Abstract
The dysregulated expression of cyclin genes can lead to the uncontrolled proliferation of cancer cells. Histone demethylase Jumonji-C domain-containing protein 5 (KDM8, JMJD5) and cyclin A1 (CCNA1) are pivotal in cell cycle progression. A promising candidate for augmenting cancer treatment is Allyl isothiocyanate (AITC), a natural dietary chemotherapeutic and epigenetic modulator. This study aimed to investigate AITC’s impact on the KDM8/CCNA1 axis to elucidate its role in oral squamous cell carcinoma (OSCC) tumorigenesis. The expression of KDM8 and CCNA1 was assessed using a tissue microarray (TMA) immunohistochemistry (IHC) assay. In vitro experiments with OSCC cell lines and in vivo experiments with patient-derived tumor xenograft (PDTX) and SAS subcutaneous xenograft tumor models were conducted to explore AITC’s effects on their expression and cell proliferation. The results showed elevated KDM8 and CCNA1 levels in the OSCC patient samples. AITC exhibited inhibitory effects on OSCC tumor growth in vitro and in vivo. Additionally, AITC downregulated KDM8 and CCNA1 expression while inducing histone H3K36me2 expression in oral cancer cells. These findings underscore AITC’s remarkable anticancer properties against oral cancer, highlighting its potential as a therapeutic option for oral cancer treatment by disrupting the cell cycle by targeting the KDM8/CCNA1 axis.
1. Introduction
Oral squamous cell carcinoma (OSCC) is a type of cancer affecting the head and neck, which has a rising trend of incidence in the world [1,2]. The development of OSCC is frequently linked to various risk factors, including but not limited to smoking, alcohol consumption, chewing habits, and infection with high-risk human papillomavirus [1,2]. Despite the considerable benefits observed in surgical procedures, radiation therapy, and chemotherapy, the 5-year survival rate for patients with OSCC remains below approximately 50% [3]. As a result, the identification of new prognostic factors and treatments is of utmost importance to impede the progression of OSCC. Due to their safety, natural compounds have recently become a focal point in the quest for anticancer drugs as potential alternatives [4]. Previous studies have indicated that CCNA1 is a cyclin-dependent protein kinase known as a serine/threonine kinase, predominantly found in the testis. It belongs to the highly conserved cyclin family, which shows periodic fluctuations in abundance during the cell cycle. In the eukaryotic cell cycle, the CCNA1 protein acts as a regulatory subunit for cyclin-dependent kinases (CDKs). CCNA1 activates CDK2 by binding specifically to it, leading to the phosphorylation of several target proteins that facilitate the progression through the G1/S and G2/M phases of the cell cycle [5,6,7]. CCNA1 plays a crucial role in regulating cell cycle progression, and its suppression in leukemia cells has been shown to impede cell growth [6]. The overexpression of CCNA1 has been linked to unfavorable prognoses in various cancers, including bladder urothelial carcinomas [8], esophageal squamous cell carcinoma [9], as well as head and neck cancer [10,11,12].
The aberration of chromatin modifications of histone tails leads to carcinogenesis [13]. Jumonji C domains protein families have been identified as major contributors to various human cancers via epigenetic remodeling [14,15]. Jumonji-C domain-containing protein 5 (JMJD5), renamed KDM8, is involved in embryonic development [16], the metabolic regulator [17], osteoclastogenesis [18], circadian rhythm regulation [19], and tumorigenesis [20]. KDM8 functions as a histone demethylase that specifically removes methyl groups from lysine 36 on histone H3 (H3K36), resulting in the modulation of gene expression. KDM8 is highly expressed in various types of cancer such as breast, lung, stomach, prostate, colon, and oral cancers [20,21,22,23,24,25,26,27]. The upregulation of KDM8 in these cancers has been linked with enhanced cell proliferation, migration, and invasion, which indicates its involvement in tumor progression. Furthermore, a research study found that the inhibition of KDM8 can impede metastasis and prompt apoptosis in oral squamous cell carcinoma through the regulation of the p53/NF-κB pathway [20]. In our previous report, we demonstrated a correlation between the high expression of KDM8 and an unfavorable prognosis in OSCC, and the downregulation of JMJD5 could inhibit proliferation in the oral cancer preclinic model [27]. Moreover, KDM8 has been revealed to positively regulate CCNA1 in cancer cell proliferation [13]. KDM8 and CCNA1 may have a function in the regulation of the p53 pathway and affect cell cycle progression and DNA damage responses by interacting with p53. Considering these discoveries, both KDM8 and CCNA1 are potential targets for cancer treatment.
Previous research has shown that natural compounds derived from plants possess chemopreventive, anticancer, and antimetastatic properties and functions [28]. Isothiocyanates (ITCs) are well-established and have been reported to exhibit anticancer effects in human cancers [29,30,31]. These compounds are present in a variety of cruciferous vegetables, including cauliflower, brussels sprouts, kale, cabbage, horseradish, and wasabi [31,32]. Allyl isothiocyanate (AITC; 3-isothiocyanato-1-propene, CH2CHCH2NCS) is responsible for the pungent flavor of mustard, horseradish, radish, and wasabi. AITC, which is a sulfur-containing organic compound, is a product of the enzymatic hydrolysis of the glucosinolate sinigrin. Research has shown that AITC can hinder cancer cell progression by impeding cell growth, proliferation, migration, and invasion [31,32]. AITC has been found to regulate DNA methylation, a process that involves the addition of a methyl group to the DNA molecule and can reduce DNA methylation in cancer cells. This leads to the reactivation of tumor suppressor genes and inhibition of cancer cell growth [33,34,35,36,37]. AITC has also been found to inhibit the activity of histone deacetylases (HDACs), which are the enzymes involved in regulating histone modification [34,38,39]. By inhibiting HDAC activity, AITC can increase histone acetylation, alter gene expression, and prevent cancer cell growth [34]. Furthermore, AITC has been shown to induce apoptosis and G2/M phase arrest in human brain malignant glioma GBM 8401 cells [40] as well as apoptotic death in human cisplatin-resistant oral cancer cells [29]. Various preclinical studies have shown that AITC can inhibit lung, breast, gastric, prostate, and bladder cancer, but this does not reflect in the pilot trials performed in humans to date [41,42,43].
The lack of sufficient tumor models that have clinical relevance poses a significant obstacle to the development