qiu, S.; Niu, Y.; et al. Deviated and early unsustainable stunted development of gut microbiota in children with autism spectrum disorder. Gut 2022, 71, 1588–1599. [Google Scholar] [CrossRef]
- Pietrucci, D.; Cerroni, R.; Unida, V.; Farcomeni, A.; Pierantozzi, M.; Mercuri, N.B.; Biocca, S.; Stefani, A.; Desideri, A. Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Park. Relat. Disord. 2019, 65, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Maki, K.A.; Burke, L.A.; Calik, M.W.; Watanabe-Chailland, M.; Sweeney, D.; Romick-Rosendale, L.E.; Green, S.J.; Fink, A.M. Sleep fragmentation increases blood pressure and is associated with alterations in the gut microbiome and fecal metabolome in rats. Physiol. Genom. 2020, 52, 280–292. [Google Scholar] [CrossRef]
- Li, P.; Lu, B.; Gong, J.; Li, L.; Chen, G.; Zhang, J.; Chen, Y.; Tian, X.; Han, B.; Guo, Y.; et al. Chickpea extract ameliorates metabolic syndrome symptoms via restoring intestinal ecology and metabolic profile in type 2 diabetic rats. Mol. Nutr. Food Res. 2021, 65, 2100007. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.; Zhou, F.; Wang, X.; Shen, B.; Kamm, M.A.; et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef]
- Foley, K.P.; Zlitni, S.; Duggan, B.M.; Barra, N.G.; Anhê, F.F.; Cavallari, J.F.; Henriksbo, B.D.; Chen, C.Y.; Huang, M.; Lau, T.C.; et al. Gut microbiota impairs insulin clearance in obese mice. Mol. Metab. 2020, 42, 101067. [Google Scholar] [CrossRef]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Xu, G.; Ran, M.; Luo, W.; Wang, H. APOE-ε4 carrier status and gut microbiota dysbiosis in patients with Alzheimer disease. Front. Neurosci. 2021, 15, 619051. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.T.; Rowan-Nash, A.D.; Sheehan, A.E.; Walsh, R.F.; Sanzari, C.M.; Korry, B.J.; Belenky, P. Reductions in anti-inflammatory gut bacteria are associated with depression in a sample of young adults. Brain Behav. Immun. 2020, 88, 308–324. [Google Scholar] [CrossRef]
- Mancini, A.; Campagna, F.; Amodio, P.; Tuohy, K.M. Gut: Liver: Brain axis: The microbial challenge in the hepatic encephalopathy. Food Funct. 2018, 9, 1373–1388. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Indias, I.; Torres, M.; Montserrat, J.M.; Sanchez-Alcoholado, L.; Cardona, F.; Tinahones, F.J.; Gozal, D.; Poroyko, V.A.; Navajas, D.; Queipo-Ortuño, M.I.; et al. Intermittent hypoxia alters gut microbiota diversity in a mouse model of sleep apnoea. Eur. Respir. J. 2015, 45, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Vals-Delgado, C.; Alcala-Diaz, J.F.; Molina-Abril, H.; Roncero-Ramos, I.; Caspers, M.P.; Schuren, F.H.; Broek, T.J.V.D.; Luque, R.; Perez-Martinez, P.; Katsiki, N.; et al. An altered microbiota pattern precedes Type 2 diabetes mellitus development: From the CORDIOPREV study. J. Adv. Res. 2022, 35, 99–108. [Google Scholar] [CrossRef]
- Pozuelo, M.; Panda, S.; Santiago, A.; Mendez, S.; Accarino, A.; Santos, J.; Guarner, F.; Azpiroz, F.; Manichanh, C. Reduction of butyrate-and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci. Rep. 2015, 5, 12693. [Google Scholar] [CrossRef]
- Nakayama, J.; Yamamoto, A.; Palermo-Conde, L.A.; Higashi, K.; Sonomoto, K.; Tan, J.; Lee, Y.-K. Impact of westernized diet on gut microbiota in children on Leyte Island. Front. Microbiol. 2017, 8, 197. [Google Scholar] [CrossRef]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef]
- Zackular, J.P.; Baxter, N.T.; Iverson, K.D.; Sadler, W.D.; Petrosino, J.F.; Chen, G.Y.; Schloss, P.D. The gut microbiome modulates colon tumorigenesis. mBio 2013, 4, e00692-13. [Google Scholar] [CrossRef]
- Jung, J.H.; Kim, G.; Byun, M.S.; Lee, J.H.; Yi, D.; Park, H.; Lee, D.Y.; The KBASE Research Group. Gut microbiome alterations in preclinical Alzheimer’s disease. PLoS ONE 2022, 17, e0278276. [Google Scholar] [CrossRef] [PubMed]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Li, X.; Zhu, Y.; Sullivan, M.A.; Deng, B.; Zhai, X.; Lu, Y. Antidepressant shugan jieyu capsule alters gut microbiota and intestinal microbiome function in rats with chronic unpredictable mild stress-induced depression. Front. Pharmacol. 2022, 13, 828595. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J. Fecal microbiota and metabolome characteristics in cirrhotic patients accompanied with hepatic encephalopathy. Res. Sq. 2019. [Google Scholar] [CrossRef]
- Davies, N.; O’Sullivan, J.M.; Plank, L.D.; Murphy, R. Gut microbial predictors of type 2 diabetes remission following bariatric surgery. Obes. Surg. 2020, 30, 3536–3548. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1. [Google Scholar] [CrossRef] [PubMed]
- Hollister, E.B.; Cain, K.C.; Shulman, R.J.; Jarrett, M.E.; Burr, R.L.; Ko, C.; Zia, J.; Han, C.J.; Heitkemper, M.M. Relationships of microbiome markers with extra-intestinal, psychological distress and gastrointestinal symptoms, and quality of life in women with irritable bowel syndrome. J. Clin. Gastroenterol. 2020, 54, 175. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; Cortés-Martín, A.; Loria-Kohen, V.; Ramírez-de-Molina, A.; García-Mantrana, I.; Collado, M.C.; Espín, J.C.; Selma, M.V. Deciphering the human gut microbiome of urolithin metabotypes: Association with enterotypes and potential cardiometabolic health implications. Mol. Nutr. Food Res. 2019, 63, 1800958. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, X.; Wu, X.; Zhang, C.; Liu, J.; Hou, S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 2021, 13, 2. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Xiayu, X.; Shi, C.; Chen, W.; Song, N.; Fu, X.; Zhou, R.; Xu, Y.-F.; Huang, L.; et al. Altered gut microbiota in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2017, 60, 1241–1257. [Google Scholar] [CrossRef]
- Vascellari, S.; Melis, M.; Palmas, V.; Pisanu, S.; Serra, A.; Perra, D.; Santoru, M.L.; Oppo, V.; Cusano, R.; Uva, P.; et al. Clinical phenotypes of Parkinson’s disease associate with distinct gut microbiota and metabolome enterotypes. Biomolecules 2021, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Pourdeh, E.F.; Ulker, I. Do all Bariatric Surgery Methods Have the Same Effects on the Gut Microbiota? In Bariatric Surgery-Past and Present; IntechOpen: London, UK, 2022. [Google Scholar]
- Villanueva-Millan, M.J.; Leite, G.; Wang, J.; Morales, W.; Parodi, G.; Pimentel, M.L.; Barlow, G.M.; Mathur, R.; Rezaie, A.; Sanchez, M.; et al. Methanogens and hydrogen sulfide producing bacteria guide distinct gut microbe profiles and irritable bowel syndrome subtypes. Am. J. Gastroenterol. 2022, 117, 2055–2066. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; García-Fuentes, E.; Cardona, F.; Queipo-Ortuño, M.I.; Tinahones, F.J. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am. J. Transl. Res. 2016, 8, 5672. [Google Scholar] [PubMed]
- Taddese, R.; Garza, D.R.; Ruiter, L.N.; de Jonge, M.I.; Belzer, C.; Aalvink, S.; Nagtegaal, I.D.; Dutilh, B.E.; Boleij, A. Growth rate alterations of human colorectal cancer cells by 157 gut bacteria. Gut Microbes 2020, 12, 1799733. [Google Scholar] [CrossRef]
- Mancuso, C.; Santangelo, R. Alzheimer’s disease and gut microbiota modifications: The long way between preclinical studies and clinical evidence. Pharmacol. Res. 2018, 129, 329–336. [Google Scholar] [CrossRef]
- Abrishamkar, P.; Rodriguez, C.V.B.; Choudry, B.; Or, S. Antidepressant usage is associated with alterations in gut microbiota diversity and abundance in Parkinson’s Disease patients. Undergrad. J. Exp. Microbiol. Immunol. 2022, 8, 1–13. [Google Scholar]
- Luo, F.; Fang, C. Association between gut microbiota and post-stroke depression in Chinese population: A meta-analysis. Heliyon 2022, 8, e12605. [Google Scholar] [CrossRef]
- Shin, N.R.; Gu, N.; Choi, H.S.; Kim, H. Combined effects of Scutellaria baicalensis with metformin on glucose tolerance of patients with type 2 diabetes via gut microbiota modulation. Am. J. Physiol.-Endocrinol. Metab. 2020, 318, E52–E61. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Wang, R.; An, Y.; Gao, W.; Bai, L.; Li, Y.; Zhao, S.; Fan, J.; Liu, E. Western diet feeding influences gut microbiota profiles in apoE knockout mice. Lipids Health Dis. 2018, 17, 159. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Torres, M.; Sanchez-Alcoholado, L.; Cardona, F.; Almendros, I.; Gozal, D.; Montserrat, J.M.; Queipo-Ortuño, M.I.; Farré, R. Normoxic recovery mimicking treatment of sleep apnea does not reverse intermittent hypoxia-induced bacterial dysbiosis and low-grade endotoxemia in mice. Sleep 2016, 39, 1891–1897. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Li, T.; Du, M.; Chang, R.; Zhan, B.; Mao, X. Beneficial effects of potentilla discolor bunge water extract on inflammatory cytokines release and gut microbiota in high-fat diet and streptozotocin-induced type 2 diabetic mice. Nutrients 2019, 11, 670. [Google Scholar] [CrossRef] [PubMed]
- Soltys, K.; Stuchlikova, M.; Hlavaty, T.; Gaalova, B.; Budis, J.; Gazdarica, J.; Krajcovicova, A.; Zelinkova, Z.; Szemes, T.; Kuba, D.; et al. Seasonal changes of circulating 25-hydroxyvitamin D correlate with the lower gut microbiome composition in inflammatory bowel disease patients. Sci. Rep. 2020, 10, 6024. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qi, Q.; Wang, Y.; Wu, H.; Jin, X.; Yao, H.; Jin, D.; Liu, Y.; Wang, C. Gut microbiota was modulated by moxibustion stimulation in rats with irritable bowel syndrome. Chin. Med. 2018, 13, 63. [Google Scholar] [CrossRef]
- Monira, S.; Nakamura, S.; Gotoh, K.; Izutsu, K.; Watanabe, H.; Alam, N.H.; Endtz, H.P.; Cravioto, A.; Ali, S.I.; Nakaya, T.; et al. Gut microbiota of healthy and malnourished children in Bangladesh. Front. Microbiol. 2011, 2, 228. [Google Scholar] [CrossRef]
- Li, L.; Li, R.; Zhu, R.; Chen, B.; Tian, Y.; Zhang, H.; Xia, B.; Jia, Q.; Wang, L.; Zhao, D.; et al. Salvianolic acid B prevents body weight gain and regulates gut microbiota and LPS/TLR4 signaling pathway in high-fat diet-induced obese mice. Food Funct. 2020, 11, 8743–8756. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, Y.; Deng, B.; Li, J.; Cao, H.; Qu, Y.; Qian, X.; Zhong, G. Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget 2016, 7, 85318. [Google Scholar] [CrossRef]
- Shen, L.; Liu, L.; Ji, H.-F. Alzheimer’s disease histological and behavioral manifestations in transgenic mice correlate with specific gut microbiome state. J. Alzheimers Dis. 2017, 56, 385–390. [Google Scholar] [CrossRef]
- Coretti, L.; Cristiano, C.; Florio, E.; Scala, G.; Lama, A.; Keller, S.; Cuomo, M.; Russo, R.; Pero, R.; Paciello, O.; et al. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci. Rep. 2017, 7, 45356. [Google Scholar] [CrossRef]
- Sun, J.; Li, H.; Jin, Y.; Yu, J.; Mao, S.; Su, K.-P.; Ling, Z.; Liu, J. Probiotic Clostridium butyricum ameliorated motor deficits in a mouse model of Parkinson’s disease via gut microbiota-GLP-1 pathway. Brain Behav. Immun. 2021, 91, 703–715. [Google Scholar] [CrossRef]
- Duan, J.; Huang, Y.; Tan, X.; Chai, T.; Wu, J.; Zhang, H.; Li, Y.; Hu, X.; Zheng, P.; Ji, P.; et al. Characterization of gut microbiome in mice model of depression with divergent response to escitalopram treatment. Transl. Psychiatry 2021, 11, 303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, F.; Shen, Y.; Chen, Y.; Ma, J. Sleep apnea is associated with the increase of certain genera of Ruminococcaceae and Lachnospiraceae in the gut microbiome of hypertensive patients. Expert Rev. Respir. Med. 2022, 16, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lou, H.; Peng, Y.; Chen, S.; Zhang, Y.; Li, X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef]
- Maukonen, J.; Kolho, K.-L.; Paasela, M.; Honkanen, J.; Klemetti, P.; Vaarala, O.; Saarela, M. Altered fecal microbiota in paediatric inflammatory bowel disease. J. Crohn’s Colitis 2015, 9, 1088–1095. [Google Scholar] [CrossRef]
- Schulz, N.; Belheouane, M.; Dahmen, B.; Ruan, V.A.; Specht, H.E.; Dempfle, A.; Herpertz-Dahlmann, B.; Baines, J.F.; Seitz, J. Gut microbiota alteration in adolescent anorexia nervosa does not normalize with short-term weight restoration. Int. J. Eat. Disord. 2021, 54, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ni, Y.; Wang, Z.; Tu, W.; Ni, L.; Zhuge, F.; Zheng, A.; Hu, L.; Zhao, Y.; Zheng, L.; et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 2020, 12, 1832857. [Google Scholar] [CrossRef] [PubMed]
- Hexun, Z.; Miyake, T.; Maekawa, T.; Mori, H.; Yasukawa, D.; Ohno, M.; Nishida, A.; Andoh, A.; Tani, M. High abundance of lachnospiraceae in the human gut microbiome is related to high immunoscores in advanced colorectal cancer. Cancer Immunol. Immunother. 2023, 72, 315–326. [Google Scholar] [CrossRef]
- Ding, X.; Xu, Y.; Zhang, X.; Zhang, L.; Duan, G.; Song, C.; Li, Z.; Yang, Y.; Wang, Y.; Wang, X.; et al. Gut microbiota changes in patients with autism spectrum disorders. J. Psychiatr. Res. 2020, 129, 149–159. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Zhang, H.-h.; Liu, J.; Lv, Y.-j.; Jiang, Y.-l.; Pan, J.-x.; Zhu, Y.-j.; Huang, M.-g.; Zhang, S.-k. Changes in intestinal microbiota of type 2 diabetes in mice in response to dietary supplementation with instant tea or matcha. Can. J. Diabetes 2020, 44, 44–52. [Google Scholar] [CrossRef]
- Guo, H.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. The Potential Therapeutic Role of Lactobacillaceae rhamnosus for Treatment of Inflammatory Bowel Disease. Foods 2023, 12, 692. [Google Scholar] [CrossRef] [PubMed]
- Taverniti, V.; Guglielmetti, S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. WJG 2014, 20, 8821. [Google Scholar] [PubMed]
- Ye, J.; Zhao, Y.; Chen, X.; Zhou, H.; Yang, Y.; Zhang, X.; Huang, Y.; Zhang, N.; Lui, E.M.; Xiao, M. Pu-erh tea ameliorates obesity and modulates gut microbiota in high fat diet fed mice. Food Res. Int. 2021, 144, 110360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, X.; Zuo, T.; Yu, J. The composition of colonic commensal bacteria according to anatomical localization in colorectal cancer. Engineering 2017, 3, 90–97. [Google Scholar] [CrossRef]
- Fu, S.-C.; Lee, C.-H.; Wang, H. Exploring the association of autism spectrum disorders and constipation through analysis of the gut microbiome. Int. J. Environ. Res. Public Health 2021, 18, 667. [Google Scholar] [CrossRef]
- Hopfner, F.; Künstner, A.; Müller, S.H.; Künzel, S.; Zeuner, K.E.; Margraf, N.G.; Deuschl, G.; Baines, J.F.; Kuhlenbäumer, G. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res. 2017, 1667, 41–45. [Google Scholar] [CrossRef]
- Chevalier, G.; Siopi, E.; Guenin-Macé, L.; Pascal, M.; Laval, T.; Rifflet, A.; Boneca, I.G.; Demangel, C.; Colsch, B.; Pruvost, A.; et al. Effect of gut microbiota on depressive-like behaviors in mice is mediated by the endocannabinoid system. Nat. Commun. 2020, 11, 6363. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, T.; Lu, W.; Duan, X.; Luo, X.; Liu, S.; Chen, Y.; Li, Y.; Chen, J.; Liao, J.; et al. Altered airway microbiota composition in patients with pulmonary hypertension. Hypertension 2020, 76, 1589–1599. [Google Scholar] [CrossRef]
- Balmasova, I.P.; Olekhnovich, E.I.; Klimina, K.M.; Korenkova, A.A.; Vakhitova, M.T.; Babaev, E.A.; Ovchinnikova, L.A.; Lomakin, Y.A.; Smirnov, I.V.; Tsarev, V.N.; et al. Drift of the subgingival periodontal microbiome during chronic periodontitis in Type 2 diabetes mellitus patients. Pathogens 2021, 10, 504. [Google Scholar] [CrossRef]
- Hourigan, S.; Chen, L.; Grigoryan, Z.; Laroche, G.; Weidner, M.; Sears, C.; Oliva-Hemker, M. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment. Pharmacol. Ther. 2015, 42, 741–752. [Google Scholar] [CrossRef]
- Shin, S.; Cho, K.Y. Altered gut microbiota and shift in Bacteroidetes between young obese and normal-weight Korean children: A cross-sectional observational study. BioMed Res. Int. 2020, 2020. [Google Scholar] [CrossRef]
- Wang, P.; Ding, S.; Sun, L.; Feng, Y.; Guo, K.; Zhu, Y.; Huang, D.; Ruan, S. Characteristics and differences of gut microbiota in patients with different traditional Chinese medicine syndromes of colorectal cancer and normal population. J. Cancer 2020, 11, 7357. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, X.; Wu, X.; Cao, H.; Li, X.; Hou, Z.; Wang, B.; Liu, J.; Ji, X.; Zhang, P.; et al. A comparison of the composition and functions of the oral and gut microbiotas in Alzheimer’s patients. Front. Cell. Infect. Microbiol. 2022, 1241, 942460. [Google Scholar] [CrossRef] [PubMed]
- Shen, L. Gut, oral and nasal microbiota and Parkinson’s disease. Microb. Cell Factories 2020, 19, 50. [Google Scholar] [CrossRef] [PubMed]
- Tien, N.T.T. Analysis of the Gut Microbiota of Japanese Alzheimer’s Disease Patients and Characterization of Their Butyrate-Producing Bacteria. 2018. Available online: https://ousar.lib.okayama-u.ac.jp/files/public/5/56307/20181212153730430590/K0005847_Fulltext.pdf (accessed on 23 September 2023).
- Chen, Z.; Xie, Y.; Zhou, F.; Zhang, B.; Wu, J.; Yang, L.; Xu, S.; Stedtfeld, R.; Chen, Q.; Liu, J.; et al. Featured gut microbiomes associated with the progression of chronic hepatitis B disease. Front. Microbiol. 2020, 11, 383. [Google Scholar] [CrossRef]
- Gonzalez-Mercado, V.J.; Lim, J.; Saligan, L.N.; Perez, N.; Rodriguez, C.; Bernabe, R.; Ozorio, S.; Pedro, E.; Sepehri, F.; Aouizerat, B. Gut microbiota and depressive symptoms at the end of CRT for rectal cancer: A cross-sectional pilot study. Depress. Res. Treat. 2021, 2021, 7967552. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, S.; Yang, Y.; Wang, Z.; Wang, B.; Zhang, B.; Yu, J.; Lu, W.; Pan, M.; Zhao, J.; et al. The diversity of gut microbiota in type 2 diabetes with or without cognitive impairment. Aging Clin. Exp. Res. 2021, 33, 589–601. [Google Scholar] [CrossRef]
- Knights, D.; Lassen, K.G.; Xavier, R.J. Advances in inflammatory bowel disease pathogenesis: Linking host genetics and the microbiome. Gut 2013, 62, 1505–1510. [Google Scholar] [CrossRef]
- Yun, Y.; Kim, H.-N.; Lee, E.-J.; Ryu, S.; Chang, Y.; Shin, H.; Kim, H.-L.; Kim, T.H.; Yoo, K.; Kim, H.Y. Fecal and blood microbiota profiles and presence of nonalcoholic fatty liver disease in obese versus lean subjects. PLoS ONE 2019, 14, e0213692. [Google Scholar] [CrossRef]
- Kim, M.-H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.-L.; Kim, H.-N. Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef]
- Park, J.-Y.; Choi, J.; Lee, Y.; Lee, J.-E.; Lee, E.-H.; Kwon, H.-J.; Yang, J.; Jeong, B.-R.; Kim, Y.-K.; Han, P.-L. Metagenome analysis of bodily microbiota in a mouse model of Alzheimer disease using bacteria-derived membrane vesicles in blood. Exp. Neurobiol. 2017, 26, 369. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, P.; Chen, Z.; Sui, X.; Xie, X.; Zhang, J. Alteration of the fecal microbiota in North-Eastern Han Chinese population with sporadic Parkinson’s disease. Neurosci. Lett. 2019, 707, 134297. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-F.; Chuang, C.-H.; Wang, Y.-P.; Lin, Y.-B.; Tu, P.-C.; Liu, P.-Y.; Wu, P.-S.; Lin, C.-Y.; Lu, C.-L. Differences in gut microbiota correlate with symptoms and regional brain volumes in patients with late-life depression. Front. Aging Neurosci. 2022, 14, 885393. [Google Scholar] [CrossRef] [PubMed]
- Xiang, K.; Zhang, J.-J.; Xu, Y.-Y.; Zhong, X.; Ni, J.; Pan, H.-F. Genetically predicted causality of 28 gut microbiome families and type 2 diabetes mellitus risk. Front. Endocrinol. 2022, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Danilova, N.; Abdulkhakov, S.; Grigoryeva, T.; Dubinkina, V.; Odintsova, V.; Tyakht, A.; Pavlenko, A.; Odintsova, A.; Abdulkhakov, R.; Govorun, V. P794 Alterations of intestinal microbiota in ulcerative colitis. J. Crohn’s Colitis 2017, 11 (Suppl. S1), S486. [Google Scholar] [CrossRef]
- Chen, H.; Ou, R.; Tang, N.; Su, W.; Yang, R.; Yu, X.; Zhang, G.; Jiao, J.; Zhou, X. Alternation of the gut microbiota in irritable bowel syndrome: An integrated analysis based on multicenter amplicon sequencing data. J. Transl. Med. 2023, 21, 117. [Google Scholar] [CrossRef]
- Smitka, K.; Prochazkova, P.; Roubalova, R.; Dvorak, J.; Papezova, H.; Hill, M.; Pokorny, J.; Kittnar, O.; Bilej, M.; Tlaskalova-Hogenova, H. Current aspects of the role of autoantibodies directed against appetite-regulating hormones and the gut microbiome in eating disorders. Front. Endocrinol. 2021, 12, 613983. [Google Scholar] [CrossRef]
- Aranaz, P.; Ramos-Lopez, O.; Cuevas-Sierra, A.; Martinez, J.A.; Milagro, F.I.; Riezu-Boj, J.I. A predictive regression model of the obesity-related inflammatory status based on gut microbiota composition. Int. J. Obes. 2021, 45, 2261–2268. [Google Scholar] [CrossRef]
- Long, S.; Yang, Y.; Shen, C.; Wang, Y.; Deng, A.; Qin, Q.; Qiao, L. Metaproteomics characterizes human gut microbiome function in colorectal cancer. NPJ Biofilms Microbiomes 2020, 6, 14. [Google Scholar] [CrossRef]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Fontana, A.; Manchia, M.; Panebianco, C.; Paribello, P.; Arzedi, C.; Cossu, E.; Garzilli, M.; Montis, M.A.; Mura, A.; Pisanu, C.; et al. Exploring the role of gut microbiota in major depressive disorder and in treatment resistance to antidepressants. Biomedicines 2020, 8, 311. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Sterbini, F.P.; Petito, V.; et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, D.; Zhang, L.; Zhao, Q.; Ma, Y.; Zhang, X.; Zhao, S.; Chen, C. Diaphragma juglandis extracts modifies the gut microbiota during prevention of type 2 diabetes in rats. J. Ethnopharmacol. 2022, 283, 114484. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Guo, J.; Chang, X.; Gui, S. Painong-San extract alleviates dextran sulfate sodium-induced colitis in mice by modulating gut microbiota, restoring intestinal barrier function and attenuating TLR4/NF-κB signaling cascades. J. Pharm. Biomed. Anal. 2022, 209, 114529. [Google Scholar] [CrossRef] [PubMed]
- Sampsell, K.; Wang, W.; Ohland, C.; Mager, L.F.; Pett, N.; Lowry, D.E.; Sales, K.M.; McNeely, M.L.; McCoy, K.D.; Culos-Reed, S.N.; et al. Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer. Cancers 2022, 14, 2722. [Google Scholar] [CrossRef] [PubMed]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The gut microbiome of Mexican children affected by obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zuo, Z.; Zhuang, J.; Qu, Z.; Han, S.; Jin, W.; Liu, J.; Han, S. Prediction model of poorly differentiated colorectal cancer (CRC) based on gut bacteria. BMC Microbiol. 2022, 22, 312. [Google Scholar]
- Jones, J.; Reinke, S.N.; Mousavi-Derazmahalleh, M.; Palmer, D.J.; Christophersen, C.T. Changes to the gut microbiome in young children showing early behavioral signs of autism. Front. Microbiol. 2022, 13, 905901. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Teng, T.; Jiang, Y.; Xiang, Y.; Fan, L.; Yu, Y.; Zhou, X.; Xie, P. Comparative analysis of gut microbiota and fecal metabolome features among multiple depressive animal models. J. Affect. Disord. 2022, 314, 103–111. [Google Scholar] [CrossRef]
- Jian, J.; Nie, M.T.; Xiang, B.; Qian, H.; Yin, C.; Zhang, X.; Zhang, M.; Zhu, X.; Xie, W.-F. Rifaximin ameliorates non-alcoholic steatohepatitis in mice through regulating gut microbiome-related bile acids. Front. Pharmacol. 2022, 13, 1099. [Google Scholar] [CrossRef] [PubMed]
- Maskarinec, G.; Raquinio, P.; Kristal, B.S.; Setiawan, V.W.; Wilkens, L.R.; Franke, A.A.; Lim, U.; Le Marchand, L.; Randolph, T.W.; Lampe, J.W.; et al. The gut microbiome and type 2 diabetes status in the Multiethnic Cohort. PLoS ONE 2021, 16, e0250855. [Google Scholar] [CrossRef] [PubMed]
- Rengarajan, S.; Vivio, E.E.; Parkes, M.; Peterson, D.A.; Roberson, E.D.; Newberry, R.D.; Ciorba, M.A.; Hsieh, C.-S. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 2020, 11, 405–420. [Google Scholar] [CrossRef]
- Shivakumar, N.; Sivadas, A.; Devi, S.; Jahoor, F.; McLaughlin, J.; Smith, C.P.; Kurpad, A.V.; Mukhopadhyay, A. Gut microbiota profiles of young South Indian children: Child sex-specific relations with growth. PLoS ONE 2021, 16, e0251803. [Google Scholar] [CrossRef] [PubMed]
- Schott, E.M.; Farnsworth, C.W.; Grier, A.; Lillis, J.A.; Soniwala, S.; Dadourian, G.H.; Bell, R.D.; Doolittle, M.L.; Villani, D.A.; Awad, H.; et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight 2018, 3, e95997. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ling, Z.; Li, L. The intestinal microbiota and colorectal cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef]
- Li, P.; Killinger, B.A.; Ensink, E.; Beddows, I.; Yilmaz, A.; Lubben, N.; Lamp, J.; Schilthuis, M.; Vega, I.E.; Woltjer, R.; et al. Gut microbiota dysbiosis is associated with elevated bile acids in Parkinson’s disease. Metabolites 2021, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Chen, H.; Li, X.; Li, D.; Sun, Y.; Yang, L.; Ma, Y.; Chen, E.C.Y. Lactobacillus paracasei IMC 502 ameliorates type 2 diabetes by mediating gut microbiota–SCFA–hormone/inflammation pathway in mice. J. Sci. Food Agric. 2023, 103, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’sullivan, O.; Ross, R.P.; O’toole, P.W.; Shanahan, F.; Cotter, P.D. Targeting the microbiota to address diet-induced obesity: A time dependent challenge. PLoS ONE 2013, 8, e65790. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, D.; Zhou, G.; Li, C. Dietary pattern, gut microbiota, and Alzheimer’s disease. J. Agric. Food Chem. 2020, 68, 12800–12809. [Google Scholar] [CrossRef] [PubMed]
- Inoue, R.; Sakaue, Y.; Sawai, C.; Sawai, T.; Ozeki, M.; Romero-Pérez, G.A.; Tsukahara, T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci. Biotechnol. Biochem. 2016, 80, 2450–2458. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Gao, Y.; Qiu, Y.; Jiang, S.; Zhang, Q.; Zhang, J.; Wang, L.; Zhang, Y.; Wang, L.; Nie, K. Gut microbiota altered in mild cognitive impairment compared with normal cognition in sporadic Parkinson’s disease. Front. Neurol. 2020, 11, 137. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. Fto deficiency reduces anxiety-and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 2019, 9, 721. [Google Scholar] [CrossRef]
- Bajaj, J.S. The role of microbiota in hepatic encephalopathy. Gut Microbes 2014, 5, 397–403. [Google Scholar] [CrossRef]
- Palm, N.W.; De Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Riezu-Boj, J.I.; Guruceaga, E.; Milagro, F.I.; Martínez, J.A. Sex-specific associations between gut Prevotellaceae and host genetics on adiposity. Microorganisms 2020, 8, 938. [Google Scholar] [CrossRef]
- Hang, Z.; Lei, T.; Zeng, Z.; Cai, S.; Bi, W.; Du, H. Composition of intestinal flora affects the risk relationship between Alzheimer’s disease/Parkinson’s disease and cancer. Biomed. Pharmacother. 2022, 145, 112343. [Google Scholar] [CrossRef]
- Sun, H.; You, Z.; Jia, L.; Wang, F. Autism spectrum disorder is associated with gut microbiota disorder in children. BMC Pediatr. 2019, 19, 516. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef]
- Wu, J.; Li, J.; Gaurav, C.; Muhammad, U.; Chen, Y.; Li, X.; Chen, J.; Wang, Z. CUMS and dexamethasone induce depression-like phenotypes in mice by differentially altering gut microbiota and triggering macroglia activation. Gen. Psychiatry 2021, 34, e100529. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Betrapally, N.S.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; White, M.B.; Unser, A.; Thacker, L.R.; Sanyal, A.J.; Kang, D.J.; et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 2015, 62, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Čipčić Paljetak, H.; Barešić, A.; Panek, M.; Perić, M.; Matijašić, M.; Lojkić, I.; Barišić, A.; Vranešić Bender, D.; Ljubas Kelečić, D.; Brinar, M.; et al. Gut microbiota in mucosa and feces of newly diagnosed, treatment-naïve adult inflammatory bowel disease and irritable bowel syndrome patients. Gut Microbes 2022, 14, 2083419. [Google Scholar] [CrossRef] [PubMed]
- Ahasan, M.S.; Waltzek, T.B.; Owens, L.; Ariel, E. Characterisation and comparison of the mucosa-associated bacterial communities across the gastrointestinal tract of stranded green turtles, Chelonia mydas. AIMS Microbiol. 2020, 6, 361. [Google Scholar]
- Orbe-Orihuela, Y.C.; Godoy-Lozano, E.E.; Lagunas-Martínez, A.; Castañeda-Márquez, A.C.; Murga-Garrido, S.; Díaz-Benítez, C.E.; Ochoa-Leyva, A.; Cornejo-Granados, F.; Cruz, M.; Estrada, K.; et al. Association of Gut Microbiota with Dietary-dependent Childhood Obesity. Arch. Med. Res. 2022, 53, 407–415. [Google Scholar] [CrossRef]
- Phipps, O.; Al-Hassi, H.O.; Quraishi, M.N.; Dickson, E.A.; Segal, J.; Steed, H.; Kumar, A.; Acheson, A.G.; Beggs, A.D.; Brookes, M.J. Oral and intravenous iron therapy differentially alter the on-and off-tumor microbiota in anemic colorectal cancer patients. Cancers 2021, 13, 1341. [Google Scholar] [CrossRef]
- Ghyselinck, J.; Verstrepen, L.; Moens, F.; Van Den Abbeele, P.; Bruggeman, A.; Said, J.; Smith, B.; Barker, L.A.; Jordan, C.; Leta, V.; et al. Influence of probiotic bacteria on gut microbiota composition and gut wall function in an in-vitro model in patients with Parkinson’s disease. Int. J. Pharm. X 2021, 3, 100087. [Google Scholar] [CrossRef]
- Kelly, J.R.; Borre, Y.; O’Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- Bajaj, J. Potential mechanisms of action of rifaximin in the management of hepatic encephalopathy and other complications of cirrhosis. Aliment. Pharmacol. Ther. 2016, 43, 11–26. [Google Scholar] [CrossRef]
- Eun, C.S.; Kwak, M.-J.; Han, D.S.; Lee, A.R.; Park, D.I.; Yang, S.-K.; Kim, Y.S.; Kim, J.F. Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC Gastroenterol. 2016, 16, 28. [Google Scholar] [CrossRef]
- Tang, B.; Hu, Y.; Chen, J.; Su, C.; Zhang, Q.; Huang, C. Oral and fecal microbiota in patients with diarrheal irritable bowel syndrome. Heliyon 2023, 9, e13114. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-T.; Meng, X.-Z.; Zhang, J.-H.; Dai, Y.-F.; Shen, Y.; Xu, X.-Y.; Wang, R.-Q.; Li, J.-L. 16S rRNA sequencing analysis of the correlation between the intestinal microbiota and body-mass of grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100699. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Lu, W.; Wang, H.; Wu, W.; Zhou, Q.; Chen, Y.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. A multiphase dietetic protocol incorporating an improved ketogenic diet enhances weight loss and alters the gut microbiome of obese people. Int. J. Food Sci. Nutr. 2022, 73, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McDowell, A.; Kim, E.K.; Seo, H.; Lee, W.H.; Moon, C.-M.; Kym, S.-M.; Lee, D.H.; Park, Y.S.; Jee, Y.-K.; et al. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Park, S.; Wu, X. Modulation of the Gut Microbiota in Memory Impairment and Alzheimer’s Disease via the Inhibition of the Parasympathetic Nervous System. Int. J. Mol. Sci. 2022, 23, 13574. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Hou, B.; Xu, B.; Ji, L.; Wu, Y. Curcumin Regulates Gut Microbiota and Exerts a Neuroprotective Effect in the MPTP Model of Parkinson’s Disease. Evid. Based Complement. Alternat. Med. 2022, 2022, 9110560. [Google Scholar] [CrossRef]
- Yu, M.; Jia, H.-M.; Qin, L.-L.; Zou, Z.-M. Gut microbiota and gut tissue metabolites involved in development and prevention of depression. J. Affect. Disord. 2022, 297, 8–17. [Google Scholar] [CrossRef]
- Wang, J.Y.; Bajaj, J.S.; Wang, J.B.; Shang, J.; Zhou, X.M.; Guo, X.L.; Zhu, X.; Na Meng, L.; Jiang, H.X.; Mi, Y.Q.; et al. Lactulose improves cognition, quality of life, and gut microbiota in minimal hepatic encephalopathy: A multicenter, randomized controlled trial. J. Dig. Dis. 2019, 20, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; He, X.; Chen, H.; He, Q.; Yao, Z.; Li, Y.; Yang, H.; Simpson, S. Oil tea improves glucose and lipid levels and alters gut microbiota in type 2 diabetic mice. Nutr. Res. 2018, 57, 67–77. [Google Scholar] [CrossRef]
- Dou, X.; Gao, N.; Yan, D.; Shan, A. Sodium butyrate alleviates mouse colitis by regulating gut microbiota dysbiosis. Animals 2020, 10, 1154. [Google Scholar] [CrossRef]
- Lian, Q.; Ding, H.; Zhu, H.; Zhang, C.; Yu, S.; Jie, H.; Zhang, Y.; Wu, B.; Liang, G.; Zhang, G.; et al. Study of Jianpi mixture on intestinal microbiota of diarrhea irritable bowel syndrome mice. Evid. Based Complement. Alternat. Med. 2020, 2020, 5241308. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Jiao, S.; Dai, Y.; An, J.; Lv, J.; Yan, X.; Wang, J.; Han, B. Probiotic Bacillus amyloliquefaciens C-1 improves growth performance, stimulates GH/IGF-1, and regulates the gut microbiota of growth-retarded beef calves. Front. Microbiol. 2018, 9, 2006. [Google Scholar] [CrossRef] [PubMed]
- Koliarakis, I.; Athanasakis, E.; Sgantzos, M.; Mariolis-Sapsakos, T.; Xynos, E.; Chrysos, E.; Souglakos, J.; Tsiaoussis, J. Intestinal microbiota in colorectal cancer surgery. Cancers 2020, 12, 3011. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Quan, M.; Zhao, H.; Jia, J. Gut microbiota changes and their correlation with cognitive and neuropsychiatric symptoms in Alzheimer’s disease. J. Alzheimers Dis. 2021, 81, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Petrak, F.; Herpertz, S.; Hirsch, J.; Röhrig, B.; Donati-Hirsch, I.; Juckel, G.; Meier, J.J.; Gatermann, S. Gut microbiota differs in composition between adults with type 1 diabetes with or without depression and healthy control participants: A case-control study. BMC Microbiol. 2022, 22, 169. [Google Scholar] [CrossRef] [PubMed]
- Badran, M.; Khalyfa, A.; Ericsson, A.; Gozal, D. Fecal microbiota transplantation from mice exposed to chronic intermittent hypoxia elicits sleep disturbances in naïve mice. Exp. Neurol. 2020, 334, 113439. [Google Scholar] [CrossRef]
- De Palma, G.; Lynch, M.D.; Lu, J.; Dang, V.T.; Deng, Y.; Jury, J.; Umeh, G.; Miranda, P.M.; Pastor, M.P.; Sidani, S.; et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017, 9, eaaf6397. [Google Scholar] [CrossRef]
- Bai, J.; Hu, Y.; Bruner, D. Composition of gut microbiota and its association with body mass index and lifestyle factors in a cohort of 7–18 years old children from the American Gut Project. Pediatr. Obes. 2019, 14, e12480. [Google Scholar] [CrossRef] [PubMed]
- Bäuerl, C.; Collado, M.C.; Diaz Cuevas, A.; Viña, J.; Pérez Martínez, G. Shifts in Gut Microbiota Composition in an APP/PSS1 Transgenic Mouse Model of Alzheimer’s Disease during Lifespan; Blackwell Science Ltd.: Oxford, UK, 2018; pp. 464–471. [Google Scholar]
- Jin, M.; Li, J.; Liu, F.; Lyu, N.; Wang, K.; Wang, L.; Liang, S.; Tao, S.; Zhu, B.; Alkasir, R. Analysis of the gut microflora in patients with Parkinson’s disease. Front. Neurosci. 2019, 13, 1184. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Xie, Z.; Bai, L.; Xiong, P.; Chen, F.; Zhu, T.; Peng, Q.; Wu, H.; Zhou, Y.; et al. Metformin modulates microbiota-derived inosine and ameliorates methamphetamine-induced anxiety and depression-like withdrawal symptoms in mice. Biomed. Pharmacother. 2022, 149, 112837. [Google Scholar] [CrossRef]
- Chassard, C.; Dapoigny, M.; Scott, K.P.; Crouzet, L.; Del’Homme, C.; Marquet, P.; Martin, J.C.; Pickering, G.; Ardid, D.; Eschalier, A.; et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012, 35, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Morissette, B.; Talbot, G.; Beaulieu, C.; Lessard, M. Growth performance of piglets during the first two weeks of lactation affects the development of the intestinal microbiota. J. Anim. Physiol. Anim. Nutr. 2018, 102, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-N.; Liu, X.-T.; Liang, Z.-H.; Wang, J.-H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837. [Google Scholar] [CrossRef]
- Tran, T.T.; Corsini, S.; Kellingray, L.; Hegarty, C.; Le Gall, G.; Narbad, A.; Müller, M.; Tejera, N.; O’Toole, P.W.; Minihane, A.-M.; et al. APOE genotype influences the gut microbiome structure and function in humans and mice: Relevance for Alzheimer’s disease pathophysiology. FASEB J. 2019, 33, 8221. [Google Scholar] [CrossRef] [PubMed]
- Onyema Oshim, I.; Regina Agbakoba, N.; Celestine Oguejiofor, O.; Anukam, K.C. Selective Microbial Biomarkers in Type-2 Diabetes with Principal Component Analysis and Receiver-operating Characteristic Curves. Int. J. Sci. Res. Dent. Med. Sci. 2021, 3, 23–34. [Google Scholar]
- Chen, H.; Ou, R.; Tang, N.; Su, W.; Yang, R.; Yu, X.; Zhang, G.; Jiao, J.; Zhou, X. An Integrated Multicenter Amplicon Sequencing Data Reveals New Evidence of the Interaction between the Gut Microbiota and Irritable Bowel Syndrome. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Lan, D.; Ji, W.; Lin, B.; Chen, Y.; Huang, C.; Xiong, X.; Fu, M.; Mipam, T.D.; Ai, Y.; Zeng, B.; et al. Correlations between gut microbiota community structures of Tibetans and geography. Sci. Rep. 2017, 7, 16982. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dong, W.; Zhao, J.; Wu, J.; Xia, J.; Xie, S.; Song, X. Gut microbiota profiling variated during colorectal cancer development in mouse. BMC Genom. 2022, 23, 848. [Google Scholar] [CrossRef]
- Tartaglione, A.M.; Villani, A.; Ajmone-Cat, M.A.; Minghetti, L.; Ricceri, L.; Pazienza, V.; De Simone, R.; Calamandrei, G. Maternal immune activation induces autism-like changes in behavior, neuroinflammatory profile and gut microbiota in mouse offspring of both sexes. Transl. Psychiatry 2022, 12, 384. [Google Scholar] [CrossRef] [PubMed]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems 2020, 5, e00561-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, D.; Lai, J.; Fu, Y.; Wu, L.; Huang, H.; Pan, Y.; Jiang, J.; Xi, C.; Che, Z.; et al. Characteristics of the gut microbiota in bipolar depressive disorder patients with distinct weight. CNS Neurosci. Ther. 2023, 29, 74–83. [Google Scholar] [CrossRef]
- Hyun, C.-K. Molecular and pathophysiological links between metabolic disorders and inflammatory bowel diseases. Int. J. Mol. Sci. 2021, 22, 9139. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Pompili, M.; Gasbarrini, A. Rifaximin Re-Treatment in Patients with Irritable Bowel Syndrome: Feels Like the First Time? Springer: Berlin/Heidelberg, Germany, 2017; pp. 2220–2222. [Google Scholar]
- Coley, E.J.; Hsiao, E.Y. Malnutrition and the microbiome as modifiers of early neurodevelopment. Trends Neurosci. 2021, 44, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Ngowi, E.E.; Wang, Y.Z.; Khattak, S.; Khan, N.H.; Mahmoud, S.S.M.; Helmy, Y.A.S.H.; Jiang, Q.; Li, T.; Duan, S.; Ji, X.; et al. Impact of the factors shaping gut microbiota on obesity. J. Appl. Microbiol. 2021, 131, 2131–2147. [Google Scholar] [CrossRef]
- Raskov, H.; Burcharth, J.; Pommergaard, H.-C. Linking gut microbiota to colorectal cancer. J. Cancer 2017, 8, 3378. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gao, Z.; Liu, C.; Liu, T.; Gao, J.; Cai, Y.; Fan, X. Alteration of Gut Microbiota: New Strategy for Treating Autism Spectrum Disorder. Front. Cell Dev. Biol. 2022, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Li, R.; Shi, W.; Yao, L. Role of the gut-microbiota-metabolite axis in the rotenone model of early-stage Parkinson’s Disease. Metab. Brain Dis. 2022, 37, 2511–2520. [Google Scholar] [CrossRef]
- Song, X.; Wang, W.; Ding, S.; Liu, X.; Wang, Y.; Ma, H. Puerarin ameliorates depression-like behaviors of with chronic unpredictable mild stress mice by remodeling their gut microbiota. J. Affect. Disord. 2021, 290, 353–363. [Google Scholar] [CrossRef]
- Lachar, J.; Bajaj, J.S. (Eds.) Changes in the microbiome in cirrhosis and relationship to complications: Hepatic encephalopathy, spontaneous bacterial peritonitis, and sepsis. In Seminars in Liver Disease; Thieme Medical Publishers: Leipzig, Germany, 2016. [Google Scholar]
- Sanchez-Alcoholado, L.; Castellano-Castillo, D.; Jordán-Martínez, L.; Moreno-Indias, I.; Cardila-Cruz, P.; Elena, D.; Muñoz-Garcia, A.J.; Queipo-Ortuño, M.I.; Jimenez-Navarro, M. Role of gut microbiota on cardio-metabolic parameters and immunity in coronary artery disease patients with and without type-2 diabetes mellitus. Front. Microbiol. 2017, 8, 1936. [Google Scholar] [CrossRef]
- Staudacher, H.M.; Scholz, M.; Lomer, M.C.; Ralph, F.S.; Irving, P.M.; Lindsay, J.O.; Fava, F.; Tuohy, K.; Whelan, K. Gut microbiota associations with diet in irritable bowel syndrome and the effect of low FODMAP diet and probiotics. Clin. Nutr. 2021, 40, 1861–1870. [Google Scholar] [CrossRef]
- Zeng, H.; Ishaq, S.L.; Zhao, F.-Q.; Wright, A.-D.G. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016, 35, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wieczorska, K.; Stolarek, M.; Stec, R. The role of the gut microbiome in colorectal cancer: Where are we? Where are we going? Clin. Colorectal. Cancer 2020, 19, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Altaib, H.; Nakamura, K.; Abe, M.; Badr, Y.; Yanase, E.; Nomura, I.; Suzuki, T. Differences in the concentration of the fecal neurotransmitters GABA and glutamate are associated with microbial composition among healthy human subjects. Microorganisms 2021, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Mohamadkhani, A. Gut microbiota and fecal metabolome perturbation in children with autism spectrum disorder. Middle East J. Dig. Dis. 2018, 10, 205. [Google Scholar] [CrossRef]
- Zhao, H.; Jin, K.; Jiang, C.; Pan, F.; Wu, J.; Luan, H.; Zhao, Z.; Chen, J.; Mou, T.; Wang, Z.; et al. A pilot exploration of multi-omics research of gut microbiome in major depressive disorders. Transl. Psychiatry 2022, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Rocco, A.; Sgamato, C.; Compare, D.; Coccoli, P.; Nardone, O.M.; Nardone, G. Gut microbes and hepatic encephalopathy: From the old concepts to new perspectives. Frontiers in Cell and Developmental Biology. 2021, 9, 748253. [Google Scholar] [CrossRef]
- Almugadam, B.S.; Liu, Y.; Chen, S.M.; Wang, C.H.; Shao, C.Y.; Ren, B.W.; Tang, L. Alterations of gut microbiota in type 2 diabetes individuals and the confounding effect of antidiabetic agents. J. Diabetes Res. 2020, 2020, 7253978. [Google Scholar] [CrossRef]
- Zhou, X.-Y.; Li, M.; Li, X.; Long, X.; Zuo, X.-L.; Hou, X.-H.; Cong, Y.-Z.; Li, Y.-Q. Visceral hypersensitive rats share common dysbiosis features with irritable bowel syndrome patients. World J. Gastroenterol. 2016, 22, 5211. [Google Scholar] [CrossRef]
- Zhai, S.; Qin, S.; Li, L.; Zhu, L.; Zou, Z.; Wang, L. Dietary butyrate suppresses inflammation through modulating gut microbiota in high-fat diet-fed mice. FEMS Microbiol. Lett. 2019, 366, fnz153. [Google Scholar] [CrossRef]
- Carson, T.L.; Byrd, D.A.; Smith, K.S.; Carter, D.; Abaskaron, M.; Little, R.B.; van Der Pol, W.J.; Lefkowitz, E.J.; Morrow, C.D.; Fruge, A.D.; et al. A case-control study of the association between the gut microbiota and colorectal cancer: Exploring the roles of diet, stress, and race. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Zhou, R.; Fan, X.; Schnabl, B. Role of the intestinal microbiome in liver fibrosis development and new treatment strategies. Transl. Res. 2019, 209, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, S.; Yuan, C.; Yu, B.; Zhang, Z.; Zhao, M.; Liu, P.; Li, X.; Cui, B. Jerusalem artichoke inulin supplementation ameliorates hepatic lipid metabolism in type 2 diabetes mellitus mice by modulating the gut microbiota and fecal metabolome. Food Funct. 2022, 13, 11503–11517. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Wang, B.; Bai, X.; Luan, Y.; Fan, Y.; Michail, S.; Sun, F. 16S rRNA and metagenomic shotgun sequencing data revealed consistent patterns of gut microbiome signature in pediatric ulcerative colitis. Sci. Rep. 2022, 12, 6421. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, J.; Chan, N.; Echavez, C.; Mathews, A. Increased body mass is associated with decreased gut microbiome diversity in Parkinson’s Disease patients. Undergrad. J. Exp. Microbiol. Immunol. 2022, 8, 1–13. [Google Scholar]
- Fernández, J.; Saettone, P.; Franchini, M.C.; Villar, C.J.; Lombó, F. Antitumor bioactivity and gut microbiota modulation of polyhydroxybutyrate (PHB) in a rat animal model for colorectal cancer. Int. J. Biol. Macromol. 2022, 203, 638–649. [Google Scholar] [CrossRef]
- Nagarajan, A. Characterization of the Metabolic and Cognitive Features of a Novel Alzheimer’s Disease Rat Model Along with Investigation of Its Gut Microbiota Change over Time. Master’s Thesis, The University of Alabama at Birmingham, Birmingham, AL, USA, 2021. [Google Scholar]
- Fattorusso, A.; Di Genova, L.; Dell’Isola, G.B.; Mencaroni, E.; Esposito, S. Autism spectrum disorders and the gut microbiota. Nutrients 2019, 11, 521. [Google Scholar] [CrossRef]
- Liu, M.; Huang, B.; Wang, L.; Lu, Q.; Liu, R. Peanut skin procyanidins ameliorate insulin resistance via modulation of gut microbiota and gut barrier in type 2 diabetic mice. J. Sci. Food Agric. 2022, 102, 5935–5947. [Google Scholar] [CrossRef]
- Cheng, S.; Hu, J.; Wu, X.; Pan, J.-A.; Jiao, N.; Li, Y.; Huang, Y.; Lin, X.; Zou, Y.; Chen, Y.; et al. Altered gut microbiome in FUT2 loss-of-function mutants in support of personalized medicine for inflammatory bowel diseases. J. Genet. Genom. 2021, 48, 771–780. [Google Scholar] [CrossRef]
- Zhao, Q.; Fu, Y.; Zhang, F.; Wang, C.; Yang, X.; Bai, S.; Xue, Y.; Shen, Q. Heat-Treated Adzuki Bean Protein Hydrolysates Reduce Obesity in Mice Fed a High-Fat Diet via Remodeling Gut Microbiota and Improving Metabolic Function. Mol. Nutr. Food Res. 2022, 66, 2100907. [Google Scholar] [CrossRef]
- Park, S.-H.; Lee, J.H.; Shin, J.; Kim, J.-S.; Cha, B.; Lee, S.; Kwon, K.S.; Shin, Y.W.; Choi, S.H. Cognitive function improvement after fecal microbiota transplantation in Alzheimer’s dementia patient: A case report. Curr. Med. Res. Opin. 2021, 37, 1739–1744. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, J.; Fujita, Y.; Shinno-Hashimoto, H.; Shan, J.; Wan, X.; Qu, Y.; Chang, L.; Wang, X.; Hashimoto, K. Effects of spleen nerve denervation on depression–like phenotype, systemic inflammation, and abnormal composition of gut microbiota in mice after administration of lipopolysaccharide: A role of brain–spleen axis. J. Affect. Disord. 2022, 317, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Burz, S.D.; Monnoye, M.; Philippe, C.; Farin, W.; Ratziu, V.; Strozzi, F.; Paillarse, J.-M.; Chêne, L.; Blottière, H.M.; Gérard, P. Fecal microbiota transplant from human to mice gives insights into the role of the gut microbiota in non-alcoholic fatty liver disease (NAFLD). Microorganisms 2021, 9, 199. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liang, L.; Yu, G.; Li, Q. Pumpkin polysaccharide modifies the gut microbiota during alleviation of type 2 diabetes in rats. Int. J. Biol. Macromol. 2018, 115, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Li, Y.; Luo, L.; Xie, F.; Xiong, Q.; Li, T.; Yang, C.; Feng, P.-M. Significant Differences in Gut Microbiota between Irritable Bowel Syndrome with Diarrhea and Healthy Controls in Southwest China. Dig. Dis. Sci. 2022, 68, 106–127. [Google Scholar] [CrossRef]
- Nirmalkar, K.; Murugesan, S.; Pizano-Zárate, M.L.; Villalobos-Flores, L.E.; García-González, C.; Morales-Hernández, R.M.; Nuñez-Hernández, J.A.; Hernández-Quiroz, F.; Romero-Figueroa, M.D.S.; Hernández-Guerrero, C.; et al. Gut microbiota and endothelial dysfunction markers in obese Mexican children and adolescents. Nutrients 2018, 10, 2009. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Liu, Q.; Xi, Y.; Wang, Q.; Liu, J.; Li, P.; Meng, X.; Liu, K.; Chen, W.; Liu, X.; Liu, Z. Mannan oligosaccharide attenuates cognitive and behavioral disorders in the 5xFAD Alzheimer’s disease mouse model via regulating the gut microbiota-brain axis. Brain Behav. Immun. 2021, 95, 330–343. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Lozupone, C.; Kang, D.-W.; Adams, J.B. Gut bacteria in children with autism spectrum disorders: Challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Health Dis. 2015, 26, 26914. [Google Scholar] [CrossRef]
- Sanada, K.; Nakajima, S.; Kurokawa, S.; Barceló-Soler, A.; Ikuse, D.; Hirata, A.; Yoshizawa, A.; Tomizawa, Y.; Salas-Valero, M.; Noda, Y.; et al. Gut microbiota and major depressive disorder: A systematic review and meta-analysis. J. Affect. Disord. 2020, 266, 1–13. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients with vs without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Muniz Pedrogo, D.A.; Kashyap, P.C. Irritable bowel syndrome: A gut microbiota-related disorder? Am. J. Physiol.-Gastrointest. Liver Physiol. 2017, 312, G52–G62. [Google Scholar] [CrossRef] [PubMed]
- Preidis, G.A.; Ajami, N.J.; Wong, M.C.; Bessard, B.C.; Conner, M.E.; Petrosino, J.F. Composition and function of the undernourished neonatal mouse intestinal microbiome. J. Nutr. Biochem. 2015, 26, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, Y.; Wang, X.; Yang, R.; Zhu, X.; Zhang, Y.; Chen, C.; Yuan, H.; Yang, Z.; Sun, L. Gut bacteria Akkermansia is associated with reduced risk of obesity: Evidence from the American Gut Project. Nutr. Metab. 2020, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I.; Dhar, R.; Pethusamy, K.; Seethy, A.; Srivastava, T.; Sah, R.; Sharma, J.; Karmakar, S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl. Biochem. Biotechnol. 2021, 193, 1780–1799. [Google Scholar] [CrossRef]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef]
- Xu, M.; Xu, X.; Li, J.; Li, F. Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Front. Psychiatry 2019, 10, 473. [Google Scholar] [CrossRef]
- Boertien, J.M.; Pereira, P.A.B.; Aho, V.T.E.; Scheperjans, F. Increasing Comparability and Utility of Gut Microbiome Studies in Parkinson’s Disease: A Systematic Review. J. Park. Dis. 2019, 9, S297–S312. [Google Scholar] [CrossRef]
- Wong, M.L.; Inserra, A.; Lewis, M.D.; Mastronardi, C.A.; Leong, L.; Choo, J.; Kentish, S.; Xie, P.; Morrison, M.; Wesselingh, S.L.; et al. Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol. Psychiatry 2016, 21, 797–805. [Google Scholar] [CrossRef]
- Mashaqi, S.; Kallamadi, R.; Matta, A.; Quan, S.F.; Patel, S.I.; Combs, D.; Estep, L.; Lee-Iannotti, J.; Smith, C.; Parthasarathy, S.; et al. Obstructive Sleep Apnea as a Risk Factor for COVID-19 Severity-The Gut Microbiome as a Common Player Mediating Systemic Inflammation via Gut Barrier Dysfunction. Cells 2022, 11, 1569. [Google Scholar] [CrossRef]
- Zhu, L.; Sha, L.; Li, K.; Wang, Z.; Wang, T.; Li, Y.; Liu, P.; Dong, X.; Dong, Y.; Zhang, X.; et al. Dietary flaxseed oil rich in omega-3 suppresses severity of type 2 diabetes mellitus via anti-inflammation and modulating gut microbiota in rats. Lipids Health Dis. 2020, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, D.M.; Riehle, K.; Mistretta, T.A.; Diaz, M.A.; Mandal, D.; Raza, S.; Weidler, E.M.; Qin, X.; Coarfa, C.; Milosavljevic, A.; et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Li, Y.; Du, Y.; Guo, L.; Chen, M.; Huang, X.; Yang, F.; Hong, J.; Kong, X. Konjaku flour reduces obesity in mice by modulating the composition of the gut microbiota. Int. J. Obes. 2019, 43, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, S.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef]
- Phan, J.; Nair, D.; Jain, S.; Montagne, T.; Flores, D.V.; Nguyen, A.; Dietsche, S.; Gombar, S.; Cotter, P. Alterations in gut microbiome composition and function in irritable bowel syndrome and increased probiotic abundance with daily supplementation. mSystems 2021, 6, e01215-21. [Google Scholar] [CrossRef]
- Nistal, E.; Saenz de Miera, L.E.; Ballesteros Pomar, M.; Sanchez-Campos, S.; Garcia-Mediavilla, M.V.; Alvarez-Cuenllas, B.; Linares, P.; Olcoz, J.L.; Arias-Loste, M.T.; García-Lobo, J.M.; et al. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev. Esp. Enferm. Dig. 2019, 111, 275–282. [Google Scholar] [CrossRef]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Yildiz, S.; Dogan, I.; Dogruman-Al, F.; Nalbantoglu, U.; Ustek, D.; Sarzhanov, F.; Yildirim, S. Association of Enteric Protist Blastocystis spp. and Gut Microbiota with Hepatic Encephalopathy. J. Gastrointestin. Liver Dis. 2016, 25, 489–497. [Google Scholar] [CrossRef]
- Ko, C.Y.; Liu, Q.Q.; Su, H.Z.; Zhang, H.P.; Fan, J.M.; Yang, J.H.; Hu, A.K.; Liu, Y.Q.; Chou, D.; Zeng, Y.M. Gut microbiota in obstructive sleep apnea-hypopnea syndrome: Disease-related dysbiosis and metabolic comorbidities. Clin. Sci. 2019, 133, 905–917. [Google Scholar] [CrossRef]
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 2014, 37, 2343–2350. [Google Scholar] [CrossRef]
- Takaishi, H.; Matsuki, T.; Nakazawa, A.; Takada, T.; Kado, S.; Asahara, T.; Kamada, N.; Sakuraba, A.; Yajima, T.; Higuchi, H.; et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med. Microbiol. 2008, 298, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Tana, C.; Umesaki, Y.; Imaoka, A.; Handa, T.; Kanazawa, M.; Fukudo, S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol. Motil. 2010, 22, 512-e115. [Google Scholar] [CrossRef] [PubMed]
- Valentine, G.C.; Hair, A.B.; Martin, C.R. Microbiome and pediatric obesity, malnutrition, and nutrition. Dev. Microbiome 2020, 157–181. [Google Scholar] [CrossRef]
- Minato, T.; Maeda, T.; Fujisawa, Y.; Tsuji, H.; Nomoto, K.; Ohno, K.; Hirayama, M. Progression of Parkinson’s disease is associated with gut dysbiosis: Two-year follow-up study. PLoS ONE 2017, 12, e0187307. [Google Scholar] [CrossRef]
- Chen, Y.; Ji, F.; Guo, J.; Shi, D.; Fang, D.; Li, L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep. 2016, 6, 34055. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Wang, C.; Wang, L.; He, T.; Hu, H.; Song, J.; Cui, C.; Qiao, J.; Qing, L.; et al. Enterotype Bacteroides Is Associated with a High Risk in Patients with Diabetes: A Pilot Study. J. Diabetes Res. 2020, 2020, 6047145. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 5828959. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.O.; Ridgway, K.; Scovell, L.; Kemsley, E.K.; Lund, E.K.; Jamieson, C.; Johnson, I.T.; Narbad, A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010, 10, 134. [Google Scholar] [CrossRef]
- Pekmez, C.T.; Dragsted, L.O.; Brahe, L.K. Gut microbiota alterations and dietary modulation in childhood malnutrition–the role of short chain fatty acids. Clin. Nutr. 2019, 38, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Guan, M.; Mayer, E.A.; Stains, J.; Liu, C.; Vora, P.; Jacobs, J.P.; Lagishetty, V.; Chang, L.; Barry, R.L.; et al. Obesity is associated with a distinct brain-gut microbiome signature that connects Prevotella and Bacteroides to the brain’s reward center. Gut Microbes 2022, 14, 2051999. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Elhefnawy, A.M.; Azouz, H.G.; Roshdy, Y.S.; Ashry, M.H.; Ibrahim, A.E.; Meheissen, M.A. Study of the gut Microbiome Profile in Children with Autism Spectrum Disorder: A Single Tertiary Hospital Experience. J. Mol. Neurosci. 2020, 70, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, Q.; Hou, Y.; Zhang, X.; Yin, Z.; Cai, X.; Wei, W.; Wang, J.; He, D.; Wang, G.; et al. Bacteroides species differentially modulate depression-like behavior via gut-brain metabolic signaling. Brain Behav. Immun. 2022, 102, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Magzal, F.; Shochat, T.; Haimov, I.; Tamir, S.; Asraf, K.; Tuchner-Arieli, M.; Even, C.; Agmon, M. Increased physical activity improves gut microbiota composition and reduces short-chain fatty acid concentrations in older adults with insomnia. Sci. Rep. 2022, 12, 2265. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Tao, J.; Xiao, S.; Jiang, S.; Shang, E.; Zhu, Z.; Qian, D.; Duan, J. Xiexin Tang improves the symptom of type 2 diabetic rats by modulation of the gut microbiota. Sci. Rep. 2018, 8, 3685. [Google Scholar] [CrossRef]
- Liu, C.; Zhan, S.; Tian, Z.; Li, N.; Li, T.; Wu, D.; Zeng, Z.; Zhuang, X. Food Additives Associated with Gut Microbiota Alterations in Inflammatory Bowel Disease: Friends or Enemies? Nutrients 2022, 14, 3049. [Google Scholar] [CrossRef]
- Valdez-Palomares, F.; Nambo-Venegas, R.; Uribe-García, J.; Mendoza-Vargas, A.; Granados-Portillo, O.; Meraz-Cruz, N.; Palacios-González, B. Intestinal microbiota fingerprint in subjects with irritable bowel syndrome responders to a low FODMAP diet. Food Funct. 2021, 12, 3206–3218. [Google Scholar] [CrossRef]
- Zhong, H.; Abdullah Deng, L.; Zhao, M.; Tang, J.; Liu, T.; Zhang, H.; Feng, F. Probiotic-fermented blueberry juice prevents obesity and hyperglycemia in high fat diet-fed mice in association with modulating the gut microbiota. Food Funct. 2020, 11, 9192–9207. [Google Scholar] [CrossRef]
- Sarhadi, V.; Lahti, L.; Saberi, F.; Youssef, O.; Kokkola, A.; Karla, T.; Tikkanen, M.; Rautelin, H.; Puolakkainen, P.; Salehi, R.; et al. Gut Microbiota and Host Gene Mutations in Colorectal Cancer Patients and Controls of Iranian and Finnish Origin. Anticancer Res. 2020, 40, 1325–1334. [Google Scholar] [CrossRef]
- D’Argenio, V.; Veneruso, I.; Gong, C.; Cecarini, V.; Bonfili, L.; Eleuteri, A.M. Gut Microbiome and Mycobiome Alterations in an In Vivo Model of Alzheimer’s Disease. Genes 2022, 13, 1564. [Google Scholar] [CrossRef]
- Chi, L.; Khan, I.; Lin, Z.; Zhang, J.; Lee, M.Y.S.; Leong, W.; Hsiao, W.W.; Zheng, Y. Fructo-oligosaccharides from Morinda officinalis remodeled gut microbiota and alleviated depression features in a stress rat model. Phytomedicine 2020, 67, 153157. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.W.; Ma, W.J.; Wang, Y.; Ma, X.H.; Xue, Y.F.; Guan, J.; Chen, X. Comparison of the effects of probiotics, rifaximin, and lactulose in the treatment of minimal hepatic encephalopathy and gut microbiota. Front. Microbiol. 2023, 14, 1091167. [Google Scholar] [CrossRef] [PubMed]
- Le, K.A.; Li, Y.; Xu, X.; Yang, W.; Liu, T.; Zhao, X.; Tang, Y.G.; Cai, D.; Go, V.L.W.; Pandol, S.; et al. Alterations in fecal Lactobacillus and Bifidobacterium species in type 2 diabetic patients in Southern China population. Front. Physiol. 2012, 3, 496. [Google Scholar]
- Fava, F.; Danese, S. Intestinal microbiota in inflammatory bowel disease: Friend of foe? World J. Gastroenterol. 2011, 17, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Iddrisu, I.; Monteagudo-Mera, A.; Poveda, C.; Pyle, S.; Shahzad, M.; Andrews, S.; Walton, G.E. Malnutrition and gut microbiota in children. Nutrients 2021, 13, 2727. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Maraninchi, M.; Henry, M.; Armougom, F.; Richet, H.; Carrieri, P.; Valero, R.; Raccah, D.; Vialettes, B.; Raoult, D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012, 36, 817–825. [Google Scholar] [CrossRef]
- Zou, S.; Fang, L.; Lee, M.H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Aho, V.T.E.; Pereira, P.A.B.; Voutilainen, S.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. Gut microbiota in Parkinson’s disease: Temporal stability and relations to disease progression. EBioMedicine 2019, 44, 691–707. [Google Scholar] [CrossRef]
- Aizawa, E.; Tsuji, H.; Asahara, T.; Takahashi, T.; Teraishi, T.; Yoshida, S.; Ota, M.; Koga, N.; Hattori, K.; Kunugi, H. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord. 2016, 202, 254–257. [Google Scholar] [CrossRef]
- Xia, X.; Chen, J.; Xia, J.; Wang, B.; Liu, H.; Yang, L.; Wang, Y.; Ling, Z. Role of probiotics in the treatment of minimal hepatic encephalopathy in patients with HBV-induced liver cirrhosis. J. Int. Med. Res. 2018, 46, 3596–3604. [Google Scholar] [CrossRef]
- Berry, D.; Reinisch, W. Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Pract. Res. Clin. Gastroenterol. 2013, 27, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.W.; Rezaie, A.; Pimentel, M. Bile acid and gut microbiota in irritable bowel syndrome. J. Neurogastroenterol. Motil. 2022, 28, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.R.; Tomas, J.; Brenner, C.; Sansonetti, P.J. Impact of high-fat diet on the intestinal microbiota and small intestinal physiology before and after the onset of obesity. Biochimie 2017, 141, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ai, D.; Pan, H.; Li, X.; Gao, Y.; Liu, G.; Xia, L.C. Identifying Gut Microbiota Associated with Colorectal Cancer Using a Zero-Inflated Lognormal Model. Front. Microbiol. 2019, 10, 826. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I.; Aguayo, G.; et al. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Caso, J.R.; MacDowell, K.S.; Gonzalez-Pinto, A.; Garcia, S.; de Diego-Adelino, J.; Carceller-Sindreu, M.; Sarramea, F.; Caballero-Villarraso, J.; Gracia-García, P.; De la Cámara, C.; et al. Gut microbiota, innate immune pathways, and inflammatory control mechanisms in patients with major depressive disorder. Transl. Psychiatry 2021, 11, 645. [Google Scholar] [CrossRef]
- Diaz-Perdigones, C.M.; Munoz-Garach, A.; Alvarez-Bermudez, M.D.; Moreno-Indias, I.; Tinahones, F.J. Gut microbiota of patients with type 2 diabetes and gastrointestinal intolerance to metformin differs in composition and functionality from tolerant patients. Biomed. Pharmacother. 2022, 145, 112448. [Google Scholar] [CrossRef]
- Sokol, H.; Jegou, S.; McQuitty, C.; Straub, M.; Leducq, V.; Landman, C.; Kirchgesner, J.; Le Gall, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Specificities of the intestinal microbiota in patients with inflammatory bowel disease and Clostridium difficile infection. Gut Microbes 2018, 9, 55–60. [Google Scholar] [CrossRef]
- Kamil, R.Z.; Murdiati, A.; Juffrie, M.; Nakayama, J.; Rahayu, E.S. Gut microbiota and short-chain fatty acid profile between normal and moderate malnutrition children in Yogyakarta, Indonesia. Microorganisms 2021, 9, 127. [Google Scholar] [CrossRef]
- Zhong, X.; Harrington, J.M.; Millar, S.R.; Perry, I.J.; O’Toole, P.W.; Phillips, C.M. Gut microbiota associations with metabolic health and obesity status in older adults. Nutrients 2020, 12, 2364. [Google Scholar] [CrossRef]
- Ni, J.J.; Li, X.S.; Zhang, H.; Xu, Q.; Wei, X.T.; Feng, G.J.; Zhao, M.; Zhang, Z.J.; Zhang, L.; Shen, G.H.; et al. Mendelian randomization study of causal link from gut microbiota to colorectal cancer. BMC Cancer 2022, 22, 1371. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, X.; Hu, X.; Wang, T.; Liang, S.; Duan, Y.; Jin, F.; Qin, B. Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 2017, 60, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Tao, Y.; Zheng, F.; Zhou, H.; Wu, H.; Shi, H.; Huang, F.; Wu, X. The alteration of gut microbiota in venlafaxine-ameliorated chronic unpredictable mild stress-induced depression in mice. Behav. Brain Res. 2023, 446, 114399. [Google Scholar] [CrossRef] [PubMed]
- Van Meijel, R.L.J.; Venema, K.; Canfora, E.E.; Blaak, E.E.; Goossens, G.H. Mild intermittent hypoxia exposure alters gut microbiota composition in men with overweight and obesity. Benef. Microbes 2022, 13, 355–363. [Google Scholar] [CrossRef]
- Alvarez-Silva, C.; Kashani, A.; Hansen, T.H.; Pinna, N.K.; Anjana, R.M.; Dutta, A.; Saxena, S.; Støy, J.; Kampmann, U.; Nielsen, T.; et al. Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Med. 2021, 13, 37. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Machiels, K.; Perrier, C.; Romero, C.; Maes, S.; Flahou, B.; Steppe, M.; Haesebrouck, F.; Sas, B.; Ducatelle, R.; et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 2013, 62, 1745–1752. [Google Scholar] [CrossRef]
- Enqi, W.; Jingzhu, S.; Lingpeng, P.; Yaqin, L. Comparison of the gut microbiota disturbance in rat models of irritable bowel syndrome induced by maternal separation and multiple early-life adversity. Front. Cell. Infect. Microbiol. 2021, 10, 581974. [Google Scholar] [CrossRef]
- Yang, M.; Yin, Y.; Wang, F.; Zhang, H.; Ma, X.; Yin, Y.; Tan, B.; Chen, J. Supplementation with Lycium barbarum Polysaccharides Reduce Obesity in High-Fat Diet-Fed Mice by Modulation of Gut Microbiota. Front. Microbiol. 2021, 12, 719967. [Google Scholar] [CrossRef]
- Huang, M.; Liu, K.; Wei, Z.; Feng, Z.; Chen, J.; Yang, J.; Zhong, Q.; Wan, G.; Kong, X.-J. Serum Oxytocin Level Correlates with Gut Microbiome Dysbiosis in Children with Autism Spectrum Disorder. Front. Neurosci. 2021, 15, 721884. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Wu, C.; Song, Y.; Qin, N.; Chen, S.-D.; Xiao, Q. Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 2018, 70, 194–202. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef] [PubMed]
- Radwan, S.; Gilfillan, D.; Eklund, B.; Radwan, H.M.; El Menofy, N.G.; Lee, J.; Kapuscinski, M.; Abdo, Z. A comparative study of the gut microbiome in Egyptian patients with Type I and Type II diabetes. PLoS ONE 2020, 15, e0238764. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Becerra, L.; Cornejo-Granados, F.; García-López, R.; Valdez-Lara, A.; Bikel, S.; Canizales-Quinteros, S.; López-Contreras, B.E.; Mendoza-Vargas, A.; Nielsen, H.; Ochoa-Leyva, A. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Fact. 2020, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Cammann, D.; Lu, Y.; Cummings, M.J.; Zhang, M.L.; Cue, J.M.; Do, J.; Ebersole, J.; Chen, X.; Oh, E.C.; Cummings, J.L.; et al. Genetic correlations between Alzheimer’s disease and gut microbiome genera. Sci. Rep. 2023, 13, 5258. [Google Scholar] [CrossRef]
- Wu, T.; Wang, H.; Lu, W.; Zhai, Q.; Zhang, Q.; Yuan, W.; Gu, Z.; Zhao, J.; Zhang, H.; Chen, W. Potential of gut microbiome for detection of autism spectrum disorder. Microb. Pathog. 2020, 149, 104568. [Google Scholar] [CrossRef]
- Zhang, P.; Huang, P.; Du, J.; He, Y.; Liu, J.; He, G.; Cui, S.; Zhang, W.; Li, G.; Chen, S. Specific gut microbiota alterations in essential tremor and its difference from Parkinson’s disease. NPJ Park. Dis. 2022, 8, 98. [Google Scholar] [CrossRef]
- Philips, C.A.; Augustine, P.; Ganesan, K.; Ranade, S.; Chopra, V.; Patil, K.; Shende, S.; Ahamed, R.; Kumbar, S.; Rajesh, S.; et al. The role of gut microbiota in clinical complications, disease severity, and treatment response in severe alcoholic hepatitis. Indian J. Gastroenterol. 2022, 41, 37–51. [Google Scholar] [CrossRef]
- Bien, J.; Palagani, V.; Bozko, P. The intestinal microbiota dysbiosis and Clostridium difficile infection: Is there a relationship with inflammatory bowel disease? Therap. Adv. Gastroenterol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- Salonen, A.; de Vos, W.M.; Palva, A. Gastrointestinal microbiota in irritable bowel syndrome: Present state and perspectives. Microbiology 2010, 156, 3205–3215. [Google Scholar] [CrossRef]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef]
- Sun, J.; Xu, J.; Yang, B.; Chen, K.; Kong, Y.; Fang, N.; Gong, T.; Wang, F.; Ling, Z.; Liu, J. Effect of Clostridium butyricum against Microglia-Mediated Neuroinflammation in Alzheimer’s Disease via Regulating Gut Microbiota and Metabolites Butyrate. Mol. Nutr. Food Res. 2020, 64, e1900636. [Google Scholar] [CrossRef] [PubMed]
- Parracho, H.M.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J. Med. Microbiol. 2005, 54 Pt 10, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Palacios, N.; Hannoun, A.; Flahive, J.; Ward, D.; Goostrey, K.; Deb, A.; Smith, K.M. Effect of Levodopa Initiation on the Gut Microbiota in Parkinson’s Disease. Front. Neurol. 2021, 12, 574529. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Hart, R.A.; Charlesworth, R.P.G.; Sharpley, C.F. Gut microbiome and depression: What we know and what we need to know. Rev. Neurosci. 2018, 29, 629–643. [Google Scholar] [CrossRef]
- Afolayan, A.; Adebusoye, L.; Cadmus, E.; Ayeni, F. Insights into the gut microbiota of Nigerian elderly with type 2 diabetes and non-diabetic elderly persons. Heliyon 2020, 6, e03971. [Google Scholar] [CrossRef]
- Chen, D.; Chen, G.; Chen, C.; Zeng, X.; Ye, H. Prebiotics effects in vitro of polysaccharides from tea flowers on gut microbiota of healthy persons and patients with inflammatory bowel disease. Int. J. Biol. Macromol. 2020, 158, 968–976. [Google Scholar] [CrossRef]
- Malinen, E.; Krogius-Kurikka, L.; Lyra, A.; Nikkilä, J.; Jääskeläinen, A.; Rinttilä, T.; Vilpponen-Salmela, T.; von Wright, A.J.; Palva, A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. WJG 2010, 16, 4532. [Google Scholar] [CrossRef]
- Gatya, M.; Fibri, D.L.N.; Utami, T.; Suroto, D.A.; Rahayu, E.S. Gut Microbiota Composition in Undernourished Children Associated with Diet and Sociodemographic Factors: A Case–Control Study in Indonesia. Microorganisms 2022, 10, 1748. [Google Scholar] [CrossRef]
- Gomez-Arango, L.F.; Barrett, H.L.; Wilkinson, S.A.; Callaway, L.K.; McIntyre, H.D.; Morrison, M.; Nitert, M.D. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Sheng, Q.; Du, H.; Cheng, X.; Cheng, X.; Tang, Y.; Pan, L.; Wang, Q.; Lin, J. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol. Lett. 2019, 18, 4834–4844. [Google Scholar] [CrossRef]
- Huang, B.; Chau, S.W.H.; Liu, Y.; Chan, J.W.Y.; Wang, J.; Ma, S.L.; Zhang, J.; Chan, P.K.S.; Yeoh, Y.K.; Chen, Z.; et al. Gut microbiome dysbiosis across early Parkinson’s disease, REM sleep behavior disorder and their first-degree relatives. Nat. Commun. 2023, 14, 2501. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jin, Y.; Zhou, W.; Xiao, T.; Wu, Z.; Su, J.; Gao, H.; Shen, P.; Zheng, B.; Luo, Q.; et al. Rifaximin Modulates the Gut Microbiota to Prevent Hepatic Encephalopathy in Liver Cirrhosis without Impacting the Resistome. Front. Cell. Infect. Microbiol. 2021, 11, 761192. [Google Scholar] [CrossRef] [PubMed]
- Durgan, D.J. Obstructive Sleep Apnea-Induced Hypertension: Role of the Gut Microbiota. Curr. Hypertens. Rep. 2017, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qiu, Y.; Gao, Y.; Zhou, R.; Hu, Q.; He, Z.; Lv, Y.; Wang, X.; Chen, W.; Deng, Y.; et al. Gut microbiota mediate melatonin signalling in association with type 2 diabetes. Diabetologia 2022, 65, 1627–1641. [Google Scholar] [CrossRef]
- Scher, J.U.; Ubeda, C.; Artacho, A.; Attur, M.; Isaac, S.; Reddy, S.M.; Marmon, S.; Neimann, A.; Brusca, S.; Patel, T.; et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015, 67, 128–139. [Google Scholar] [CrossRef]
- Monteleone, A.M.; Troisi, J.; Fasano, A.; Dalle Grave, R.; Marciello, F.; Serena, G.; Calugi, S.; Scala, G.; Corrivetti, G.; Cascino, G.; et al. Multi-omics data integration in anorexia nervosa patients before and after weight regain: A microbiome-metabolomics investigation. Clin. Nutr. 2021, 40, 1137–1146. [Google Scholar] [CrossRef]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Andreo-Martinez, P.; Garcia-Martinez, N.; Sanchez-Samper, E.P.; Martinez-Gonzalez, A.E. An approach to gut microbiota profile in children with autism spectrum disorder. Environ. Microbiol. Rep. 2020, 12, 115–135. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.I.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef]
- Mashaqi, S.; Gozal, D. Obstructive Sleep Apnea and Systemic Hypertension: Gut Dysbiosis as the Mediator? J. Clin. Sleep Med. 2019, 15, 1517–1527. [Google Scholar] [CrossRef]
- Gong, X.; Xiong, L.; Bi, C.; Zhang, B. Diosmetin ameliorate type 2 diabetic mellitus by up-regulating Corynebacterium glutamicum to regulate IRS/PI3K/AKT-mediated glucose metabolism disorder in KK-Ay mice. Phytomedicine 2021, 87, 153582. [Google Scholar] [CrossRef] [PubMed]
- Gracey, M.; Suharjono, S.; Sunoto, S.; Stone, D.E. Microbial Contamination of the Gut; another Feature of Malnutrition. Paediatr. Indones. 1975, 15, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.I.; Niang, E.H.A.; Ndongo, S.; Raoult, D.; Fournier, P.E.; Fenollar, F. Corynebacterium bouchesdurhonense sp. nov., and Corynebacterium provencense sp. nov., two new species isolated from obese patients. New Microbes New Infect. 2019, 31, 100581. [Google Scholar] [CrossRef]
- Hasan, R.; Bose, S.; Roy, R.; Paul, D.; Rawat, S.; Nilwe, P.; Chauhan, N.K.; Choudhury, S. Tumor tissue-specific bacterial biomarker panel for colorectal cancer: Bacteroides massiliensis, Alistipes species, Alistipes onderdonkii, Bifidobacterium pseudocatenulatum, Corynebacterium appendicis. Arch. Microbiol. 2022, 204, 348. [Google Scholar] [CrossRef]
- Chang, C.H.; Lin, C.H.; Lane, H.Y. d-glutamate and Gut Microbiota in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2676. [Google Scholar] [CrossRef]
- Aitbali, Y.; Ba-M’hamed, S.; Elhidar, N.; Nafis, A.; Soraa, N.; Bennis, M. Glyphosate based- herbicide exposure affects gut microbiota, anxiety and depression-like behaviors in mice. Neurotoxicol. Teratol. 2018, 67, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Suzuki, F.; Imamura, M.; Murashima, N.; Yanase, M.; Mine, T.; Fujisawa, M.; Sato, I.; Yoshiji, H.; Okita, K.; et al. Rifaximin-altered gut microbiota components associated with liver/neuropsychological functions in patients with hepatic encephalopathy: An exploratory data analysis of phase II/III clinical trials. Hepatol. Res. 2019, 49, 404–418. [Google Scholar] [CrossRef]
- Nie, Q.; Hu, J.; Chen, H.; Geng, F.; Nie, S. Arabinoxylan ameliorates type 2 diabetes by regulating the gut microbiota and metabolites. Food Chem. 2022, 371, 131106. [Google Scholar] [CrossRef]
- Gobert, A.P.; Sagrestani, G.; Delmas, E.; Wilson, K.T.; Verriere, T.G.; Dapoigny, M.; Del’homme, C.; Bernalier-Donadille, A. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci. Rep. 2016, 6, 39399. [Google Scholar] [CrossRef]
- Karlsson, C.L.; Onnerfalt, J.; Xu, J.; Molin, G.; Ahrne, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef]
- Li, C.; Wang, N.; Zheng, G.; Yang, L. Oral Administration of Resveratrol-Selenium-Peptide Nanocomposites Alleviates Alzheimer’s Disease-like Pathogenesis by Inhibiting Abeta Aggregation and Regulating Gut Microbiota. ACS Appl. Mater. Interfaces 2021, 13, 46406–46420. [Google Scholar] [CrossRef] [PubMed]
- Heberling, C.A.; Dhurjati, P.S.; Sasser, M. Hypothesis for a systems connectivity model of Autism Spectrum Disorder pathogenesis: Links to gut bacteria, oxidative stress, and intestinal permeability. Med. Hypotheses 2013, 80, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Murros, K.E.; Huynh, V.A.; Takala, T.M.; Saris, P.E.J. Desulfovibrio Bacteria Are Associated with Parkinson’s Disease. Front. Cell. Infect. Microbiol. 2021, 11, 652617. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Zhang, H.; Chen, X.; Zhang, Y.; Wu, J.; Zhao, L.; Wang, D.; Pu, J.; Ji, P.; et al. Toward a Deeper Understanding of Gut Microbiome in Depression: The Promise of Clinical Applicability. Adv. Sci. 2022, 9, e2203707. [Google Scholar] [CrossRef] [PubMed]
- Elbere, I.; Silamikelis, I.; Dindune, I.I.; Kalnina, I.; Ustinova, M.; Zaharenko, L.; Silamikele, L.; Rovite, V.; Gudra, D.; Konrade, I.; et al. Baseline gut microbiome composition predicts metformin therapy short-term efficacy in newly diagnosed type 2 diabetes patients. PLoS ONE 2020, 15, e0241338. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Petito, V.; Graziani, C.; Schiavoni, E.; Sterbini, F.P.; Poscia, A.; Gaetani, E.; Franceschi, F.; Cammarota, G.; Sanguinetti, M.; et al. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: Time for microbial marker of gastrointestinal disorders. Dig. Dis. 2018, 36, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Chumpitazi, B.P.; Hollister, E.B.; Oezguen, N.; Tsai, C.M.; McMeans, A.R.; Luna, R.A.; Savidge, T.C.; Versalovic, J.; Shulman, R.J. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes 2014, 5, 165–175. [Google Scholar] [CrossRef]
- Toe, L.C.; Kerckhof, F.-M.; De Bodt, J.; Morel, F.B.; Ouedraogo, J.-B.; Kolsteren, P.; Van de Wiele, T. A prebiotic-enhanced lipid-based nutrient supplement (LNSp) increases Bifidobacterium relative abundance and enhances short-chain fatty acid production in simulated colonic microbiota from undernourished infants. FEMS Microbiol. Ecol. 2020, 96, fiaa105. [Google Scholar] [CrossRef]
- Naderpoor, N.; Mousa, A.; Gomez-Arango, L.F.; Barrett, H.L.; Dekker Nitert, M.; de Courten, B. Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. J. Clin. Med. 2019, 8, 452. [Google Scholar] [CrossRef]
- Huo, R.X.; Wang, Y.J.; Hou, S.B.; Wang, W.; Zhang, C.Z.; Wan, X.H. Gut mucosal microbiota profiles linked to colorectal cancer recurrence. World J. Gastroenterol. 2022, 28, 1946–1964. [Google Scholar] [CrossRef]
- Chiang, H.L.; Lin, C.H. Altered Gut Microbiome and Intestinal Pathology in Parkinson’s Disease. J. Mov. Disord. 2019, 12, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020, 10, 5450. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.V.; Charbonneau, M.R.; Salih, T.; Barratt, M.J.; Venkatesh, S.; Ilkaveya, O.; Subramanian, S.; Manary, M.J.; Trehan, I.; Jorgensen, J.M.; et al. Gut bacteria that rescue growth impairments transmitted by immature microbiota from undernourished children. Science 2016, 351, 854. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Baker, S.S.; Nugent, C.A.; Tsompana, M.; Cai, L.; Wang, Y.; Buck, M.J.; Genco, R.J.; Baker, R.D.; Zhu, R.; et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol Genom. 2018, 50, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 2019, 15, 1357–1366. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Ito, M.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Yoshida, T.; Hanada, H.; Takeuchi, I.; et al. Short chain fatty acids-producing and mucin-degrading intestinal bacteria predict the progression of early Parkinson’s disease. NPJ Park. Dis. 2022, 8, 65. [Google Scholar] [CrossRef]
- Zhang, Q.; Yun, Y.; An, H.; Zhao, W.; Ma, T.; Wang, Z.; Yang, F. Gut Microbiome Composition Associated With Major Depressive Disorder and Sleep Quality. Front. Psychiatry 2021, 12, 645045. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Hylemon, P.B.; Ridlon, J.M.; Heuman, D.M.; Daita, K.; White, M.B.; Monteith, P.; Noble, N.A.; Sikaroodi, M.; Gillevet, P.M.; et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G675–G685. [Google Scholar] [CrossRef]
- Tang, S.S.; Liang, C.H.; Liu, Y.L.; Wei, W.; Deng, X.R.; Shi, X.Y.; Wang, L.M.; Zhang, L.J.; Yuan, H.J. Intermittent hypoxia is involved in gut microbial dysbiosis in type 2 diabetes mellitus and obstructive sleep apnea-hypopnea syndrome. World J. Gastroenterol. 2022, 28, 2320–2333. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Zheng, H.; Li, J.; Wu, H.; Qin, S.; Luo, L.; Huang, S. Gut microbiota-bile acid crosstalk in diarrhea-irritable bowel syndrome. BioMed Res. Int. 2020, 2020, 3828249. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Shen, N.; Liu, Y. Associations between the Gut Microbiome and Migraines in Children Aged 7–18 Years: An Analysis of the American Gut Project Cohort. Pain Manag. Nurs. 2023, 24, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Pinart, M.; Dotsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut Microbiome Composition in Obese and Non-Obese Persons: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wei, H.; Liu, W.; Coker, O.O.; Gou, H.; Liu, C.; Zhao, L.; Li, C.; Zhou, Y.; Wang, G.; et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 2022, 71, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4, e00321-18. [Google Scholar] [CrossRef]
- Lubomski, M.; Davis, R.L.; Sue, C.M. The gut microbiota: A novel therapeutic target in Parkinson’s disease? Park. Relat. Disord. 2019, 66, 265–266. [Google Scholar] [CrossRef]
- Philips, C.A.; Ahamed, R.; Rajesh, S.; Singh, S.; Tharakan, A.; Abduljaleel, J.K.; Augustine, P. Clinical outcomes and gut microbiota analysis of severe alcohol-associated hepatitis patients undergoing healthy donor fecal transplant or pentoxifylline therapy: Single-center experience from Kerala. Gastroenterol. Rep. 2022, 10, goac074. [Google Scholar] [CrossRef]
- Xu, T.; Ge, Y.; Du, H.; Li, Q.; Xu, X.; Yi, H.; Wu, X.; Kuang, T.; Fan, G.; Zhang, Y. Berberis kansuensis extract alleviates type 2 diabetes in rats by regulating gut microbiota composition. J. Ethnopharmacol. 2021, 273, 113995. [Google Scholar] [CrossRef]
- Balish, E.; Warner, T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am. J. Pathol. 2002, 160, 2253–2257. [Google Scholar] [CrossRef]
- Ji, M.; Huang, H.; Lan, X. Correlation between intestinal microflora in irritable bowel syndrome and severity. Dis. Markers 2022, 2022, 1031844. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Wang, Y.; Yang, T.; Zhan, X.; Li, Z.; Hu, H.; Li, T.; Chen, J. Vitamin A deficiency impacts the structural segregation of gut microbiota in children with persistent diarrhea. J. Clin. Biochem. Nutr. 2016, 59, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Pan, A.; Zhang, X.; Cai, Y.Y.; Wang, Q.; Huang, F.Q.; Alolga, R.N.; Li, J.; Qi, L.W.; Liu, Q. Cordyceps Improves Obesity and its Related Inflammation via Modulation of Enterococcus cecorum Abundance and Bile Acid Metabolism. Am. J. Chin. Med. 2022, 50, 817–838. [Google Scholar] [CrossRef]
- de Almeida, C.V.; Taddei, A.; Amedei, A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap. Adv. Gastroenterol. 2018, 11, 1756284818783606. [Google Scholar] [CrossRef]
- Fan, H.X.; Sheng, S.; Zhang, F. New hope for Parkinson’s disease treatment: Targeting gut microbiota. CNS Neurosci. Ther. 2022, 28, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.M.; Kim, J.K.; Joo, M.K.; Shin, Y.J.; Lee, K.E.; Lee, C.K.; Kim, H.J.; Kim, D.H. Enterococcus faecium and Pediococcus acidilactici deteriorate Enterobacteriaceae-induced depression and colitis in mice. Sci. Rep. 2022, 12, 9389. [Google Scholar] [CrossRef] [PubMed]
- Loguercio, C.; Del Vecchio Blanco, C.; Coltorti, M. Enterococcus lactic acid bacteria strain SF68 and lactulose in hepatic encephalopathy: A controlled study. J. Int. Med. Res. 1987, 15, 335–343. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Krogh Pedersen, H.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef]
- Lee, K.N.; Lee, O.Y. Intestinal microbiota in pathophysiology and management of irritable bowel syndrome. World J. Gastroenterol. WJG 2014, 20, 8886. [Google Scholar]
- Million, M.; Angelakis, E.; Maraninchi, M.; Henry, M.; Giorgi, R.; Valero, R.; Vialettes, B.; Raoult, D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int. J. Obes. 2013, 37, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Radilla-Vazquez, R.B.; Parra-Rojas, I.; Martinez-Hernandez, N.E.; Marquez-Sandoval, Y.F.; Illades-Aguiar, B.; Castro-Alarcon, N. Gut Microbiota and Metabolic Endotoxemia in Young Obese Mexican Subjects. Obes. Facts. 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bostanciklioglu, M. The role of gut microbiota in pathogenesis of Alzheimer’s disease. J. Appl. Microbiol. 2019, 127, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Carissimi, C.; Laudadio, I.; Palone, F.; Fulci, V.; Cesi, V.; Cardona, F.; Alfonsi, C.; Cucchiara, S.; Isoldi, S.; Stronati, L. Functional analysis of gut microbiota and immunoinflammation in children with autism spectrum disorders. Dig. Liver Dis. 2019, 51, 1366–1374. [Google Scholar] [CrossRef]
- Sun, M.F.; Shen, Y.Q. Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef]
- Du Toit, A. Gut microbiota and depression. Nat. Rev. Microbiol. 2022, 20, 190. [Google Scholar] [CrossRef]
- Liu, Q.; Duan, Z.P.; Ha, D.K.; Bengmark, S.; Kurtovic, J.; Riordan, S.M. Synbiotic modulation of gut flora: Effect on minimal hepatic encephalopathy in patients with cirrhosis. Hepatology 2004, 39, 1441–1449. [Google Scholar] [CrossRef]
- Bhute, S.S.; Suryavanshi, M.V.; Joshi, S.M.; Yajnik, C.S.; Shouche, Y.S.; Ghaskadbi, S.S. Gut Microbial Diversity Assessment of Indian Type-2-Diabetics Reveals Alterations in Eubacteria, Archaea, and Eukaryotes. Front. Microbiol. 2017, 8, 214. [Google Scholar] [CrossRef]
- Jones, R.B.; Alderete, T.L.; Kim, J.S.; Millstein, J.; Gilliland, F.D.; Goran, M.I. High intake of dietary fructose in overweight/obese teenagers associated with depletion of Eubacterium and Streptococcus in gut microbiome. Gut Microbes 2019, 10, 712–719. [Google Scholar] [CrossRef]
- Ryu, S.W.; Kim, J.S.; Oh, B.S.; Choi, W.J.; Yu, S.Y.; Bak, J.E.; Park, S.H.; Kang, S.W.; Lee, J.; Jung, W.Y.; et al. Gut Microbiota Eubacterium callanderi Exerts Anti-Colorectal Cancer Activity. Microbiol. Spectr. 2022, 10, e0253122. [Google Scholar] [CrossRef]
- Shabbir, U.; Arshad, M.S.; Sameen, A.; Oh, D.H. Crosstalk between Gut and Brain in Alzheimer’s Disease: The Role of Gut Microbiota Modulation Strategies. Nutrients 2021, 13, 690. [Google Scholar] [PubMed]
- Merli, M.; Iebba, V.; Giusto, M. What is new about diet in hepatic encephalopathy. Metab. Brain Dis. 2016, 31, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Navab-Moghadam, F.; Sedighi, M.; Khamseh, M.E.; Alaei-Shahmiri, F.; Talebi, M.; Razavi, S.; Amirmozafari, N. The association of type II diabetes with gut microbiota composition. Microb. Pathog. 2017, 110, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shen, J.; Ran, Z.H. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: A meta-analysis and systematic review of the literature. Gastroenterol. Res. Pract. 2014, 2014, 872725. [Google Scholar] [CrossRef]
- Maharshak, N.; Ringel, Y.; Katibian, D.; Lundqvist, A.; Sartor, R.B.; Carroll, I.M.; Ringel-Kulka, T. Fecal and mucosa-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Dig. Dis. Sci. 2018, 63, 1890–1899. [Google Scholar] [CrossRef]
- Maioli, T.U.; Borras-Nogues, E.; Torres, L.; Barbosa, S.C.; Martins, V.D.; Langella, P.; Azevedo, V.A.; Chatel, J.-M. Possible Benefits of Faecalibacterium prausnitzii for Obesity-Associated Gut Disorders. Front. Pharmacol. 2021, 12, 740636. [Google Scholar] [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Chiu, H.T.; Alshawsh, M.A. Faecalibacterium prausnitzii Ameliorates Colorectal Tumorigenesis and Suppresses Proliferation of HCT116 Colorectal Cancer Cells. Biomedicines 2022, 10, 1128. [Google Scholar] [CrossRef]
- Leblhuber, F.; Ehrlich, D.; Steiner, K.; Geisler, S.; Fuchs, D.; Lanser, L.; Kurz, K. The Immunopathogenesis of Alzheimer’s Disease Is Related to the Composition of Gut Microbiota. Nutrients 2021, 13, 361. [Google Scholar] [CrossRef]
- Hua, X.; Zhu, J.; Yang, T.; Guo, M.; Li, Q.; Chen, J.; Li, T. The Gut Microbiota and Associated Metabolites Are Altered in Sleep Disorder of Children with Autism Spectrum Disorders. Front. Psychiatry 2020, 11, 855. [Google Scholar] [CrossRef]
- Ye, X.; Wang, D.; Zhu, H.; Wang, D.; Li, J.; Tang, Y.; Wu, J. Gut Microbiota Changes in Patients with Major Depressive Disorder Treated with Vortioxetine. Front. Psychiatry 2021, 12, 641491. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Gillevet, P.M.; Patel, N.R.; Ahluwalia, V.; Ridlon, J.M.; Kettenmann, B.; Schubert, C.M.; Sikaroodi, M.; Heuman, D.M.; Crossey, M.M.E.; et al. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab. Brain Dis. 2012, 27, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Si, X.; Yang, L.; Wang, H.; Sun, Y.; Liu, N. Association between intestinal microbiota and inflammatory bowel disease. Anim. Model. Exp. Med. 2022, 5, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Wiredu Ocansey, D.K.; Hang, S.; Yuan, X.; Qian, H.; Zhou, M.; Valerie Olovo, C.; Zhang, X.; Mao, F. The diagnostic and prognostic potential of gut bacteria in inflammatory bowel disease. Gut Microbes 2023, 15, 2176118. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Wang, Y.; Zhang, Q.; Zou, R.; Guo, M.; Zheng, H. Characteristics of gut microbiota in people with obesity. PLoS ONE 2021, 16, e0255446. [Google Scholar] [CrossRef]
- Kelly, D.; Yang, L.; Pei, Z. Gut Microbiota, Fusobacteria, and Colorectal Cancer. Diseases 2018, 6, 109. [Google Scholar] [CrossRef]
- Yan, C.; Diao, Q.; Zhao, Y.; Zhang, C.; He, X.; Huang, R.; Li, Y. Fusobacterium nucleatum infection-induced neurodegeneration and abnormal gut microbiota composition in Alzheimer’s disease-like rats. Front. Neurosci. 2022, 16, 884543. [Google Scholar] [CrossRef]
- Talukdar, R.; Sarkar, P.; Jakkampudi, A.; Sarkar, S.; Aslam, M.; Jandhyala, M.; Deepika, G.; Unnisa, M.; Reddy, D.N. The gut microbiome in pancreatogenic diabetes differs from that of Type 1 and Type 2 diabetes. Sci. Rep. 2021, 11, 10978. [Google Scholar] [CrossRef]
- Brand, E.C.; Klaassen, M.A.; Gacesa, R.; Vila, A.V.; Ghosh, H.; De Zoete, M.R.; Boomsma, D.I.; Hoentjen, F.; Horjus Talabur Horje, C.S.; van de Meeberg, P.C.; et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology 2021, 160, 1970–1985. [Google Scholar] [CrossRef]
- Selma, M.V.; Romo-Vaquero, M.; Garcia-Villalba, R.; Gonzalez-Sarrias, A.; Tomas-Barberan, F.A.; Espin, J.C. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Funct. 2016, 7, 1769–1774. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; Fernandez-Villalba, E.; Gil-Martinez, A.L.; Cuenca-Bermejo, L.; Espin, J.C.; Herrero, M.T.; Selma, M.V. Urolithins: Potential biomarkers of gut dysbiosis and disease stage in Parkinson’s patients. Food Funct. 2022, 13, 6306–6316. [Google Scholar] [CrossRef]
- Chen, J.J.; Zheng, P.; Liu, Y.Y.; Zhong, X.G.; Wang, H.Y.; Guo, Y.J.; Xie, P. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 2018, 14, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Wasilewska, J.; Klukowski, M.; Debkowska, K.; Kilon, J.; Citko, D.; Flisiak, M.; Oleksinska, M.; Kaczmarski, M. Helicobacter pylori seroprevalence in children with sleep-disordered breathing. Int. J. Pediatr. Otorhinolaryngol. 2016, 87, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Hosseininasab Nodoushan, S.A.; Nabavi, A. The Interaction of Helicobacter pylori Infection and Type 2 Diabetes Mellitus. Adv. Biomed. Res. 2019, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Jiang, L.; Ruan, G.; Liu, T.; Yang, H. Helicobacter pylori may participate in the development of inflammatory bowel disease by modulating the intestinal microbiota. Chin. Med. J. 2022, 135, 634–638. [Google Scholar] [CrossRef]
- Ng, Q.X.; Foo, N.X.; Loke, W.; Koh, Y.Q.; Seah, V.J.M.; Soh, A.Y.S.; Yeo, W.S. Is there an association between Helicobacter pylori infection and irritable bowel syndrome? A meta-analysis. World J. Gastroenterol. 2019, 25, 5702. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, B.; Yakoob, J.; Abbas, Z.; Azmat, R.; Fatima, S.S.; Awan, S. Distribution of Helicobacter pylori infection and abnormal body-mass index (BMI) in a developing country. J. Infect. Dev. Ctries 2018, 12, 342–346. [Google Scholar] [CrossRef]
- Dai, K.; Song, Y.; Zhang, D.; Wei, Y.; Jiang, S.; Xu, F.; Wang, H.; Zhang, X.; Shao, X. Thinned peach polyphenols alleviate obesity in high fat mice by affecting gut microbiota. Food Res. Int. 2022, 157, 111255. [Google Scholar] [CrossRef]
- Dahmus, J.D.; Kotler, D.L.; Kastenberg, D.M.; Kistler, C.A. The gut microbiome and colorectal cancer: A review of bacterial pathogenesis. J. Gastrointest. Oncol. 2018, 9, 769–777. [Google Scholar] [CrossRef]
- Doulberis, M.; Kotronis, G.; Gialamprinou, D.; Polyzos, S.A.; Papaefthymiou, A.; Katsinelos, P.; Kountouras, J. Alzheimer’s disease and gastrointestinal microbiota; impact of Helicobacter pylori infection involvement. Int. J. Neurosci. 2021, 131, 289–301. [Google Scholar] [CrossRef]
- Cao, X.; Lin, P.; Jiang, P.; Li, C. Characteristics of the gastrointestinal microbiome in children with autism spectrum disorder: A systematic review. Shanghai Arch Psychiatry 2013, 25, 342–353. [Google Scholar]
- McGee, D.J.; Lu, X.H.; Disbrow, E.A. Stomaching the Possibility of a Pathogenic Role for Helicobacter pylori in Parkinson’s Disease. J. Park. Dis. 2018, 8, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.F.; Chen, M.H.; Wang, Y.P.; Liu, P.Y.; Hou, M.C.; Lee, F.Y.; Lu, C.L. Increased risk of short-term depressive disorder after Helicobacter pylori eradication: A population-based nested cohort study. Helicobacter 2021, 26, e12824. [Google Scholar] [CrossRef]
- Schulz, C.; Schutte, K.; Malfertheiner, P. Does H. pylori eradication therapy benefit patients with hepatic encephalopathy?: Systematic review. J. Clin. Gastroenterol. 2014, 48, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Nardone, O.M.; de Sire, R.; Petito, V.; Testa, A.; Villani, G.; Scaldaferri, F.; Castiglione, F. Inflammatory bowel diseases and sarcopenia: The role of inflammation and gut microbiota in the development of muscle failure. Front. Immunol. 2021, 12, 694217. [Google Scholar] [CrossRef] [PubMed]
- Chumpitazi, B.P.; Hoffman, K.L.; Smith, D.P.; McMeans, A.R.; Musaad, S.; Versalovic, J.; Petrosino, J.F.; Shulman, R.J. Fructan-sensitive children with irritable bowel syndrome have distinct gut microbiome signatures. Aliment. Pharmacol. Ther. 2021, 53, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Osaki, T.; Oikawa, S. Use of T-RFLP and seven restriction enzymes to compare the faecal microbiota of obese and lean Japanese healthy men. Benef. Microbes 2015, 6, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Rajani, C.; Xu, H.; Zheng, X. Gut microbiota alterations are distinct for primary colorectal cancer and hepatocellular carcinoma. Protein Cell 2021, 12, 374–393. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Yeom, M.J.; Ahn, S.; Oh, J.Y.; Ji, S.; Kim, T.H.; Park, H.J. Acupuncture inhibits neuroinflammation and gut microbial dysbiosis in a mouse model of Parkinson’s disease. Brain Behav. Immun. 2020, 89, 641–655. [Google Scholar] [CrossRef]
- Bloom, P.P.; Luevano, J.M., Jr.; Miller, K.J.; Chung, R.T. Deep stool microbiome analysis in cirrhosis reveals an association between short-chain fatty acids and hepatic encephalopathy. Ann. Hepatol. 2021, 25, 100333. [Google Scholar] [CrossRef]
- Tiwana, H.; Wilson, C.; Walmsley, R.; Wakefield, A.; Smith, M.; Cox, N.; Hudson, M.J.; Ebringer, A. Antibody responses to gut bacteria in ankylosing spondylitis, rheumatoid arthritis, Crohn’s disease and ulcerative colitis. Rheumatol. Int. 1997, 17, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Ganji, L.; Alebouyeh, M.; Shirazi, M.H.; Eshraghi, S.S.; Mirshafiey, A.; Daryani, N.E.; Zali, M.R. Dysbiosis of fecal microbiota and high frequency of Citrobacter, Klebsiella spp., and Actinomycetes in patients with irritable bowel syndrome and gastroenteritis. Gastroenterol. Hepatol. Bed Bench 2016, 9, 325. [Google Scholar] [PubMed]
- Chen, Y.R.; Zheng, H.M.; Zhang, G.X.; Chen, F.L.; Chen, L.D.; Yang, Z.C. High Oscillospira abundance indicates constipation and low BMI in the Guangdong Gut Microbiome Project. Sci. Rep. 2020, 10, 9364. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, H.; Jiang, Q.; Zhu, Y.Z. The gut microbiome as non-invasive biomarkers for identifying overweight people at risk for osteoarthritis. Microb. Pathog. 2021, 157, 104976. [Google Scholar] [CrossRef]
- Cong, J.; Zhu, H.; Liu, D.; Li, T.; Zhang, C.; Zhu, J.; Lv, H.; Liu, K.; Hao, C.; Tian, Z.; et al. A Pilot Study: Changes of Gut Microbiota in Post-surgery Colorectal Cancer Patients. Front. Microbiol. 2018, 9, 2777. [Google Scholar] [CrossRef]
- Yildirim, S.; Nalbantoglu, O.U.; Bayraktar, A.; Ercan, F.B.; Gundogdu, A.; Velioglu, H.A.; Göl, M.F.; Soylu, A.E.; Koç, F.; Gülpınar, E.A.; et al. Stratification of the Gut Microbiota Composition Landscape across the Alzheimer’s Disease Continuum in a Turkish Cohort. mSystems 2022, 7, e0000422. [Google Scholar] [CrossRef]
- Ye, F.; Gao, X.; Wang, Z.; Cao, S.; Liang, G.; He, D.; Lv, Z.; Wang, L.; Xu, P.; Zhang, Q. Comparison of gut microbiota in autism spectrum disorders and neurotypical boys in China: A case-control study. Synth. Syst. Biotechnol. 2021, 6, 120–126. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Demirkan, A.; Twa, G.; Cohen, G.; Dean, M.N.; Standaert, D.G.; Sampson, T.R.; Payami, H. Metagenomics of Parkinson’s disease implicates the gut microbiome in multiple disease mechanisms. Nat. Commun. 2022, 13, 6958. [Google Scholar] [CrossRef]
- Lin, P.; Ding, B.; Feng, C.; Yin, S.; Zhang, T.; Qi, X.; Lv, H.; Guo, X.; Dong, K.; Zhu, Y.; et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J. Affect. Disord. 2017, 207, 300–304. [Google Scholar] [CrossRef]
- Halawa, M.R.; El-Salam, M.A.; Mostafa, B.M.; Sallout, S.S. The Gut Microbiome, Lactobacillus acidophilus; Relation with Type 2 Diabetes Mellitus. Curr. Diabetes Rev. 2019, 15, 480–485. [Google Scholar] [CrossRef]
- Wang, W.; Chen, L.; Zhou, R.; Wang, X.; Song, L.; Huang, S.; Wang, G.; Xia, B. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 2014, 52, 398–406. [Google Scholar] [CrossRef]
- Szablewski, L. Human Gut Microbiota in Health and Alzheimer’s Disease. J. Alzheimers Dis. 2018, 62, 549–560. [Google Scholar] [CrossRef]
- De Angelis, M.; Francavilla, R.; Piccolo, M.; De Giacomo, A.; Gobbetti, M. Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Macbeth, W.A.; Kass, E.H.; McDermott, W.V., Jr. Treatment of Hepatic Encephalopathy by Alteration of Intestinal Flora with Lactobacillus acidophilus. Lancet 1965, 1, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Tellez Corral, M.A.; Herrera Daza, E.; Cuervo Jimenez, H.K.; Bravo Becerra, M.d.M.; Villamil, J.C.; Hidalgo Martinez, P.; Roa Molina, N.S.; Otero, L.; Cortés, M.E.; Parra Giraldo, C.M. Cryptic Oral Microbiota: What Is Its Role as Obstructive Sleep Apnea-Related Periodontal Pathogens? Int. J. Environ. Res. Public Health 2023, 20, 1740. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef]
- Wasen, C.; Simonsen, E.; Ekwudo, M.N.; Profant, M.R.; Cox, L.M. The emerging role of the microbiome in Alzheimer’s disease. Int. Rev. Neurobiol. 2022, 167, 101–139. [Google Scholar]
- Lee, Y.; Park, J.Y.; Lee, E.H.; Yang, J.; Jeong, B.R.; Kim, Y.K.; Seoh, J.Y.; Lee, S.; Han, P.L.; Kim, E.J. Rapid Assessment of Microbiota Changes in Individuals with Autism Spectrum Disorder Using Bacteria-derived Membrane Vesicles in Urine. Exp. Neurobiol. 2017, 26, 307–317. [Google Scholar] [CrossRef]
- Tetz, G.; Brown, S.M.; Hao, Y.; Tetz, V. Parkinson’s disease and bacteriophages as its overlooked contributors. Sci. Rep. 2018, 8, 10812. [Google Scholar] [CrossRef]
- Ganci, M.; Suleyman, E.; Butt, H.; Ball, M. Associations between self-reported psychological symptom severity and gut microbiota: Further support for the microgenderome. BMC Psychiatry 2022, 22, 307. [Google Scholar] [CrossRef]
- Chen, Z.; Radjabzadeh, D.; Chen, L.; Kurilshikov, A.; Kavousi, M.; Ahmadizar, F.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Association of Insulin Resistance and Type 2 Diabetes with Gut Microbial Diversity: A Microbiome-Wide Analysis from Population Studies. JAMA Netw. Open 2021, 4, e2118811. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, F.; Ye, H.; Lu, B. Integrative analysis of the gut microbiome and metabolome in a rat model with stress induced irritable bowel syndrome. Sci. Rep. 2021, 11, 17596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Qian, Y.; Xie, Y.H.; Jiang, S.S.; Kang, Z.R.; Chen, Y.; Chen, Z.; Fang, J. Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. Int. J. Cancer 2021, 149, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Abraham, D.; Feher, J.; Scuderi, G.L.; Szabo, D.; Dobolyi, A.; Cservenak, M.; Juhasz, J.; Ligeti, B.; Pongor, S.; Gomez-Cabrera, M.C.; et al. Exercise and probiotics attenuate the development of Alzheimer’s disease in transgenic mice: Role of microbiome. Exp. Gerontol. 2019, 115, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Abujamel, T.S.; Al-Otaibi, N.M.; Abuaish, S.; AlHarbi, R.H.; Assas, M.B.; Alzahrani, S.A.; Alotaibi, S.M.; El-Ansary, A.; Aabed, K. Different Alterations in Gut Microbiota between Bifidobacterium longum and Fecal Microbiota Transplantation Treatments in Propionic Acid Rat Model of Autism. Nutrients 2022, 14, 608. [Google Scholar] [CrossRef]
- Chen, T.J.; Feng, Y.; Liu, T.; Wu, T.T.; Chen, Y.J.; Li, X.; Li, Q.; Wu, Y.C. Fisetin Regulates Gut Microbiota and Exerts Neuroprotective Effect on Mouse Model of Parkinson’s Disease. Front. Neurosci. 2020, 14, 549037. [Google Scholar] [CrossRef]
- Tung, T.H.; Tung, Y.T.; Lin, I.H.; Shih, C.K.; Nguyen, N.T.K.; Shabrina, A.; Huang, S.Y. Fish Oil, but Not Olive Oil, Ameliorates Depressive-Like Behavior and Gut Microbiota Dysbiosis in Rats under Chronic Mild Stress. Biomolecules 2019, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Lucking, E.F.; O’Connor, K.M.; Strain, C.R.; Fouhy, F.; Bastiaanssen, T.F.S.; Burns, D.P.; Golubeva, A.V.; Stanton, C.; Clarke, G.; Cryan, J.F.; et al. Chronic intermittent hypoxia disrupts cardiorespiratory homeostasis and gut microbiota composition in adult male guinea-pigs. EBioMedicine 2018, 38, 191–205. [Google Scholar] [CrossRef]
- Torquati, L.; Gajanand, T.; Cox, E.R.; Willis, C.R.G.; Zaugg, J.; Keating, S.E.; Coombes, J.S. Effects of exercise intensity on gut microbiome composition and function in people with type 2 diabetes. Eur. J. Sport Sci. 2022, 23, 530–541. [Google Scholar] [CrossRef]
- Ghavami, S.B.; Rostami, E.; Sephay, A.A.; Shahrokh, S.; Balaii, H.; Aghdaei, H.A.; Zali, M.R. Alterations of the human gut Methanobrevibacter smithii as a biomarker for inflammatory bowel diseases. Microb. Pathog. 2018, 117, 285–289. [Google Scholar] [CrossRef]
- Huang, H.L.; Chen, H.T.; Luo, Q.L.; Xu, H.M.; He, J.; Li, Y.Q.; Zhou, Y.L.; Yao, F.; Nie, Y.Q.; Zhou, Y.J. Relief of irritable bowel syndrome by fecal microbiota transplantation is associated with changes in diversity and composition of the gut microbiota. J. Dig. Dis. 2019, 20, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.Y.; Yu, J. Altered Gut Archaea Composition and Interaction with Bacteria Are Associated with Colorectal Cancer. Gastroenterology 2020, 159, 1459–1470.e5. [Google Scholar] [CrossRef] [PubMed]
- Garcez, M.L.; Jacobs, K.R.; Guillemin, G.J. Microbiota Alterations in Alzheimer’s Disease: Involvement of the Kynurenine Pathway and Inflammation. Neurotox. Res. 2019, 36, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Newell, C.; Bomhof, M.R.; Reimer, R.A.; Hittel, D.S.; Rho, J.M.; Shearer, J. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol. Autism. 2016, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Alvarez-Mon, M.A.; García-Montero, C.; Fraile-Martinez, O.; Guijarro, L.G.; Lahera, G.; Monserrat, J.; Valls, P.; Mora, F.; Rodríguez-Jiménez, R.; et al. Gut Microbiota Metabolites in Major Depressive Disorder-Deep Insights into Their Pathophysiological Role and Potential Translational Applications. Metabolites 2022, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, T.; Sun, J.; Huang, Y.; Liu, T.; Ye, Z.; Hu, J.; Zhang, G.; Chen, H.; He, Y.; et al. Calorie restriction ameliorates hyperglycemia, modulates the disordered gut microbiota, and mitigates metabolic endotoxemia and inflammation in type 2 diabetic rats. J. Endocrinol. Investig. 2023, 46, 699–711. [Google Scholar] [CrossRef]
- Pisani, A.; Rausch, P.; Bang, C.; Ellul, S.; Tabone, T.; Marantidis Cordina, C.; Zahra, G.; Franke, A.; Ellul, P. Dysbiosis in the gut microbiota in patients with inflammatory bowel disease during remission. Microbiol. Spectr. 2022, 10, e00616-22. [Google Scholar] [CrossRef]
- Tap, J.; Derrien, M.; Törnblom, H.; Brazeilles, R.; Cools-Portier, S.; Doré, J.; Störsrud, S.; Le Nevé, B.; Öhman, L.; Simrén, M. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology 2017, 152, 111–123.e8. [Google Scholar] [CrossRef]
- Xu, M.; Mo, X.; Huang, H.; Chen, X.; Liu, H.; Peng, Z.; Chen, L.; Rong, S.; Yang, W.; Xu, S.; et al. Yeast beta-glucan alleviates cognitive deficit by regulating gut microbiota and metabolites in Abeta(1)(−)(42)-induced AD-like mice. Int. J. Biol. Macromol. 2020, 161, 258–270. [Google Scholar] [CrossRef]
- Levi Mortera, S.; Vernocchi, P.; Basadonne, I.; Zandona, A.; Chierici, M.; Durighello, M.; Marzano, V.; Gardini, S.; Gasbarrini, A.; Urbani, A.; et al. A metaproteomic-based gut microbiota profiling in children affected by autism spectrum disorders. J. Proteom. 2022, 251, 104407. [Google Scholar] [CrossRef]
- O’donovan, S.M.; Crowley, E.K.; Brown, J.R.; O’sullivan, O.; O’leary, O.F.; Timmons, S.; Nolan, Y.M.; Clarke, D.J.; Hyland, N.P.; Joyce, S.A.; et al. Nigral overexpression of alpha-synuclein in a rat Parkinson’s disease model indicates alterations in the enteric nervous system and the gut microbiome. Neurogastroenterol. Motil. 2020, 32, e13726. [Google Scholar] [CrossRef] [PubMed]
- Iebba, V.; Guerrieri, F.; Di Gregorio, V.; Levrero, M.; Gagliardi, A.; Santangelo, F.; Sobolev, A.P.; Circi, S.; Giannelli, V.; Mannina, L.; et al. Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci. Rep. 2018, 8, 8210. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Melnik, A.V.; Xue, J.; Poulsen, O.; Meehan, M.J.; Humphrey, G.; Jiang, L.; Ackermann, G.; McDonald, D.; Zhou, D.; et al. Intermittent Hypoxia and Hypercapnia, a Hallmark of Obstructive Sleep Apnea, Alters the Gut Microbiome and Metabolome. mSystems 2018, 3, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Liu, F.; Zhang, B.; Dong, K.; Lu, M.; Jiang, R.; Xu, Y.; Diao, L.; Zhao, J.; Tang, H. Liraglutide-induced structural modulation of the gut microbiota in patients with type 2 diabetes mellitus. PeerJ 2021, 9, e11128. [Google Scholar] [CrossRef]
- Cremon, C.; Guglielmetti, S.; Gargari, G.; Taverniti, V.; Castellazzi, A.M.; Valsecchi, C.; Tagliacarne, C.; Fiore, W.; Bellini, M.; Bertani, L.; et al. Effect of Lactobacillus paracasei CNCM I-1572 on symptoms, gut microbiota, short chain fatty acids, and immune activation in patients with irritable bowel syndrome: A pilot randomized clinical trial. United Eur. Gastroenterol. J. 2018, 6, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.W.; Lee, W.H.; Tu, S.J.; Huang, W.C.; Chen, H.M.; Sun, T.H.; Tsai, M.C.; Wang, C.C.; Chen, H.Y.; Huang, C.C.; et al. Enterotype-based Analysis of Gut Microbiota along the Conventional Adenoma-Carcinoma Colorectal Cancer Pathway. Sci. Rep. 2019, 9, 10923. [Google Scholar] [CrossRef]
- Hang, Z.; Cai, S.; Lei, T.; Zhang, X.; Xiao, Z.; Wang, D.; Li, Y.; Bi, W.; Yang, Y.; Deng, S.; et al. Transfer of Tumor-Bearing Mice Intestinal Flora Can Ameliorate Cognition in Alzheimer’s Disease Mice. J. Alzheimers Dis. 2022, 86, 1287–1300. [Google Scholar] [CrossRef]
- Zhai, Q.; Cen, S.; Jiang, J.; Zhao, J.; Zhang, H.; Chen, W. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: A pilot study of Chinese children. Environ. Res. 2019, 171, 501–509. [Google Scholar] [CrossRef]
- Zhang, M.; Li, A.; Yang, Q.; Li, J.; Wang, L.; Liu, X.; Huang, Y.; Liu, L. Beneficial Effect of Alkaloids from Sophora alopecuroides L. on CUMS-Induced Depression Model Mice via Modulating Gut Microbiota. Front. Cell. Infect. Microbiol. 2021, 11, 665159. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, X.; Ding, D.; Lu, Y. Characteristics of the intestinal flora in patients with peripheral neuropathy associated with type 2 diabetes. J. Int. Med. Res. 2020, 48, 300060520936806. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, F.; Mao, L.; Feng, T.; Wang, K.; Xu, M.; Lv, B.; Wang, X. Bifico relieves irritable bowel syndrome by regulating gut microbiota dysbiosis and inflammatory cytokines. Eur. J. Nutr. 2023, 62, 139–155. [Google Scholar] [CrossRef]
- Wu, T.R.; Lin, C.S.; Chang, C.J.; Lin, T.L.; Martel, J.; Ko, Y.F.; Ojcius, D.M.; Lu, C.C.; Young, J.D.; Lai, H.C. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 2019, 68, 248–262. [Google Scholar] [CrossRef]
- Sheng, Q.S.; He, K.X.; Li, J.J.; Zhong, Z.F.; Wang, F.X.; Pan, L.L.; Lin, J.J. Comparison of Gut Microbiome in Human Colorectal Cancer in Paired Tumor and Adjacent Normal Tissues. Onco Targets Ther. 2020, 13, 635–646. [Google Scholar] [CrossRef]
- Sun, Z.Z.; Li, X.Y.; Wang, S.; Shen, L.; Ji, H.F. Bidirectional interactions between curcumin and gut microbiota in transgenic mice with Alzheimer’s disease. Appl. Microbiol. Biotechnol. 2020, 104, 3507–3515. [Google Scholar] [CrossRef]
- Gomez-Nguyen, A.; Basson, A.R.; Dark-Fleury, L.; Hsu, K.; Osme, A.; Menghini, P.; Pizarro, T.T.; Cominelli, F. Parabacteroides distasonis induces depressive-like behavior in a mouse model of Crohn’s disease. Brain Behav. Immun. 2021, 98, 245–250. [Google Scholar] [CrossRef]
- Guarner, F. The intestinal flora in inflammatory bowel disease: Normal or abnormal? Curr. Opin. Gastroenterol. 2005, 21, 414–418. [Google Scholar] [PubMed]
- Shen, H.; Han, J.; Li, Y.; Lu, C.; Zhou, J.; Li, Y.; Su, X. Different host-specific responses in thyroid function and gut microbiota modulation between diet-induced obese and normal mice given the same dose of iodine. Appl. Microbiol. Biotechnol. 2019, 103, 3537–3547. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A. Activation of aryl hydrocarbon receptor (AhR) in Alzheimer’s disease: Role of tryptophan metabolites generated by gut host-microbiota. J. Mol. Med. 2023, 101, 201–222. [Google Scholar] [CrossRef]
- Dogra, N.; Mani, R.J.; Katare, D.P. The Gut-Brain Axis: Two Ways Signaling in Parkinson’s Disease. Cell. Mol. Neurobiol. 2022, 42, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.Y.; Fan, J.M.; Hu, A.K.; Su, H.Z.; Yang, J.H.; Huang, L.M.; Yan, F.; Zhang, H.; Zeng, Y. Disruption of sleep architecture in Prevotella enterotype of patients with obstructive sleep apnea-hypopnea syndrome. Brain Behav. 2019, 9, e01287. [Google Scholar] [CrossRef] [PubMed]
- Leite, A.Z.; Rodrigues, N.C.; Gonzaga, M.I.; Paiolo, J.C.C.; de Souza, C.A.; Stefanutto, N.A.V.; Omori, W.P.; Pinheiro, D.G.; Brisotti, J.L.; Junior, E.M.; et al. Detection of Increased Plasma Interleukin-6 Levels and Prevalence of Prevotella copri and Bacteroides vulgatus in the Feces of Type 2 Diabetes Patients. Front. Immunol. 2017, 8, 1107. [Google Scholar] [CrossRef] [PubMed]
- Altomare, A.; Putignani, L.; Del Chierico, F.; Cocca, S.; Angeletti, S.; Ciccozzi, M.; Tripiciano, C.; Dalla Piccola, B.; Cicala, M.; Guarino, M.P.L. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig. Liver Dis. 2019, 51, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Liu, R.; Lee, A.; Long, Y.; Du, L.; Lai, S.; Chen, X.; Wang, L.; Si, J.; Owyang, C.; et al. Altered intestinal microbiota with increased abundance of Prevotella is associated with high risk of diarrhea-predominant irritable bowel syndrome. Gastroenterol. Res. Pract. 2018, 2018, 6961783. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, A.C.; Akter, S.; Hanson, M.M.; Busi, S.B.; Parker, T.W.; Schehr, R.J.; Hankins, M.A.; Ahner, C.E.; Davis, J.W.; Franklin, C.L.; et al. Differential susceptibility to colorectal cancer due to naturally occurring gut microbiota. Oncotarget 2015, 6, 33689–33704. [Google Scholar] [CrossRef]
- Guo, M.; Peng, J.; Huang, X.; Xiao, L.; Huang, F.; Zuo, Z. Gut Microbiome Features of Chinese Patients Newly Diagnosed with Alzheimer’s Disease or Mild Cognitive Impairment. J. Alzheimers Dis. 2021, 80, 299–310. [Google Scholar] [CrossRef]
- Mertsalmi, T.H.; Aho, V.T.E.; Pereira, P.A.B.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. More than constipation–bowel symptoms in Parkinson’s disease and their connection to gut microbiota. Eur. J. Neurol. 2017, 24, 1375–1383. [Google Scholar] [CrossRef]
- Dhiman, R.K. Gut microbiota, inflammation and hepatic encephalopathy: A puzzle with a solution in sight. J. Clin. Exp. Hepatol. 2012, 2, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Kobyliak, N.; Falalyeyeva, T.; Mykhalchyshyn, G.; Kyriienko, D.; Komissarenko, I. Effect of alive probiotic on insulin resistance in type 2 diabetes patients: Randomized clinical trial. Diabetes Metab. Syndr. 2018, 12, 617–624. [Google Scholar] [CrossRef]
- Brahe, L.K.; Astrup, A.; Larsen, L.H. Can We Prevent Obesity-Related Metabolic Diseases by Dietary Modulation of the Gut Microbiota? Adv. Nutr. 2016, 7, 90–101. [Google Scholar] [CrossRef]
- Lacroix, S.; Pechereau, F.; Leblanc, N.; Boubertakh, B.; Houde, A.; Martin, C.; Flamand, N.; Silvestri, C.; Raymond, F.; Di Marzo, V.; et al. Rapid and Concomitant Gut Microbiota and Endocannabinoidome Response to Diet-Induced Obesity in Mice. mSystems. 2019, 4, e00407-19. [Google Scholar] [CrossRef]
- Wagner, J.; Short, K.; Catto-Smith, A.G.; Cameron, D.J.; Bishop, R.F.; Kirkwood, C.D. Identification and characterisation of Pseudomonas 16S ribosomal DNA from ileal biopsies of children with Crohn’s disease. PLoS ONE 2008, 3, e3578. [Google Scholar] [CrossRef] [PubMed]
- Kerckhoffs, A.P.; Ben-Amor, K.; Samsom, M.; van der Rest, M.E.; de Vogel, J.; Knol, J.; Akkermans, L.M.A. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J. Med. Microbiol. 2011, 60, 236–245. [Google Scholar] [CrossRef]
- Petriz, B.A.; Castro, A.P.; Almeida, J.A.; Gomes, C.P.; Fernandes, G.R.; Kruger, R.H.; Pereira, R.W.; Franco, O.L. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genom. 2014, 15, 511. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, X.; Jiang, R.; Yan, X.; Ling, Z. Roles and Mechanisms of Gut Microbiota in Patients with Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 650047. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.; Mazumder, A.; Grace, T. Role of the Gut Microbiome in Autism Spectrum Disorders. Adv. Exp. Med. Biol. 2019, 1118, 253–269. [Google Scholar]
- Wang, Y.; Tong, Q.; Ma, S.R.; Zhao, Z.X.; Pan, L.B.; Cong, L.; Han, P.; Peng, R.; Yu, H.; Lin, Y.; et al. Oral berberine improves brain dopa/dopamine levels to ameliorate Parkinson’s disease by regulating gut microbiota. Signal. Transduct. Target Ther. 2021, 6, 77. [Google Scholar] [CrossRef]
- Waclawikova, B.; El Aidy, S. Role of Microbiota and Tryptophan Metabolites in the Remote Effect of Intestinal Inflammation on Brain and Depression. Pharmaceuticals 2018, 11, 63. [Google Scholar] [CrossRef]
- Jang, S.; Kim, Y.; Lee, C.; Kwon, B.; Noh, J.; Jee, J.J.; Yoon, S.S.; Koh, H.; Park, S. The effect of formula-based nutritional treatment on colitis in a murine model. J. Korean Med. Sci. 2021, 36, e342. [Google Scholar] [CrossRef]
- Seo, K.H.; Kim, D.H.; Yokoyama, W.H.; Kim, H. Synbiotic Effect of Whole Grape Seed Flour and Newly Isolated Kefir Lactic Acid Bacteria on Intestinal Microbiota of Diet-Induced Obese Mice. J. Agric. Food Chem. 2020, 68, 13131–13137. [Google Scholar] [CrossRef]
- Romero-Garmendia, I.; Garcia-Etxebarria, K. Host Genetics and Microbiota Interactions in Colorectal Cancer: Shared or Independent Risk? Microorganisms 2022, 10, 2129. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Roh, Y.J.; Choi, Y.J.; Lee, S.J.; Jin, Y.J.; Song, H.J.; Hong, J.T.; Hwang, D.Y. Dysbiosis of Fecal Microbiota in Tg2576 Mice for Alzheimer’s Disease during Pathological Constipation. Int. J. Mol. Sci. 2022, 23, 14928. [Google Scholar] [CrossRef] [PubMed]
- Getachew, B.; Tizabi, Y. Effects of C-Terminal Domain of the Heavy Chain of Tetanus Toxin on Gut Microbiota in a Rat Model of Depression. Clin. Pharmacol. Transl. Med. 2019, 3, 152–159. [Google Scholar] [PubMed]
- Faden, H. The Role of Faecalibacterium, Roseburia, and Butyrate in Inflammatory Bowel Disease. Dig. Dis. 2022, 40, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Ruusunen, A.; Rocks, T.; Jacka, F.; Loughman, A. The gut microbiome in anorexia nervosa: Relevance for nutritional rehabilitation. Psychopharmacology 2019, 236, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.; Barrett, H.; Gomez-Arango, L.; McIntyre, H.D.; Callaway, L.; Dekker Nitert, M. Ketonuria Is Associated with Changes to the Abundance of Roseburia in the Gut Microbiota of Overweight and Obese Women at 16 Weeks Gestation: A Cross-Sectional Observational Study. Nutrients 2019, 11, 1836. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Fan, H.; Tang, X.; Zhai, H.; Zhang, Z. Diversified pattern of the human colorectal cancer microbiome. Gut Pathog. 2013, 5, 2. [Google Scholar] [CrossRef]
- Jiang, L.; Li, J.C.; Tang, B.S.; Guo, J.F. Associations between Gut Microbiota and Parkinson’s Disease: A Bidirectional Mendelian Randomization Analysis. Eur. J. Neurol. 2023. [Google Scholar] [CrossRef]
- Qu, W.; Liu, S.; Zhang, W.; Zhu, H.; Tao, Q.; Wang, H.; Yan, H. Impact of traditional Chinese medicine treatment on chronic unpredictable mild stress-induced depression-like behaviors: Intestinal microbiota and gut microbiome function. Food Funct. 2019, 10, 5886–5897. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef]
- Salas-Perez, F.; Assmann, T.S.; Ramos-Lopez, O.; Martinez, J.A.; Riezu-Boj, J.I.; Milagro, F.I. Crosstalk between Gut Microbiota and Epigenetic Markers in Obesity Development: Relationship between Ruminococcus, BMI, and MACROD2/SEL1L2 Methylation. Nutrients 2023, 15, 1550. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism. 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Lukic, I.; Getselter, D.; Ziv, O.; Oron, O.; Reuveni, E.; Koren, O.; Elliott, E. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl. Psychiatry 2019, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Silamikele, L.; Saksis, R.; Silamikelis, I.; Kotovica, P.P.; Briviba, M.; Kalnina, I.; Kalniņa, Z.; Fridmanis, D.; Kloviņš, J. Spatial variation of the gut microbiome in response to long-term metformin treatment in high-fat diet-induced type 2 diabetes mouse model of both sexes. Gut Microbes 2023, 15, 2188663. [Google Scholar] [CrossRef]
- Ma, H.-Q.; Yu, T.-T.; Zhao, X.-J.; Zhang, Y.; Zhang, H.-J. Fecal microbial dysbiosis in Chinese patients with inflammatory bowel disease. World J. Gastroenterol. 2018, 24, 1464. [Google Scholar] [CrossRef]
- Mohamad, N.E.; Yeap, S.K.; Ky, H.; Liew, N.W.C.; Beh, B.K.; Boo, S.Y.; Ho, W.Y.; Sharifuddin, S.A.; Long, K.; Alitheen, N.B. Pineapple Vinegar Regulates Obesity-Related Genes and Alters the Gut Microbiota in High-Fat Diet (HFD) C57BL/6 Obese Mice. Evid. Based Complement. Alternat. Med. 2020, 2020, 1257962. [Google Scholar] [CrossRef]
- Getachew, B.; Aubee, J.I.; Schottenfeld, R.S.; Csoka, A.B.; Thompson, K.M.; Tizabi, Y. Ketamine interactions with gut-microbiota in rats: Relevance to its antidepressant and anti-inflammatory properties. BMC Microbiol. 2018, 18, 222. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Guo, R.; Yu, W.; Zhang, F.; Wu, F.; Shang, J. The Alteration in Composition and Function of Gut Microbiome in Patients with Type 2 Diabetes. J. Diabetes Res. 2020, 2020, 8842651. [Google Scholar] [CrossRef]
- Cheung, Y.B.; Xu, Y.; Mangani, C.; Fan, Y.-M.; Dewey, K.G.; Salminen, S.J.; Maleta, K.; Ashorn, P. Gut microbiota in Malawian infants in a nutritional supplementation trial. Trop. Med. Int. Health 2016, 21, 283–290. [Google Scholar] [CrossRef]
- Nie, Q.; Chen, H.; Hu, J.; Fan, S.; Nie, S. Dietary compounds and traditional Chinese medicine ameliorate type 2 diabetes by modulating gut microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 848–863. [Google Scholar] [CrossRef]
- Correa, T.A.F.; Rogero, M.M.; Hassimotto, N.M.A.; Lajolo, F.M. The Two-Way Polyphenols-Microbiota Interactions and Their Effects on Obesity and Related Metabolic Diseases. Front. Nutr. 2019, 6, 188. [Google Scholar] [CrossRef] [PubMed]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol. Rep. 2016, 35, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Kuo, P.H.; Hsu, C.Y.; Chiu, Y.H.; Liu, Y.W.; Lu, M.L.; Chen, C.H. Psychophysiological Effects of Lactobacillus plantarum PS128 in Patients with Major Depressive Disorder: A Preliminary 8-Week Open Trial. Nutrients 2021, 13, 3731. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.Y.; Zolnik, C.P.; Wang, Z.; Qiu, Y.; Usyk, M.; Wang, T.; Kizer, J.R.; Landay, A.L.; Kurland, I.J.; Anastos, K.; et al. Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine 2018, 37, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Zareef, R.; Younis, N.; Mahfouz, R. Inflammatory bowel disease: A key role for microbiota? Meta Gene 2020, 25, 100713. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Sun, H.; Jiang, F.; Shen, Y.; Wei, P.; Shen, X. Characterization of the gut microbiota in Chinese children with overweight and obesity using 16S rRNA gene sequencing. PeerJ 2021, 9, e11439. [Google Scholar] [CrossRef]
- Liu, C.; Shao, W.; Gao, M.; Liu, J.; Guo, Q.; Jin, J.; Meng, F. Changes in intestinal flora in patients with type 2 diabetes on a low-fat diet during 6 months of follow-up. Exp. Ther. Med. 2020, 20, 40. [Google Scholar] [CrossRef]
- Zhong, Y.-B.; Kang, Z.-P.; Wang, M.-X.; Long, J.; Wang, H.-Y.; Huang, J.-Q.; Wei, S.-Y.; Zhou, W.; Zhao, H.-M.; Liu, D.-Y. Curcumin ameliorated dextran sulfate sodium-induced colitis via regulating the homeostasis of DCs and Treg and improving the composition of the gut microbiota. J. Funct. Foods 2021, 86, 104716. [Google Scholar] [CrossRef]
- Lin, C.; Li, B.; Tu, C.; Chen, X.; Guo, M. Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma. BioMed Res. Int. 2022, 2022, 3140070. [Google Scholar] [CrossRef]
- Wu, Y.; Niu, X.; Li, P.; Tong, T.; Wang, Q.; Zhang, M.; Li, Y.; Liu, J.; Li, Z. Lactobacillaceae improve cognitive dysfunction via regulating gut microbiota and suppressing Abeta deposits and neuroinflammation in APP/PS1 mice. Arch. Microbiol. 2023, 205, 118. [Google Scholar] [CrossRef]
- Singh, Y.; El-Hadidi, M.; Admard, J.; Wassouf, Z.; Schulze-Hentrich, J.M.; Kohlhofer, U.; Quintanilla-Martinez, L.; Huson, D.; Riess, O.; Casadei, N. Enriched Environmental Conditions Modify the Gut Microbiome Composition and Fecal Markers of Inflammation in Parkinson’s Disease. Front. Neurosci. 2019, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Hu, Y.; Ma, H.; Zou, Z.; Xiao, Y.; Yang, Y.; Feng, M.; Li, X.; Ye, X. Rhizoma Coptidis alkaloids alleviate hyperlipidemia in B6 mice by modulating gut microbiota and bile acid pathways. Biochim. Biophys. Acta 2016, 1862, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Katsidzira, L.; Ocvirk, S.; Wilson, A.; Li, J.; Mahachi, C.B.; Soni, D.; DeLany, J.; Nicholson, J.K.; Zoetendal, E.G.; O’keefe, S.J.D. Differences in Fecal Gut Microbiota, Short-Chain Fatty Acids and Bile Acids Link Colorectal Cancer Risk to Dietary Changes Associated with Urbanization among Zimbabweans. Nutr. Cancer 2019, 71, 1313–1324. [Google Scholar] [CrossRef]
- Yao, Y.; Qi, X.; Jia, Y.; Ye, J.; Chu, X.; Wen, Y.; Cheng, B.; Cheng, S.; Liu, L.; Liang, C.; et al. Evaluating the interactive effects of dietary habits and human gut microbiome on the risks of depression and anxiety. Psychol. Med. 2022, 53, 3047–3055. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.I.; Gill, A.L.; Mooney, R.A.; Gill, S.R. Modulation of Gut Microbiota Metabolism in Obesity-Related Type 2 Diabetes Reduces Osteomyelitis Severity. Microbiol. Spectr. 2022, 10, e0017022. [Google Scholar] [CrossRef]
- Azimirad, M.; Krutova, M.; Balaii, H.; Kodori, M.; Shahrokh, S.; Azizi, O.; Yadegar, A.; Aghdaei, H.A.; Zali, M.R. Coexistence of Clostridioides difficile and Staphylococcus aureus in gut of Iranian outpatients with underlying inflammatory bowel disease. Anaerobe 2020, 61, 102113. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Delzenne, N.M. Interplay between obesity and associated metabolic disorders: New insights into the gut microbiota. Curr. Opin. Pharmacol. 2009, 9, 737–743. [Google Scholar] [CrossRef]
- Pistollato, F.; Sumalla Cano, S.; Elio, I.; Masias Vergara, M.; Giampieri, F.; Battino, M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 2016, 74, 624–634. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Wilkowska, A.; Szalach, L.P.; Cubala, W.J. Gut Microbiota in Depression: A Focus on Ketamine. Front. Behav. Neurosci. 2021, 15, 693362. [Google Scholar] [CrossRef]
- Sun, X.; Chi, X.; Zhao, Y.; Liu, S.; Xing, H. Characteristics and Clinical Significance of Intestinal Microbiota in Patients with Chronic Hepatitis B Cirrhosis and Type 2 Diabetes Mellitus. J. Diabetes Res. 2022, 2022, 1826181. [Google Scholar] [CrossRef]
- Heidarian, F.; Noormohammadi, Z.; Aghdaei, H.A.; Alebouyeh, M. Relative abundance of streptococcus spp. and its association with disease activity in inflammatory bowel disease patients compared with controls. Arch. Clin. Infect. Dis. 2017, 12, e57291. [Google Scholar] [CrossRef]
- Hong, S.N.; Rhee, P.-L. Unraveling the ties between irritable bowel syndrome and intestinal microbiota. World J. Gastroenterol. WJG 2014, 20, 2470. [Google Scholar] [CrossRef]
- Wang, M.; Doenyas, C.; Wan, J.; Zeng, S.; Cai, C.; Zhou, J.; Liu, Y.; Yin, Z.; Zhou, W. Virulence factor-related gut microbiota genes and immunoglobulin A levels as novel markers for machine learning-based classification of autism spectrum disorder. Comput. Struct. Biotechnol. J. 2021, 19, 545–554. [Google Scholar] [CrossRef]
- Li, Z.; Lu, G.; Luo, E.; Wu, B.; Li, Z.; Guo, J.; Xia, Z.; Zheng, C.; Su, Q.; Zeng, Y.; et al. Oral, Nasal, and Gut Microbiota in Parkinson’s Disease. Neuroscience 2022, 480, 65–78. [Google Scholar] [CrossRef]
- Smith, R.P.; Easson, C.; Lyle, S.M.; Kapoor, R.; Donnelly, C.P.; Davidson, E.J.; Parikh, E.; Lopez, J.V.; Tartar, J.L. Gut microbiome diversity is associated with sleep physiology in humans. PLoS ONE 2019, 14, e0222394. [Google Scholar] [CrossRef]
- Bai, Z.; Huang, X.; Wu, G.; Ye, H.; Huang, W.; Nie, Q.; Chen, H.; Yin, J.; Chen, Y.; Nie, S. Polysaccharides from red kidney bean alleviating hyperglycemia and hyperlipidemia in type 2 diabetic rats via gut microbiota and lipid metabolic modulation. Food Chem. 2023, 404, 134598. [Google Scholar] [CrossRef]
- Loh, G.; Blaut, M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 2012, 3, 544–555. [Google Scholar] [CrossRef]
- Khoo, X.H.; Chong, C.W.; Talha, A.M.; Philip, K.; Teh, C.S.J.; Isa, A.M.; Wong, M.S.; Chew, D.C.H.; Wong, Z.; Jusoh, N.S.; et al. The impact of diet and ethnicity on Gut Microbiota Variation in Irritable Bowel Syndrome: A Multi-centre Study. J. Gastroenterol. Hepatol. 2023, 38, 1259–1268. [Google Scholar] [CrossRef]
- Chavez-Carbajal, A.; Nirmalkar, K.; Perez-Lizaur, A.; Hernandez-Quiroz, F.; Ramirez-Del-Alto, S.; Garcia-Mena, J.; Hernández-Guerrero, C. Gut Microbiota and Predicted Metabolic Pathways in a Sample of Mexican Women Affected by Obesity and Obesity Plus Metabolic Syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]
- Natalello, G.; Bosello, S.L.; Paroni Sterbini, F.; Posteraro, B.; De Lorenzis, E.; Canestrari, G.B.; Gigante, L.; Verardi, L.; Ferraccioli, G.; Sanguinetti, M.; et al. Gut microbiota analysis in systemic sclerosis according to disease characteristics and nutritional status. Clin. Exp. Rheumatol. 2020, 38 (Suppl. S125), 73–84. [Google Scholar] [PubMed]
- Zhang, M.; Lv, Y.; Hou, S.; Liu, Y.; Wang, Y.; Wan, X. Differential Mucosal Microbiome Profiles across Stages of Human Colorectal Cancer. Life 2021, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, R.; Liu, M.; Zhou, Q.; Zhai, Y.-H.; Fan, L.-H.; Lan, Y.-Z.; Zhu, X.-D. Metagenomic analysis of Tongxie Yaofang therapy for rat models of ulcerative colitis with liver depression and spleen deficiency syndrome. All Life 2023, 16, 1–12. [Google Scholar] [CrossRef]
- Bischoff, S.C.; Nguyen, N.K.; Seethaler, B.; Beisner, J.; Kügler, P.; Stefan, T. Gut Microbiota Patterns Predicting Long-Term Weight Loss Success in Individuals with Obesity Undergoing Nonsurgical Therapy. Nutrients 2022, 14, 3182. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Shao, L.; Heizhati, M.; Wu, T.; Yao, X.; Wang, Y.; Wang, L.; Li, N. Oropharyngeal Microbiome in Obstructive Sleep Apnea: Decreased Diversity and Abundance. J. Clin. Sleep Med. 2019, 15, 1777–1788. [Google Scholar] [CrossRef]
- Tai, N.; Wong, F.S.; Wen, L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev. Endocr. Metab. Disord. 2015, 16, 55–65. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Fischer, A.; Siegmund, B.; Kupz, A.; Niebergall, J.; Fuchs, D.; Jahn, H.-K.; Freudenberg, M.; Loddenkemper, C.; Batra, A.; et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS ONE 2007, 2, e662. [Google Scholar] [CrossRef]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 2013, 21, E607-15. [Google Scholar] [CrossRef]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7, 11590. [Google Scholar] [CrossRef]
- Wang, J.; Ye, F.; Cheng, X.; Zhang, X.; Liu, F.; Liu, G.; Ni, M.; Qiao, S.; Zhou, W.; Zhang, Y. The Effects of LW-AFC on Intestinal Microbiome in Senescence-Accelerated Mouse Prone 8 Strain, a Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2016, 53, 907–919. [Google Scholar] [CrossRef]
- Afroz, K.F.; Reyes, N.; Young, K.; Parikh, K.; Misra, V.; Alvina, K. Author Correction: Altered gut microbiome and autism like behavior are associated with parental high salt diet in male mice. Sci. Rep. 2022, 12, 5686. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Huan, F.; Wang, J.; Xie, X.; Yu, G.; Wang, X.; Jiang, L.; Gao, R.; Xiao, H.; Ding, H.; et al. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine Induced Parkinson’s Disease in Mouse: Potential Association between Neurotransmitter Disturbance and Gut Microbiota Dysbiosis. ACS Chem. Neurosci. 2020, 11, 3366–3376. [Google Scholar] [CrossRef]
- Lai, W.T.; Zhao, J.; Xu, S.X.; Deng, W.F.; Xu, D.; Wang, M.B.; He, F.S.; Liu, Y.H.; Guo, Y.Y.; Ye, S.W.; et al. Shotgun metagenomics reveals both taxonomic and tryptophan pathway differences of gut microbiota in bipolar disorder with current major depressive episode patients. J. Affect. Disord. 2021, 278, 311–319. [Google Scholar] [CrossRef]
- Cai, Y.; Juszczak, H.M.; Cope, E.K.; Goldberg, A.N. The microbiome in obstructive sleep apnea. Sleep 2021, 44, zsab061. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, Q.; Cao, J.; Xu, Y.; Pei, Z.; Fan, H.; Yuan, Y.; Shen, X.; Li, C. A sea cucumber (Holothuria leucospilota) polysaccharide improves the gut microbiome to alleviate the symptoms of type 2 diabetes mellitus in Goto-Kakizaki rats. Food Chem. Toxicol. 2020, 135, 110886. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Christman, L.M.; Li, R.; Gu, L. Synergic interactions between polyphenols and gut microbiota in mitigating inflammatory bowel diseases. Food Funct. 2020, 11, 4878–4891. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Bachar, D.; Yasir, M.; Musso, D.; Djossou, F.; Melenotte, C.; Robert, C.; Davoust, B.; Gaborit, B.; Azhar, E.; et al. Comparison of the gut microbiota of obese individuals from different geographic origins. New Microbes New Infect. 2019, 27, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.; Knorr, D.A.; Haptonstall, K.M. Alzheimer’s disease and symbiotic microbiota: An evolutionary medicine perspective. Ann. N. Y. Acad. Sci. 2019, 1449, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Scassellati, C.; Marizzoni, M.; Cattane, N.; Lopizzo, N.; Mombelli, E.; Riva, M.A.; Cattaneo, A. The Complex Molecular Picture of Gut and Oral Microbiota-Brain-Depression System: What We Know and What We Need to Know. Front. Psychiatry 2021, 12, 722335. [Google Scholar] [CrossRef]
- Bajer, L.; Kverka, M.; Kostovcik, M.; Macinga, P.; Dvorak, J.; Stehlikova, Z.; Brezina, J.; Wohl, P.; Spicak, J.; Drastich, P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017, 23, 4548. [Google Scholar] [CrossRef]
- de Souza, A.Z.; Zambom, A.Z.; Abboud, K.Y.; Reis, S.K.; Tannihao, F.; Guadagnini, D.; Saad, M.J.; Prada, P.O. Oral supplementation with L-glutamine alters gut microbiota of obese and overweight adults: A pilot study. Nutrition 2015, 31, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Liu, G.; Lin, Y.; Guo, C.L.; Han, J.; Chu, E.S.H.; Shi, C.; Li, Y.; Zhang, H.; Hu, C.; et al. Altered gut microbiome composition by appendectomy contributes to colorectal cancer. Oncogene 2023, 42, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, W.; Zhang, J.; He, Y.; Wang, J. Analysis of gut microbiota profiles and microbe-disease associations in children with autism spectrum disorders in China. Sci. Rep. 2018, 8, 13981. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Li, A.; Huang, T.; Lai, J.; Li, J.; Sublette, M.E.; Lu, H.; Lu, Q.; Du, Y.; Hu, Z.; et al. Gut Microbiota Changes in Patients with Bipolar Depression. Adv. Sci. 2019, 6, 1900752. [Google Scholar] [CrossRef] [PubMed]
- Salamone, D.; Rivellese, A.A.; Vetrani, C. The relationship between gut microbiota, short-chain fatty acids and type 2 diabetes mellitus: The possible role of dietary fibre. Acta Diabetol. 2021, 58, 1131–1138. [Google Scholar] [CrossRef]
- Zuo, T.; Kamm, M.A.; Colombel, J.-F.; Ng, S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 440–452. [Google Scholar] [CrossRef]
- Chen, J.; He, X.; Huang, J. Diet effects in gut microbiome and obesity. J. Food Sci. 2014, 79, R442–R451. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, J.; Liu, T.; Xing, J.; Zhang, H.; Wang, D.; Tang, D. Bidirectional effects of intestinal microbiota and antibiotics: A new strategy for colorectal cancer treatment and prevention. J. Cancer Res. Clin. Oncol. 2022, 148, 2387–2404. [Google Scholar] [CrossRef]
- Allaband, C.; Lingaraju, A.; Martino, C.; Russell, B.; Tripathi, A.; Poulsen, O.; Machado, A.C.D.; Zhou, D.; Xue, J.; Elijah, E.; et al. Intermittent hypoxia and hypercapnia alter diurnal rhythms of luminal gut microbiome and metabolome. mSystems 2021, 6, e00116-21. [Google Scholar] [CrossRef]
- Remely, M.; Hippe, B.; Zanner, J.; Aumueller, E.; Brath, H.; Haslberger, A.G. Gut Microbiota of Obese, Type 2 Diabetic Individuals is Enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after Weight Loss. Endocr. Metab. Immune Disord. Drug Targets 2016, 16, 99–106. [Google Scholar] [CrossRef]
- Zhang, T.; Ji, X.; Lu, G.; Zhang, F. The potential of Akkermansia muciniphila in inflammatory bowel disease. Appl. Microbiol. Biotechnol. 2021, 105, 5785–5794. [Google Scholar] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [PubMed]
- Osman, M.A.; Neoh, H.-M.; Ab Mutalib, N.-S.; Chin, S.-F.; Mazlan, L.; Raja Ali, R.A.; Zakaria, A.D.; Ngiu, C.S.; Ang, M.Y.; Jamal, R. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 2021, 11, 2925. [Google Scholar] [PubMed]
- Ou, Z.; Deng, L.; Lu, Z.; Wu, F.; Liu, W.; Huang, D.; Peng, Y. Protective effects of Akkermansia muciniphila on cognitive deficits and amyloid pathology in a mouse model of Alzheimer’s disease. Nutr. Diabetes 2020, 10, 12. [Google Scholar]
- Fang, X.; Li, F.J.; Hong, D.J. Potential Role of Akkermansia muciniphila in Parkinson’s Disease and Other Neurological/Autoimmune Diseases. Curr. Med. Sci. 2021, 41, 1172–1177. [Google Scholar]
- Ding, Y.; Bu, F.; Chen, T.; Shi, G.; Yuan, X.; Feng, Z.; Duan, Z.; Wang, R.; Zhang, S.; Wang, Q.; et al. A next-generation probiotic: Akkermansia muciniphila ameliorates chronic stress-induced depressive-like behavior in mice by regulating gut microbiota and metabolites. Appl. Microbiol. Biotechnol. 2021, 105, 8411–8426. [Google Scholar] [PubMed]
- Xu, J.; Lian, F.; Zhao, L.; Zhao, Y.; Chen, X.; Zhang, X.; Guo, Y.; Zhang, C.; Zhou, Q.; Xue, Z.; et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015, 9, 552–562. [Google Scholar]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar]
- Lotankar, M.; Mokkala, K.; Houttu, N.; Koivuniemi, E.; Sorensen, N.; Nielsen, H.B.; Munukka, E.; Lahti, L.; Laitinen, K. Distinct Diet-Microbiota-Metabolism Interactions in Overweight and Obese Pregnant Women: A Metagenomics Approach. Microbiol. Spectr. 2022, 10, e0089321. [Google Scholar]
- Sun, T.; Liu, S.; Zhou, Y.; Yao, Z.; Zhang, D.; Cao, S.; Wei, Z.; Tan, B.; Li, Y.; Lian, Z.; et al. Evolutionary biologic changes of gut microbiota in an ‘adenoma-carcinoma sequence’ mouse colorectal cancer model induced by 1, 2-Dimethylhydrazine. Oncotarget 2017, 8, 444–457. [Google Scholar]
- Li, Z.; Zhu, H.; Guo, Y.; Du, X.; Qin, C. Gut microbiota regulate cognitive deficits and amyloid deposition in a model of Alzheimer’s disease. J. Neurochem. 2020, 155, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.; Kovtun, A.; Danilenko, V. Bacterial Genes of Metabolite Biomarkers of Autism Spectrum Disorders in Gut Microbiota of Young Children: Detection by Real-Time PCR and In Silico Analysis. Russ. J. Genet. 2020, 56, 1260–1268. [Google Scholar] [CrossRef]
- Avagliano, C.; Coretti, L.; Lama, A.; Pirozzi, C.; De Caro, C.; De Biase, D.; Turco, L.; Mollica, M.P.; Paciello, O.; Calignano, A.; et al. Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice-Neuroprotective and Anti-Inflammatory Effects of Butyrate. Int. J. Mol. Sci. 2022, 23, 6367. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Gao, M.; Liu, Z.; Zhang, Y.; Tu, H.; Lei, L.; Wu, P.; Zhang, A.; Yang, C.; Li, G.; et al. Gut Microbiome Composition Linked to Inflammatory Factors and Cognitive Functions in First-Episode, Drug-Naive Major Depressive Disorder Patients. Front. Neurosci. 2021, 15, 800764. [Google Scholar] [CrossRef]
- Han, Y.; Xu, J.; Yan, Y.; Zhao, X. Dynamics of the gut microbiota in rats after hypobaric hypoxia exposure. PeerJ 2022, 10, e14090. [Google Scholar] [CrossRef]
- Kulkarni, P.; Devkumar, P.; Chattopadhyay, I. Could dysbiosis of inflammatory and anti-inflammatory gut bacteria have an implications in the development of type 2 diabetes? A pilot investigation. BMC Res. Notes 2021, 14, 52. [Google Scholar]
- Hou, Y.; Dong, L.; Lu, X.; Shi, H.; Xu, B.; Zhong, W.; Ma, L.; Wang, S.; Yang, C.; He, X.; et al. Distinctions Between Fecal and Intestinal Mucosal Microbiota in Subgroups of Irritable Bowel Syndrome. Dig. Dis. Sci. 2022, 67, 5580–5592. [Google Scholar] [CrossRef]
- Del Chierico, F.; Abbatini, F.; Russo, A.; Quagliariello, A.; Reddel, S.; Capoccia, D.; Caccamo, R.; Ginanni Corradini, S.; Nobili, V.; De Peppo, F.; et al. Gut Microbiota Markers in Obese Adolescent and Adult Patients: Age-Dependent Differential Patterns. Front. Microbiol. 2018, 9, 1210. [Google Scholar] [CrossRef]
- Song, C.H.; Kim, N.; Nam, R.H.; Choi, S.I.; Jang, J.Y.; Lee, H.N. Changes in Gut Microbiome upon Orchiectomy and Testosterone Administration in AOM/DSS-Induced Colon Cancer Mouse Model. Cancer Res. Treat. 2023, 55, 196–218. [Google Scholar] [CrossRef]
- Wagner, V.E.; Dey, N.; Guruge, J.; Hsiao, A.; Ahern, P.P.; Semenkovich, N.P.; Blanton, L.V.; Cheng, J.; Griffin, N.; Stappenbeck, T.S.; et al. Effects of a gut pathobiont in a gnotobiotic mouse model of childhood undernutrition. Sci. Transl. Med. 2016, 8, 366ra164. [Google Scholar] [CrossRef]
- Shen, W.D.; Lin, X.; Liu, H.M.; Li, B.Y.; Qiu, X.; Lv, W.Q.; Zhu, X.Z.; Greenbaum, J.; Liu, R.K.; Shen, J.; et al. Gut microbiota accelerates obesity in peri-/post-menopausal women via Bacteroides fragilis and acetic acid. Int. J. Obes. 2022, 46, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Cao, S.; Liu, S.; Yao, Z.; Sun, T.; Li, Y.; Li, J.; Zhang, D.; Zhou, Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget 2016, 7, 46158–46172. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Xu, F.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zhao, H.; Zheng, H. Changes in the Gut Microbiota of Children with Autism Spectrum Disorder. Autism Res. 2020, 13, 1614–1625. [Google Scholar] [CrossRef]
- Luna, R.A.; Foster, J.A. Gut brain axis: Diet microbiota interactions and implications for modulation of anxiety and depression. Curr. Opin. Biotechnol. 2015, 32, 35–41. [Google Scholar] [CrossRef]
- Schafer, D.F.; Jones, E.A. Hepatic encephalopathy and the gamma-aminobutyric-acid neurotransmitter system. Lancet 1982, 1, 18–20. [Google Scholar] [CrossRef]
- Ren, Y.; Hao, L.; Liu, J.; Wang, P.; Ding, Q.; Chen, C.; Song, Y. Alterations in the Gut Microbiota in Pregnant Women with Pregestational Type 2 Diabetes Mellitus. mSystems 2023, 8, e0114622. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Caldarelli, G.; Gili, T. Network analysis of a complex disease: The gut microbiota in the inflammatory bowel disease case. arXiv 2022, arXiv:220807763. [Google Scholar]
- Di Luccia, B.; Ahern, P.P.; Griffin, N.W.; Cheng, J.; Guruge, J.L.; Byrne, A.E.; Rodionov, D.A.; Leyn, S.A.; Osterman, A.L.; Ahmed, T.; et al. Combined Prebiotic and Microbial Intervention Improves Oral Cholera Vaccination Responses in a Mouse Model of Childhood Undernutrition. Cell Host Microbe 2020, 27, 899–908.e5. [Google Scholar] [CrossRef]
- West, K.A.; Yin, X.; Rutherford, E.M.; Wee, B.; Choi, J.; Chrisman, B.S.; Dunlap, K.L.; Hannibal, R.L.; Hartono, W.; Lin, M.; et al. Multi-angle meta-analysis of the gut microbiome in Autism Spectrum Disorder: A step toward understanding patient subgroups. Sci. Rep. 2022, 12, 17034. [Google Scholar] [CrossRef]
- Bi, M.; Feng, L.; He, J.; Liu, C.; Wang, Y.; Jiang, H.; Liu, S.-J. Emerging insights between gut microbiome dysbiosis and Parkinson’s disease: Pathogenic and clinical relevance. Ageing Res. Rev. 2022, 82, 101759. [Google Scholar] [CrossRef]
- Liu, W.; Du, Q.; Han, D.; Zhang, H. The Gut Microbiome and Obstructive Sleep Apnea Syndrome in Children. Sleep Med. 2022, 100, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ma, C.; Han, L.; Nawaz, M.; Gao, F.; Zhang, X.; Yu, P.; Zhao, C.; Li, L.; Zhou, A.; et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010, 61, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Franzosa, E.A.; Lloyd-Price, J.; McIver, L.J.; Schwager, R.; Poon, T.W.; Ananthakrishnan, A.N.; Andrews, E.; Barron, G.; Lake, K.; et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018, 3, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Miyata, N.; Takakura, S.; Yoshihara, K.; Asano, Y.; Kimura-Todani, T.; Yamashita, M.; Zhang, X.T.; Watanabe, N.; Mikami, K. The Gut Microbiome Derived from Anorexia Nervosa Patients Impairs Weight Gain and Behavioral Performance in Female Mice. Endocrinology 2019, 160, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Nicolucci, A.C.; Hume, M.P.; Martinez, I.; Mayengbam, S.; Walter, J.; Reimer, R.A. Prebiotics Reduce Body Fat and Alter Intestinal Microbiota in Children Who Are Overweight or with Obesity. Gastroenterology 2017, 153, 711–722. [Google Scholar] [CrossRef]
- Liang, S.; Sin, Z.Y.; Yu, J.; Zhao, S.; Xi, Z.; Bruzzone, R.; Tun, H.M. Multi-cohort analysis of depression-associated gut bacteria sheds insight on bacterial biomarkers across populations. Cell. Mol. Life Sci. 2022, 80, 9. [Google Scholar] [CrossRef]
- Qian, X.; Si, Q.; Lin, G.; Zhu, M.; Lu, J.; Zhang, H.; Wang, G.; Chen, W. Bifidobacterium adolescentis Is Effective in Relieving Type 2 Diabetes and May Be Related to Its Dominant Core Genome and Gut Microbiota Modulation Capacity. Nutrients 2022, 14, 2479. [Google Scholar] [CrossRef]
- Kowalska-Duplaga, K.; Gosiewski, T.; Kapusta, P.; Sroka-Oleksiak, A.; Wędrychowicz, A.; Pieczarkowski, S.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Fyderek, K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci. Rep. 2019, 9, 18880. [Google Scholar] [CrossRef]
- Iribarren, C.; Magnusson, M.K.; Vigsnaes, L.K.; Aziz, I.; Amundsen, I.D.; Suligoj, T.; Juge, N.; Patel, P.; Sapnara, M.; Johnsen, L.; et al. The Effects of Human Milk Oligosaccharides on Gut Microbiota, Metabolite Profiles and Host Mucosal Response in Patients with Irritable Bowel Syndrome. Nutrients 2021, 13, 3836. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Sikes, M.; Bruno-Barcena, J.M. The intestinal microbiota, gastrointestinal environment and colorectal cancer: A putative role for probiotics in prevention of colorectal cancer? Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G401–G424. [Google Scholar] [CrossRef]
- Mulle, J.G.; Sharp, W.G.; Cubells, J.F. The gut microbiome: A new frontier in autism research. Curr. Psychiatry Rep. 2013, 15, 337. [Google Scholar] [CrossRef] [PubMed]
- Alfonsetti, M.; Castelli, V.; d’Angelo, M. Are We What We Eat? Impact of Diet on the Gut–Brain Axis in Parkinson’s Disease. Nutrients 2022, 14, 380. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xie, J.P.; Deng, K.; Li, X.; Yuan, Y.; Xuan, Q.; Xie, J.; He, X.-M.; Wang, Q.; Li, J.-J.; et al. Prophylactic Effects of Bifidobacterium adolescentis on Anxiety and Depression-Like Phenotypes after Chronic Stress: A Role of the Gut Microbiota-Inflammation Axis. Front. Behav. Neurosci. 2019, 13, 126. [Google Scholar] [CrossRef]
- Bloom, P.P.; Donlan, J.; Torres Soto, M.; Daidone, M.; Hohmann, E.; Chung, R.T. Fecal microbiota transplant improves cognition in hepatic encephalopathy and its effect varies by donor and recipient. Hepatol. Commun. 2022, 6, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Suceveanu, A.I.; Stoian, A.P.; Parepa, I.; Voinea, C.; Hainarosie, R.; Manuc, D.; Nitipir, C.; Mazilu, L.; Suceveanu, A.P. Gut microbiota patterns in obese and type 2 diabetes (T2D) patients from romanian black sea coast region. Rev. Chim. 2018, 69, 2260–2267. [Google Scholar] [CrossRef]
- Lee, K.J.; Tack, J. Altered intestinal microbiota in irritable bowel syndrome. Neurogastroenterol. Motil. 2010, 22, 493–498. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, K.; Wu, W.; Lv, L.; Bian, X.; Yang, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; et al. Administration of Bifidobacterium bifidum CGMCC 15068 modulates gut microbiota and metabolome in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer (CAC) in mice. Appl. Microbiol. Biotechnol. 2020, 104, 5915–5928. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.; Park, S.J.; Park, G.; Shin, H.; Park, M.S.; Kim, J. Administration of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI Improves Cognitive and Memory Function in the Mouse Model of Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 709091. [Google Scholar] [CrossRef]
- Settanni, C.R.; Bibbò, S.; Ianiro, G.; Rinninella, E.; Cintoni, M.; Mele, M.C.; Cammarota, G.; Gasbarrini, A. Gastrointestinal involvement of autism spectrum disorder: Focus on gut microbiota. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 599–622. [Google Scholar] [CrossRef]
- Bullich, C.; Keshavarzian, A.; Garssen, J.; Kraneveld, A.; Perez-Pardo, P. Gut Vibes in Parkinson’s Disease: The Microbiota-Gut-Brain Axis. Mov. Disord. Clin. Pract. 2019, 6, 639–651. [Google Scholar] [CrossRef]
- Soverini, M.; Turroni, S.; Biagi, E.; Quercia, S.; Brigidi, P.; Candela, M.; Rampelli, S. Variation of Carbohydrate-Active Enzyme Patterns in the Gut Microbiota of Italian Healthy Subjects and Type 2 Diabetes Patients. Front. Microbiol. 2017, 8, 2079. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.P.; Schippa, S.; Zamboni, I.; Penta, M.; Chiarini, F.; Seganti, L.; Osborn, J.; Falconieri, P.; Borrelli, O.; Cucchiara, S. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006, 55, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Malinen, E.; Rinttila, T.; Kajander, K.; Matto, J.; Kassinen, A.; Krogius, L.; Saarela, M.; Korpela, R.; Palva, A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am. J. Gastroenterol. 2005, 100, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; White, J.R.; Padgett, R.N.; Perrotta, B.G.; Jenkins, G.D.; Chia, N.; Chen, J. Gut microbiome meta-analysis reveals dysbiosis is independent of body mass index in predicting risk of obesity-associated CRC. BMJ Open Gastroenterol. 2019, 6, e000247. [Google Scholar] [CrossRef]
- Ha, S.; Oh, D.; Lee, S.; Park, J.; Ahn, J.; Choi, S.; Cheon, K.-A. Altered Gut Microbiota in Korean Children with Autism Spectrum Disorders. Nutrients 2021, 13, 3300. [Google Scholar] [CrossRef]
- Bolliri, C.; Fontana, A.; Cereda, E.; Barichella, M.; Cilia, R.; Ferri, V.; Caronni, S.; Calandrella, D.; Morelli, L.; Pezzoli, G. Gut Microbiota in Monozygotic Twins Discordant for Parkinson’s Disease. Ann. Neurol. 2022, 92, 631–636. [Google Scholar] [CrossRef]
- Galley, J.D.; Mashburn-Warren, L.; Blalock, L.C.; Lauber, C.L.; Carroll, J.E.; Ross, K.M.; Hobel, C.; Coussons-Read, M.; Schetter, C.D.; Gur, T.L. Maternal anxiety, depression and stress affects offspring gut microbiome diversity and bifidobacterial abundances. Brain Behav. Immun. 2023, 107, 253–264. [Google Scholar] [CrossRef]
- Badran, M.; Khalyfa, A.; Ericsson, A.C.; Puech, C.; McAdams, Z.; Bender, S.B.; Gozal, D. Gut microbiota mediate vascular dysfunction in a murine model of sleep apnoea: Effect of probiotics. Eur. Respir. J. 2023, 61, 2200002. [Google Scholar] [CrossRef]
- Yao, S.; Zhao, Z.; Wang, W.; Liu, X. Bifidobacterium Longum: Protection against Inflammatory Bowel Disease. J. Immunol. Res. 2021, 2021, 8030297. [Google Scholar] [CrossRef]
- Fanigliulo, L.; Comparato, G.; Aragona, G.; Cavallaro, L.; Iori, V.; Maino, M.; Cavestro, G.M.; Soliani, P.; Sianesi, M.; Franzè, A.; et al. Role of gut microflora and probiotic effects in the irritable bowel syndrome. Acta Biomed. 2006, 77, 85–89. [Google Scholar]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; De Caro, C.; Comegna, M.; et al. Corrigendum: Gut Microbiota Features in Young Children with Autism Spectrum Disorders. Front. Microbiol. 2019, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Zou, R.; Song, L.; Zhang, X.; Jiang, B.; Wang, G.; Lee, Y.-K.; Zhao, J.; Zhang, H.; Chen, W. Ingestion of Bifidobacterium longum subspecies infantis strain CCFM687 regulated emotional behavior and the central BDNF pathway in chronic stress-induced depressive mice through reshaping the gut microbiota. Food Funct. 2019, 10, 7588–7598. [Google Scholar] [CrossRef] [PubMed]
- Malaguarnera, M.; Greco, F.; Barone, G.; Gargante, M.P.; Malaguarnera, M.; Toscano, M.A. Bifidobacterium longum with fructo-oligosaccharide (FOS) treatment in minimal hepatic encephalopathy: A randomized, double-blind, placebo-controlled study. Dig. Dis. Sci. 2007, 52, 3259–3265. [Google Scholar] [CrossRef]
- Olson, C.A.; Iniguez, A.J.; Yang, G.E.; Fang, P.; Pronovost, G.N.; Jameson, K.G.; Rendon, T.K.; Paramo, J.; Barlow, J.T.; Ismagilov, R.F.; et al. Alterations in the gut microbiota contribute to cognitive impairment induced by the ketogenic diet and hypoxia. Cell Host Microbe 2021, 29, 1378–1392. [Google Scholar] [CrossRef]
- Velizarova, M.; Yanachkova, V.; Boneva, T.; Giragosyan, S.; Mihaleva, I.; Andreeva-Gateva, P.; Svinarov, D.; Dimova, I. Relationship between Vitamin D status and microbiome changes in Bulgarian patients with type 2 diabetes mellitus. Biotechnol. Biotechnol. Equip. 2023, 37, 2209662. [Google Scholar] [CrossRef]
- Devkota, S.; Chang, E.B. Interactions between diet, bile acid metabolism, gut microbiota, and inflammatory bowel diseases. Dig. Dis. 2015, 33, 351–356. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Lagishetty, V.; Hauer, M.C.; Labus, J.S.; Dong, T.S.; Toma, R.; Vuyisich, M.; Naliboff, B.D.; Lackner, J.M.; Gupta, A.; et al. Multi-omics profiles of the intestinal microbiome in irritable bowel syndrome and its bowel habit subtypes. Microbiome 2023, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Jian, C.; Luukkonen, P.; Sadevirta, S.; Yki-Jarvinen, H.; Salonen, A. Impact of short-term overfeeding of saturated or unsaturated fat or sugars on the gut microbiota in relation to liver fat in obese and overweight adults. Clin. Nutr. 2021, 40, 207–216. [Google Scholar] [CrossRef]
- Verhaar, B.J.H.; Hendriksen, H.M.A.; de Leeuw, F.A.; Doorduijn, A.S.; van Leeuwenstijn, M.; Teunissen, C.E.; Barkhof, F.; Scheltens, P.; Kraaij, R.; van Duijn, C.M.; et al. Gut Microbiota Composition Is Related to AD Pathology. Front. Immunol. 2021, 12, 794519. [Google Scholar] [CrossRef]
- Kittana, M.; Ahmadani, A.; Al Marzooq, F.; Attlee, A. Dietary Fat Effect on the Gut Microbiome, and Its Role in the Modulation of Gastrointestinal Disorders in Children with Autism Spectrum Disorder. Nutrients 2021, 13, 3818. [Google Scholar] [CrossRef]
- Stevens, B.R.; Pepine, C.J.; Richards, E.M.; Kim, S.; Raizada, M.K. Depressive hypertension: A proposed human endotype of brain/gut microbiome dysbiosis. Am. Heart J. 2021, 239, 27–37. [Google Scholar] [CrossRef]
- Di Lodovico, L.; Mondot, S.; Dore, J.; Mack, I.; Hanachi, M.; Gorwood, P. Anorexia nervosa and gut microbiota: A systematic review and quantitative synthesis of pooled microbiological data. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 106, 110114. [Google Scholar] [CrossRef] [PubMed]
- Parnell, J.A.; Reimer, R.A. Prebiotic fiber modulation of the gut microbiota improves risk factors for obesity and the metabolic syndrome. Gut Microbes 2012, 3, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chang, Y.; Zheng, Q.; Zhang, R.; Hu, C.; Jia, W. Altered intestinal microbiota associated with colorectal cancer. Front. Med. 2019, 13, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Cordero, E.M.; Cuevas-Budhart, M.A.; Perez Moran, D.; Trejo Villeda, M.A.; Gomez-Del-Pulgar, G.f.-M.M. Relationship Between the Gut Microbiota and Alzheimer’s Disease: A Systematic Review. J. Alzheimers Dis. 2022, 87, 519–528. [Google Scholar] [CrossRef]
- Matarazzo, I.; Toniato, E.; Robuffo, I. Psychobiome Feeding Mind: Polyphenolics in Depression and Anxiety. Curr. Top. Med Chem. 2018, 18, 2108–2115. [Google Scholar] [CrossRef]
- Million, M.; Alou, M.T.; Khelaifia, S.; Bachar, D.; Lagier, J.C.; Dione, N.; Brah, S.; Hugon, P.; Lombard, V.; Armougom, F.; et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci. Rep. 2016, 6, 26051. [Google Scholar] [CrossRef]
- Leung, J.; Burke, B.; Ford, D.; Garvin, G.; Korn, C.; Sulis, C.; Bhadelia, N. Possible association between obesity and Clostridium difficile infection. Emerg. Infect. Dis. 2013, 19, 1791–1798. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Azimirad, M.; Alebouyeh, M.; Amoli, H.A.; Hosseini, P.; Ghasemian-Safaei, H.; Moghim, S. The rate and importance of Clostridium difficile in colorectal cancer patients. Gastroenterol. Hepatol. Bed Bench 2019, 12, 358–363. [Google Scholar]
- Toh, M.C.; Allen-Vercoe, E. The human gut microbiota with reference to autism spectrum disorder: Considering the whole as more than a sum of its parts. Microb. Ecol. Health Dis. 2015, 26, 26309. [Google Scholar] [CrossRef]
- Evrensel, A.; Ceylan, M.E. The Gut-Brain Axis: The Missing Link in Depression. Clin. Psychopharmacol. Neurosci. 2015, 13, 239–244. [Google Scholar] [CrossRef]
- Riordan, S.M.; Williams, R. Gut flora and hepatic encephalopathy in patients with cirrhosis. N. Engl. J. Med. 2010, 362, 1140–1142. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef] [PubMed]
- Maukonen, J.; Satokari, R.; Matto, J.; Soderlund, H.; Mattila-Sandholm, T.; Saarela, M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J. Med. Microbiol. 2006, 55 Pt 5, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Coelho, D.; Blachier, F. Review of the association between meat consumption and risk of colorectal cancer. Nutr. Res. 2013, 33, 983–994. [Google Scholar] [CrossRef]
- Kleessen, B.; Kroesen, A.J.; Buhr, H.J.; Blaut, M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 2002, 37, 1034–1041. [Google Scholar] [CrossRef]
- Palacios, T.; Coulson, S.; Butt, H.; Vitetta, L. The gastrointestinal microbiota and multi-strain probiotic therapy: In children and adolescent obesity. Adv. Integr. Med. 2014, 1, 2–8. [Google Scholar] [CrossRef]
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children with Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 218–230. [Google Scholar] [CrossRef]
- Karnachuk, O.V.; Ikkert, O.P.; Avakyan, M.R.; Knyazev, Y.V.; Volochaev, M.N.; Zyusman, V.S.; Panov, V.L.; Kadnikov, V.V.; Mardanov, A.V.; Ravin, N.V. Desulfovibrio desulfuricans AY5 Isolated from a Patient with Autism Spectrum Disorder Binds Iron in Low-Soluble Greigite and Pyrite. Microorganisms 2021, 9, 2558. [Google Scholar] [CrossRef]
- Wu, S.; Hammarstedt-Nordenvall, L.; Jangard, M.; Cheng, L.; Radu, S.A.; Angelidou, P.; Zha, Y.; Hamsten, M.; Engstrand, L.; Du, J.; et al. Tonsillar Microbiota: A Cross-Sectional Study of Patients with Chronic Tonsillitis or Tonsillar Hypertrophy. Msystems 2021, 6, e01302-20. [Google Scholar] [CrossRef] [PubMed]
- El-Salhy, M.; Casen, C.; Valeur, J.; Hausken, T.; Hatlebakk, J.G. Responses to faecal microbiota transplantation in female and male patients with irritable bowel syndrome. World J. Gastroenterol. 2021, 27, 2219–2237. [Google Scholar] [CrossRef] [PubMed]
- McLeod, A.; Penalver Bernabe, B.; Xia, Y.; Sanchez-Flack, J.; Lamar, M.; Schiffer, L.; Castellanos, K.; Fantuzzi, G.; Maki, P.; Fitzgibbon, M.; et al. Comparing the gut microbiome of obese, African American, older adults with and without mild cognitive impairment. PLoS ONE 2023, 18, e0280211. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.W.; Kim, M.J.; Kim, J.; Lee, H.G.; Ryoo, S.B.; Ku, J.L.; Jeong, S.Y.; Park, K.J.; Kim, D.; Kim, J.F.; et al. Enterotypical Prevotella and three novel bacterial biomarkers in preoperative stool predict the clinical outcome of colorectal cancer. Microbiome 2022, 10, 203. [Google Scholar] [CrossRef]
- Bosch, J.A.; Nieuwdorp, M.; Zwinderman, A.H.; Deschasaux, M.; Radjabzadeh, D.; Kraaij, R.; Davids, M.; de Rooij, S.R.; Lok, A. The gut microbiota and depressive symptoms across ethnic groups. Nat. Commun. 2022, 13, 7129. [Google Scholar] [CrossRef]
- Badran, M.; Mashaqi, S.; Gozal, D. The gut microbiome as a target for adjuvant therapy in obstructive sleep apnea. Expert Opin. Ther. Targets 2020, 24, 1263–1282. [Google Scholar] [CrossRef]
- Yu, S.; Ge, X.; Xu, H.; Tan, B.; Tian, B.; Shi, Y.; Dai, Y.; Li, Y.; Hu, S.; Qian, J. Gut microbiome and mycobiome in inflammatory bowel disease patients with Clostridioides difficile infection. Front. Cell. Infect. Microbiol. 2023, 13, 1129043. [Google Scholar] [CrossRef]
- Shin, S.Y.; Park, S.; Moon, J.M.; Kim, K.; Kim, J.W.; Chun, J.; Lee, T.H.; Choi, C.H.; The Microbiome Research Group of the Korean Society for Neurogastroenterology and Motility. Compositional Changes in the Gut Microbiota of Responders and Non-responders to Probiotic Treatment among Patients with Diarrhea-predominant Irritable Bowel Syndrome: A Post Hoc Analysis of a Randomized Clinical Trial. J. Neurogastroenterol. Motil. 2022, 28, 642–654. [Google Scholar] [CrossRef]
- Ondee, T.; Pongpirul, K.; Janchot, K.; Kanacharoen, S.; Lertmongkolaksorn, T.; Wongsaroj, L.; Somboonna, N.; Ngamwongsatit, N.; Leelahavanichkul, A. Lactiplantibacillus plantarum dfa1 Outperforms Enterococcus faecium dfa1 on Anti-Obesity in High Fat-Induced Obesity Mice Possibly through the Differences in Gut Dysbiosis Attenuation, despite the Similar Anti-Inflammatory Properties. Nutrients 2021, 14, 80. [Google Scholar] [CrossRef]
- Kovtun, A.S.; Averina, O.V.; Alekseeva, M.G.; Danilenko, V.N. Antibiotic resistance genes in the gut microbiota of children with autistic spectrum disorder as possible predictors of the disease. Microb. Drug Resist. 2020, 26, 1307–1320. [Google Scholar] [CrossRef]
- Loguercio, C.; Abbiati, R.; Rinaldi, M.; Romano, A.; Del Vecchio Blanco, C.; Coltorti, M. Long-term effects of Enterococcus faecium SF68 versus lactulose in the treatment of patients with cirrhosis and grade 1–2 hepatic encephalopathy. J. Hepatol. 1995, 23, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Munir, S.S.; Sert Kuniyoshi, F.H.; Singh, P.; Covassin, N. Is the Gut Microbiome Implicated in the Excess Risk of Hypertension Associated with Obstructive Sleep Apnea? A Contemporary Review. Antioxidants 2023, 12, 866. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-J.; Tsai, W.-C.; Hung, W.-C.; Hung, W.-W.; Chang, C.-C.; Dai, C.-Y.; Tsai, Y.-C. Gut microbiota and subclinical cardiovascular disease in patients with type 2 diabetes mellitus. Nutrients 2021, 13, 2679. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.M. The role of Escherichia coli in inflammatory bowel disease. Gut 2007, 56, 610–612. [Google Scholar] [CrossRef]
- Liu, H.N.; Wu, H.; Chen, Y.Z.; Chen, Y.J.; Shen, X.Z.; Liu, T.T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig. Liver Dis. 2017, 49, 331–337. [Google Scholar] [CrossRef]
- Veziant, J.; Villeger, R.; Barnich, N.; Bonnet, M. Gut Microbiota as Potential Biomarker and/or Therapeutic Target to Improve the Management of Cancer: Focus on Colibactin-Producing Escherichia coli in Colorectal Cancer. Cancers 2021, 13, 2215. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wei, J.; Zhao, X.; Liu, Y.; Chen, Z.; Wei, K.; Lu, J.; Chen, W.; Jiang, M.; Li, S.; et al. Neuroprotective effects of an engineered Escherichia coli Nissle 1917 on Parkinson’s disease in mice by delivering GLP-1 and modulating gut microbiota. Bioeng. Transl. Med. 2022, 8, e10351. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.-W.; Kim, J.-K.; Han, M.; Kim, D.-H. Lacticaseibacillus paracasei NK112 mitigates Escherichia coli-induced depression and cognitive impairment in mice by regulating IL-6 expression and gut microbiota. Benef. Microbes 2021, 12, 541–551. [Google Scholar] [CrossRef]
- Manzhalii, E.; Moyseyenko, V.; Kondratiuk, V.; Molochek, N.; Falalyeyeva, T.; Kobyliak, N. Effect of a specific Escherichia coli Nissle 1917 strain on minimal/mild hepatic encephalopathy treatment. World J. Hepatol. 2022, 14, 634–646. [Google Scholar] [CrossRef]
- Morales, E.J.D.C. Obstructive Sleep Apnea and the Gut Microbiome. Master’s Thesis, The University of Arizona, Tucson, AZ, USA, 2018. [Google Scholar]
- Ge, X.; Zhang, A.; Li, L.; Sun, Q.; He, J.; Wu, Y.; Tan, R.; Pan, Y.; Zhao, J.; Xu, Y.; et al. Application of machine learning tools: Potential and useful approach for the prediction of type 2 diabetes mellitus based on the gut microbiome profile. Exp. Ther. Med. 2022, 23, 305. [Google Scholar] [CrossRef]
- Balamurugan, R.; George, G.; Kabeerdoss, J.; Hepsiba, J.; Chandragunasekaran, A.M.; Ramakrishna, B.S. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br. J. Nutr. 2010, 103, 335–338. [Google Scholar] [CrossRef]
- Bojovic, K.; Ignjatovic Eth, I.; Sokovic Bajic, S.; Vojnovic Milutinovic, D.; Tomic, M.; Golic, N.; Tolinački, M. Gut Microbiota Dysbiosis Associated with Altered Production of Short Chain Fatty Acids in Children with Neurodevelopmental Disorders. Front. Cell. Infect. Microbiol. 2020, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lai, J.; Lu, H.; Ng, C.; Huang, T.; Zhang, H.; Ding, K.; Wang, Z.; Jiang, J.; Hu, J.; et al. Gut Microbiota in Bipolar Depression and Its Relationship to Brain Function: An Advanced Exploration. Front. Psychiatry 2019, 10, 784. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Chumpitazi, B.P.; Cope, J.L.; Hollister, E.B.; Tsai, C.M.; McMeans, A.R.; Luna, R.A.; Versalovic, J.; Shulman, R.J. Randomised clinical trial: Gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment. Pharmacol. Ther. 2015, 42, 418–427. [Google Scholar] [CrossRef]
- Ueda, A.; Shinkai, S.; Shiroma, H.; Taniguchi, Y.; Tsuchida, S.; Kariya, T.; Kawahara, T.; Kobayashi, Y.; Kohda, N.; Ushida, K.; et al. Identification of Faecalibacterium prausnitzii strains for gut microbiome-based intervention in Alzheimer’s-type dementia. Cell Rep. Med. 2021, 2, 100398. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.-H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Xu, H.; Fu, Z.; Wang, F.; Huang, W.; Wu, K.; Li, C.; Liu, Y.; Zou, J.; et al. Changes in the oral and nasal microbiota in pediatric obstructive sleep apnea. J. Oral Microbiol. 2023, 15, 2182571. [Google Scholar] [CrossRef]
- Sedighi, M.; Razavi, S.; Navab-Moghadam, F.; Khamseh, M.E.; Alaei-Shahmiri, F.; Mehrtash, A.; Amirmozafari, N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017, 111, 362–369. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, X.; Zhu, X.; Jiao, J.; Wu, Y.; Li, Y.; Zhao, L. Fusobacterium nucleatum aggravates ulcerative colitis through promoting gut microbiota dysbiosis and dysmetabolism. J. Periodontol. 2023, 94, 405–418. [Google Scholar] [CrossRef]
- Kushak, R.I.; Sengupta, A.; Winter, H.S. Interactions between the intestinal microbiota and epigenome in individuals with autism spectrum disorder. Dev. Med. Child Neurol. 2022, 64, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Kountouras, J.; Polyzos, S.A.; Deretzi, G. Helicobacter pylori associated with obstructive sleep apnea might contribute to sleep, cognition, and driving performance disturbances in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2015, 13, 1547. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.S.; Mohammed, M.H.; Ghosh, T.S.; Kanungo, S.; Nair, G.B.; Mande, S.S. Metagenome of the gut of a malnourished child. Gut Pathog. 2011, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Lender, N.; Talley, N.J.; Enck, P.; Haag, S.; Zipfel, S.; Morrison, M.; Holtmann, G.J. Review article: Associations between Helicobacter pylori and obesity—An ecological study. Aliment. Pharmacol. Ther. 2014, 40, 24–31. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Ahmadi Amoli, H.; Ghasemian-Safaei, H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol. Hepatol. Bed Bench 2018, 11, 101–109. [Google Scholar]
- Elfil, M.; Kamel, S.; Kandil, M.; Koo, B.B.; Schaefer, S.M. Implications of the Gut Microbiome in Parkinson’s Disease. Mov. Disord. 2020, 35, 921–933. [Google Scholar] [CrossRef]
- Mobini, R.; Tremaroli, V.; Ståhlman, M.; Karlsson, F.; Levin, M.; Ljungberg, M.; Sohlin, M.; Bertéus Forslund, H.; Perkins, R.; Bäckhed, F. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: A randomized controlled trial. Diabetes Obes. Metab. 2017, 19, 579–589. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, C.; Huang, J.; Kuai, X.; Shao, X. The potential therapeutic role of Lactobacillus reuteri for treatment of inflammatory bowel disease. Am. J. Transl. Res. 2020, 12, 1569–1583. [Google Scholar]
- Niv, E.; Naftali, T.; Hallak, R.; Vaisman, N. The efficacy of Lactobacillus reuteri ATCC 55730 in the treatment of patients with irritable bowel syndrome—A double blind, placebo-controlled, randomized study. Clin. Nutr. 2005, 24, 925–931. [Google Scholar] [CrossRef]
- Bell, H.N.; Rebernick, R.J.; Goyert, J.; Singhal, R.; Kuljanin, M.; Kerk, S.A.; Huang, W.; Das, N.K.; Andren, A.; Solanki, S.; et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 2022, 40, 185–200.e6. [Google Scholar] [CrossRef]
- Liu, G.; Tan, F.H.; Lau, S.A.; Jaafar, M.H.; Chung, F.Y.; Azzam, G.; Liong, M.; Li, Y. Lactic acid bacteria feeding reversed the malformed eye structures and ameliorated gut microbiota profiles of Drosophila melanogaster Alzheimer’s disease model. J. Appl. Microbiol. 2022, 132, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259.e6. [Google Scholar] [CrossRef]
- Du, Y.; Li, Y.; Xu, X.; Li, R.; Zhang, M.; Cui, Y.; Zhang, L.; Wei, Z.; Wang, S.; Tuo, H. Probiotics for constipation and gut microbiota in Parkinson’s disease. Park. Relat. Disord. 2022, 103, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, R.; Zhang, S.; Zhang, X.; Zhou, H.; Wen, T.; Wang, J. Depression-like symptoms due to Dcf1 deficiency are alleviated by intestinal transplantation of Lactobacillus murine and Lactobacillus reuteri. Biochem. Biophys. Res. Commun. 2022, 593, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Flores, R.; Morán-Villota, S.; Cervantes-Barragán, L.; López-Macias, C.; Uribe, M. Manipulation of microbiota with probiotics as an alternative for treatment of hepatic encephalopathy. Nutrition 2020, 73, 110693. [Google Scholar] [CrossRef]
- Zhang, W.-Y.; Zhang, H.-H.; Yu, C.-H.; Fang, J.; Ying, H.-Z. Ethanol extract of Atractylodis macrocephalae Rhizoma ameliorates insulin resistance and gut microbiota in type 2 diabetic db/db mice. J. Funct. Foods 2017, 39, 139–151. [Google Scholar] [CrossRef]
- Ghoshal, U.; Shukla, R.; Srivastava, D.; Ghoshal, U.C. Irritable Bowel Syndrome, Particularly the Constipation-Predominant Form, Involves an Increase in Methanobrevibacter smithii, Which Is Associated with Higher Methane Production. Gut Liver 2016, 10, 932–938. [Google Scholar] [CrossRef]
- Tidjani Alou, M.; Million, M.; Traore, S.I.; Mouelhi, D.; Khelaifia, S.; Bachar, D.; Caputo, A.; Delerce, J.; Brah, S.; Alhousseini, D.; et al. Gut Bacteria Missing in Severe Acute Malnutrition, Can We Identify Potential Probiotics by Culturomics? Front. Microbiol. 2017, 8, 899. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, S.; Li, S.; Zhang, Q.; Cai, Y.; Li, P.; Li, H.; Shen, B.; Liao, Q.; Hong, Y.; et al. Indoleacrylic acid produced by Parabacteroides distasonis alleviates type 2 diabetes via activation of AhR to repair intestinal barrier. BMC Biol. 2023, 21, 90. [Google Scholar] [CrossRef]
- Herndon, C.C.; Wang, Y.P.; Lu, C.L. Targeting the gut microbiota for the treatment of irritable bowel syndrome. Kaohsiung J. Med. Sci. 2020, 36, 160–170. [Google Scholar] [CrossRef]
- Song, H.; Shen, X.; Chu, Q.; Zheng, X. Pomegranate fruit pulp polyphenols reduce diet-induced obesity with modulation of gut microbiota in mice. J. Sci. Food Agric. 2022, 102, 1968–1977. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Li, X.; Wang, M.; Zhang, Z.; Qiao, L.; Gao, Y.; Xu, X.; Zhi, J.; Li, Y.; Li, Z.; et al. Gut Microbiota Alteration Influences Colorectal Cancer Metastasis to the Liver by Remodeling the Liver Immune Microenvironment. Gut Liver 2022, 16, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.V.; Kovtun, A.S.; Polyakova, S.I.; Savilova, A.M.; Rebrikov, D.V.; Danilenko, V.N. The bacterial neurometabolic signature of the gut microbiota of young children with autism spectrum disorders. J. Med. Microbiol. 2020, 69, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, X.; Xu, X.; Ming, J.; Wang, Z.; Gao, B.; Xing, Y.; Zhou, J.; Fu, J.; Liu, T.; et al. The fecal microbiota is already altered in normoglycemic individuals who go on to have type 2 diabetes. Front. Cell. Infect. Microbiol. 2021, 11, 598672. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Koido, S. Intestinal microbiota and ulcerative colitis. J. Infect. Chemother. 2015, 21, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Goll, R.; Johnsen, P.H.; Hjerde, E.; Diab, J.; Valle, P.C.; Hilpusch, F.; Cavanagh, J.P. Effects of fecal microbiota transplantation in subjects with irritable bowel syndrome are mirrored by changes in gut microbiome. Gut Microbes 2020, 12, 1794263. [Google Scholar] [CrossRef] [PubMed]
- Dhopatkar, N.; Keeler, J.L.; Mutwalli, H.; Whelan, K.; Treasure, J.; Himmerich, H. Gastrointestinal symptoms, gut microbiome, probiotics and prebiotics in anorexia nervosa: A review of mechanistic rationale and clinical evidence. Psychoneuroendocrinology 2023, 147, 105959. [Google Scholar] [CrossRef]
- Liu, W.; Fang, X.; Zhou, Y.; Dou, L.; Dou, T. Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 2022, 24, 104892. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, N.; Park, J.H.; Kim, Y.S.; Lee, J.; Kim, H.W.; Choi, Y.J.; Shin, C.M.; Park, Y.S.; Lee, D.H.; et al. Comparisons of Gut Microbiota among Healthy Control, Patients with Conventional Adenoma, Sessile Serrated Adenoma, and Colorectal Cancer. J. Cancer Prev. 2017, 22, 108–114. [Google Scholar] [CrossRef]
- Kong, X.; Liu, J.; Cetinbas, M.; Sadreyev, R.; Koh, M.; Huang, H.; Adeseye, A.; He, P.; Zhu, J.; Russell, H.; et al. New and Preliminary Evidence on Altered Oral and Gut Microbiota in Individuals with Autism Spectrum Disorder (ASD): Implications for ASD Diagnosis and Subtyping Based on Microbial Biomarkers. Nutrients 2019, 11, 2128. [Google Scholar] [CrossRef]
- Kovtun, A.S.; Averina, O.V.; Angelova, I.Y.; Yunes, R.A.; Zorkina, Y.A.; Morozova, A.Y.; Pavlichenko, A.V.; Syunyakov, T.S.; Karpenko, O.A.; Kostyuk, G.P.; et al. Alterations of the Composition and Neurometabolic Profile of Human Gut Microbiota in Major Depressive Disorder. Biomedicines 2022, 10, 2162. [Google Scholar] [CrossRef] [PubMed]
- Alsharairi, N.A. The Therapeutic Role of Short-Chain Fatty Acids Mediated Very Low-Calorie Ketogenic Diet-Gut Microbiota Relationships in Paediatric Inflammatory Bowel Diseases. Nutrients 2022, 14, 4113. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.; Alifirova, V.; Saltykova, I.; Zhukova, I.; Zhukova, N.; Dorofeeva, Y.B.; Tyakht, A.V.; Altukhov, I.A.; Kostryukova, E.S.; Titova, M.A.; et al. Comparison study of gut microbiota in case of Parkinson’s disease and other neurological disorders. Bull. Sib. Med. 2017, 15, 113–125. [Google Scholar] [CrossRef]
- Bakir-Gungor, B.; Hacilar, H.; Jabeer, A.; Nalbantoglu, O.U.; Aran, O.; Yousef, M. Inflammatory bowel disease biomarkers of human gut microbiota selected via different feature selection methods. PeerJ 2022, 10, e13205. [Google Scholar] [CrossRef]
- Lyra, A.; Rinttila, T.; Nikkila, J.; Krogius-Kurikka, L.; Kajander, K.; Malinen, E.; Mättö, J.; Mäkelä, L.; Palva, A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J. Gastroenterol. 2009, 15, 5936–5945. [Google Scholar] [CrossRef]
- Wang, Y.; Lim, Y.Y.; He, Z.; Wong, W.T.; Lai, W.F. Dietary phytochemicals that influence gut microbiota: Roles and actions as anti-Alzheimer agents. Crit. Rev. Food Sci. Nutr. 2022, 62, 5140–5166. [Google Scholar] [CrossRef]
- Wan, Y.; Zuo, T.; Xu, Z.; Zhang, F.; Zhan, H.; Dorothy, C.; Leung, T.-F.; Yeoh, Y.K.; Chan, F.K.L.; Chan, R.; et al. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 2022, 71, 910–918. [Google Scholar] [CrossRef]
- Kwak, M.S.; Cha, J.M.; Shin, H.P.; Jeon, J.W.; Yoon, J.Y. Development of a Novel Metagenomic Biomarker for Prediction of Upper Gastrointestinal Tract Involvement in Patients with Crohn’s Disease. Front. Microbiol. 2020, 11, 1162. [Google Scholar] [CrossRef]
- Hynonen, U.; Rasinkangas, P.; Satokari, R.; Paulin, L.; de Vos, W.M.; Pietilä, T.E.; Kant, R.; Palva, A. Isolation and whole genome sequencing of a Ruminococcus-like bacterium, associated with irritable bowel syndrome. Anaerobe 2016, 39, 60–67. [Google Scholar] [CrossRef]
- Togo, A.H.; Diop, A.; Bittar, F.; Maraninchi, M.; Valero, R.; Armstrong, N.; Dubourg, G.; Labas, N.; Richez, M.; Delerce, J.; et al. Correction to: Description of Mediterraneibacter massiliensis, gen. nov., sp. nov., a new genus isolated from the gut microbiota of an obese patient and reclassification of Ruminococcus faecis, Ruminococcus lactaris, Ruminococcus torques, Ruminococcus gnavus and Clostridium glycyrrhizinilyticum as Mediterraneibacter faecis comb. nov., Mediterraneibacter lactaris comb. nov., Mediterraneibacter torques comb. nov., Mediterraneibacter gnavus comb. nov. and Mediterraneibacter glycyrrhizinilyticus comb. nov. Antonie Van Leeuwenhoek 2018, 111, 2129–2130. [Google Scholar]
- Matto, J.; Maunuksela, L.; Kajander, K.; Palva, A.; Korpela, R.; Kassinen, A.; Saarela, M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome—A longitudinal study in IBS and control subjects. FEMS Immunol. Med. Microbiol. 2005, 43, 213–222. [Google Scholar] [CrossRef]
- Han, K.; Ji, L.; Wang, C.; Shao, Y.; Chen, C.; Liu, L.; Feng, M.; Yang, F.; Wu, X.; Li, X.; et al. The host genetics affects gut microbiome diversity in Chinese depressed patients. Front. Genet. 2022, 13, 976814. [Google Scholar] [CrossRef] [PubMed]
- Yukawa-Muto, Y.; Kamiya, T.; Fujii, H.; Mori, H.; Toyoda, A.; Sato, I.; Konishi, Y.; Hirayama, A.; Hara, E.; Fukuda, S.; et al. Distinct responsiveness to rifaximin in patients with hepatic encephalopathy depends on functional gut microbial species. Hepatol. Commun. 2022, 6, 2090–2104. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.Y.; Yao, F.; Zhou, C.R.; Huang, X.Y.; Wang, Q.; Long, H.; Wu, Q.M. Role of gut microbiome in regulating the effectiveness of metformin in reducing colorectal cancer in type 2 diabetes. World J. Clin. Cases 2020, 8, 6213–6228. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Beatty, J.K.; Bhargava, A.; Buret, A.G. Post-infectious irritable bowel syndrome: Mechanistic insights into chronic disturbances following enteric infection. World J. Gastroenterol. 2014, 20, 3976–3985. [Google Scholar] [CrossRef]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef]
- Mehal, W.Z. The Gordian Knot of dysbiosis, obesity and NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 637–644. [Google Scholar] [CrossRef]
- Gao, R.; Gao, Z.; Huang, L.; Qin, H. Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 757–769. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Saidara, E.; Kharrazian, D. Reaction of Amyloid-beta Peptide Antibody with Different Infectious Agents Involved in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 847–860. [Google Scholar] [CrossRef]
- Mehta, K.K.; Bhat, R.; Markande, A.R. Effects of gut bacteria and their amyloids on mental health and neurodegeneration in Parkinson’s disease. J. Appl. Biol. Biotechnol. 2023, 11, 38–46. [Google Scholar] [CrossRef]
- Strugala, G.J.; Rauws, A.G.; Elbers, R. Intestinal first pass metabolism of amygdalin in the rat in vitro. Biochem. Pharmacol. 1986, 35, 2123–2128. [Google Scholar] [CrossRef] [PubMed]
- Manhardt, M.A. Correlation of Injected Amygdalin and Urinary Thiocyanate in JAX C57 BL/KsJ Mice. Master’s Thesis, Loyola University Chicago, Chicago, IL, USA, 1981. [Google Scholar]
- Niimura, T.; Tokieda, T.; Yamaha, T. Partial purification and some properties of cyclamate sulfamatase. J. Biochem. 1974, 75, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.; Allison, C. Utilisation of protein by human gut bacteria. FEMS Microbiol. Ecol. 1986, 2, 19–24. [Google Scholar] [CrossRef]
- Macfarlane, G.; Cummings, J.; Allison, C. Protein degradation by human intestinal bacteria. Microbiology 1986, 132, 1647–1656. [Google Scholar] [CrossRef]
- Elshahed, M.S.; Miron, A.; Aprotosoaie, A.C.; Farag, M.A. Pectin in diet: Interactions with the human microbiome, role in gut homeostasis, and nutrient-drug interactions. Carbohydr. Polym. 2021, 255, 117388. [Google Scholar] [CrossRef]
- Romano, K.A.; Vivas, E.I.; Amador-Noguez, D.; Rey, F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 2015, 6, e02481-14. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Yang, M.; Ma, J.; Ruan, J.; Ye, Y.; Fu, P.P.-C.; Lin, G. Intestinal and hepatic biotransformation of pyrrolizidine alkaloid N-oxides to toxic pyrrolizidine alkaloids. Arch. Toxicol. 2019, 93, 2197–2209. [Google Scholar] [CrossRef]
- Widjaja, F.; Wesseling, S.; Rietjens, I.M.C.M. Physiologically based kinetic modelling predicts the in vivo relative potency of riddelliine N-oxide compared to riddelliine in rat to be dose dependent. Arch. Toxicol. 2022, 96, 135–151. [Google Scholar] [CrossRef]
- Juste, C.; Gérard, P. Cholesterol-to-coprostanol conversion by the gut microbiota: What we know, suspect, and ignore. Microorganisms 2021, 9, 1881. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.; Chandrasekaran, A.; Reuning, R.; Hui, J.; Rawal, B. Reduction of digoxin to 20R-dihydrodigoxin by cultures of Eubacterium lentum. Appl. Environ. Microbiol. 1986, 51, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Heinze, T.M.; Chen, S.; Cerniglia, C.E.; Chen, H. Anaerobic metabolism of 1-amino-2-naphthol-based azo dyes (Sudan dyes) by human intestinal microflora. Appl. Environ. Microbiol. 2007, 73, 7759–7762. [Google Scholar] [CrossRef] [PubMed]
- Gingell, R.; Bridges, J.; Williams, R. The role of the gut flora in the metabolism of prontosil and neoprontosil in the rat. Xenobiotica 1971, 1, 143–156. [Google Scholar] [CrossRef]
- Klotz, U.; Maier, K.; Fischer, C.; Heinkel, K. Therapeutic efficacy of sulfasalazine and its metabolites in patients with ulcerative colitis and Crohn’s disease. N. Engl. J. Med. 1980, 303, 1499–1502. [Google Scholar] [CrossRef] [PubMed]
- Strong, H.; Renwick, A.; George, C.; Liu, Y.; Hill, M. The reduction of sulphinpyrazone and sulindac by intestinal bacteria. Xenobiotica 1987, 17, 685–696. [Google Scholar] [CrossRef]
- Xin, Z.; Fengwei, T.; Gang, W.; Xiaoming, L.; Qiuxiang, Z.; Hao, Z.; Wei, C. Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiol. Ecol. 2012, 82, 703–712. [Google Scholar] [CrossRef]
- LinWu, S.-W.; Syu, C.-J.; Chen, Y.-L.; Wang, A.H.-J.; Peng, F.-C. Characterization of Escherichia coli nitroreductase NfsB in the metabolism of nitrobenzodiazepines. Biochem. Pharmacol. 2009, 78, 96–103. [Google Scholar] [CrossRef]
- Feng, R.; Shou, J.-W.; Zhao, Z.-X.; He, C.-Y.; Ma, C.; Huang, M.; Fu, J.; Tan, X.-S.; Li, X.-Y.; Wen, B.-Y. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci. Rep. 2015, 5, 12155. [Google Scholar] [CrossRef]
- Jourova, L.; Anzenbacher, P.; Matuskova, Z.; Vecera, R.; Strojil, J.; Kolar, M.; Nobilis, M.; Hermanova, P.; Hudcovic, T.; Kozakova, H. Gut microbiota metabolizes nabumetone in vitro: Consequences for its bioavailability in vivo in the rodents with altered gut microbiome. Xenobiotica 2019, 49, 1296–1302. [Google Scholar] [CrossRef]
- Yan, A.; Culp, E.; Perry, J.; Lau, J.T.; MacNeil, L.T.; Surette, M.G.; Wright, G.D. Transformation of the anticancer drug doxorubicin in the human gut microbiome. ACS Infect. Dis. 2018, 4, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Catala, D.M.; Spenkelink, A.; Rietjens, I.M.; Beekmann, K. An in vitromodel to quantify interspecies differences in kinetics for intestinal microbial bioactivation and detoxification of zearalenone. Toxicol. Rep. 2020, 7, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, V.; Kostiuk, B.; Unterweger, D.; Diaz-Satizabal, L.; Ogg, S.; Pukatzki, S. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 2015, 9, e0004031. [Google Scholar] [CrossRef] [PubMed]
- Deloménie, C.; Fouix, S.; Longuemaux, S.; Brahimi, N.m.; Bizet, C.; Picard, B.; Denamur, E.; Dupret, J.-M. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: Evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol. 2001, 183, 3417–3427. [Google Scholar] [CrossRef]
- Li, F.; Jin, J.; Rietjens, I.M.; Xing, F. Interindividual differences in in vitro human intestinal microbial conversion of 3-acetyl-DON and 15-acetyl-DON. Toxins 2022, 14, 199. [Google Scholar] [CrossRef]
- Humblot, C.; Murkovic, M.; Rigottier-Gois, L.; Bensaada, M.; Bouclet, A.; Andrieux, C.; Anba, J.; Rabot, S. β-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo [4, 5-f] quinoline in rats. Carcinogenesis 2007, 28, 2419–2425. [Google Scholar] [CrossRef]
- van Kessel, S.P.; Frye, A.K.; El-Gendy, A.O.; Castejon, M.; Keshavarzian, A.; van Dijk, G.; El Aidy, S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat. Commun. 2019, 10, 310. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol-and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef]
- García-Villalba, R.; Beltrán, D.; Espín, J.C.; Selma, M.V.; Tomás-Barberán, F.A. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agric. Food Chem. 2013, 61, 8797–8806. [Google Scholar] [CrossRef]
- Goldin, B.R.; Peppercorn, M.A.; Goldman, P. Contributions of host and intestinal microflora in the metabolism of L-dopa by the rat. J. Pharmacol. Exp. Ther. 1973, 186, 160–166. [Google Scholar]
- Clavel, T.; Henderson, G.; Engst, W.; Dore, J.; Blaut, M. Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol. Ecol. 2006, 55, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mao, B.; Cui, S.; Zhang, Q.; Zhao, J.; Tang, X.; Chen, W. Absorption, metabolism, bioactivity, and biotransformation of epigallocatechin gallate. Crit. Rev. Food Sci. Nutr. 2023, 1–21. [Google Scholar] [CrossRef]
- Malet-Martino, M.-C.; Martino, R.; De Forni, M.; Andremont, A.; Hartmann, O.; Armand, J. Flucytosine conversion to fluorouracil in humans: Does a correlation with gut flora status exist? A report of two cases using fluorine-19 magnetic resonance spectroscopy. Infection 1991, 19, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; de Bruijn, W.J.; Bruins, M.E.; Vincken, J.-P. Reciprocal interactions between epigallocatechin-3-gallate (EGCG) and human gut microbiota in vitro. J. Agric. Food Chem. 2020, 68, 9804–9815. [Google Scholar] [CrossRef] [PubMed]
- Espín, J.C.; González-Sarrías, A.; Tomás-Barberán, F.A. The gut microbiota: A key factor in the therapeutic effects of (poly) phenols. Biochem. Pharmacol. 2017, 139, 82–93. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, A.; Xie, G.; Chi, Y.; Zhao, L.; Li, H.; Wang, C.; Bao, Y.; Jia, W.; Luther, M. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci. Transl. Med. 2013, 5, 172ra122. [Google Scholar] [CrossRef]
- Oku, T.; Nakamura, S. Digestion, absorption, fermentation, and metabolism of functional sugar substitutes and their available energy. Pure Appl. Chem. 2002, 74, 1253–1261. [Google Scholar] [CrossRef]
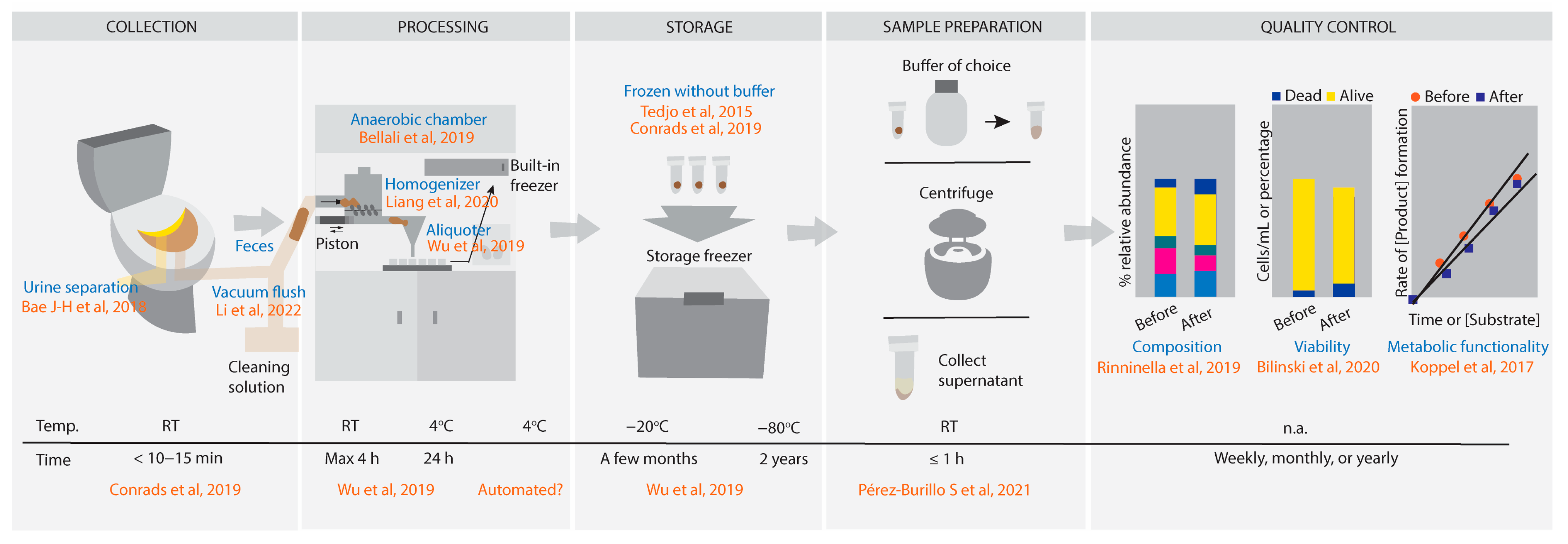
| Processing | |||
|---|---|---|---|
| Temperature | Time | Remarks | Reference |
| RT | 3 h | After defecation. Stability not indicated. | [62] |
| 20 ± 2 °C | <24 h | Non-significant effects at <24 h, but deletrious effects above this temperature. | [69] |
| >20 °C | >24 h | Samples should not be exposed to >20 °C if time from transformation to transplants exceeds 24 h. | |
| 37 °C | >24 h | Extinction and proliferation of genus specific to each individual. Metabolomic fingerprints deviates even more as time increases. | |
| RT | 4 h | After stool sample collection, it is acceptable for samples to reach the lab in this condition without preservation [54,78,79]. | [22] |
| 4 °C | 24 to 48 h | Keep the sample in cold storage when transported from participant to laboratory staff handling. | |
| RT (with stabilizer) | Up to 4 weeks | Stabilizers such as RNALater, 95% ethanol, Omnigene-Gut (DNA Genotek, Stittsville, Ottawa, ON, Canada) and FTA card (Sigma-Aldrich, St. Louis, MO, USA) can protect stool DNA from degradation [80]. | |
| 4 °C (without stabilizer) | Up to a week | Fecal materials are processed without stabilizer [80]. | |
| RT | Short-term | Short-term RT storage showed minimal effect on microbiome and metabolome profiles. | [55] |
| Storage | |||
| Temperature | Time | Remarks | Reference |
| −20 °C | Until further use | Storage condition. Stability not indicated. | [62] |
| 4 °C | Short-term | Ideal for short shipment. | [69] |
| −80 °C then rapid thaw at 37 °C | 3 months | Stable fecal sample transplants in maltodextrin–trehalose solutions within this observation period. | |
| RT | Greater than 15 min | Storage with or without buffer showed considerable divergence compared to −80 °C samples. | [74] |
| 4 °C | 24 h | Does not alter microbiota compared to −80 °C samples. | |
| −20 °C | 72 h | Does not alter microbiota compared to −80 °C samples. | |
| −80 °C | n.a. | Freezing significantly decreases overall viability to around 25% but does not significantly change viable microbiota composition compared to fresh anaerobically processed specimen. | [50] |
| −80 °C (with stabilizer) | 5 years | Long term stool storage with RNAlater had only limited effects on human fecal microbiota composition. | [71] |
| −20 to −80 °C | As soon as possible | Once samples reach the laboratory, they should be stored in this condition. Stability not indicated. | [22] |
| −20 °C | A few months | Stable composition in microbial community. | |
| −80 °C | 2 years to long-term storage | Stable composition in microbial community. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Widjaja, F.; Rietjens, I.M.C.M. From-Toilet-to-Freezer: A Review on Requirements for an Automatic Protocol to Collect and Store Human Fecal Samples for Research Purposes. Biomedicines 2023, 11, 2658. https://doi.org/10.3390/biomedicines11102658
Widjaja F, Rietjens IMCM. From-Toilet-to-Freezer: A Review on Requirements for an Automatic Protocol to Collect and Store Human Fecal Samples for Research Purposes. Biomedicines. 2023; 11(10):2658. https://doi.org/10.3390/biomedicines11102658
Chicago/Turabian StyleWidjaja, Frances, and Ivonne M. C. M. Rietjens. 2023. "From-Toilet-to-Freezer: A Review on Requirements for an Automatic Protocol to Collect and Store Human Fecal Samples for Research Purposes" Biomedicines 11, no. 10: 2658. https://doi.org/10.3390/biomedicines11102658
APA StyleWidjaja, F., & Rietjens, I. M. C. M. (2023). From-Toilet-to-Freezer: A Review on Requirements for an Automatic Protocol to Collect and Store Human Fecal Samples for Research Purposes. Biomedicines, 11(10), 2658. https://doi.org/10.3390/biomedicines11102658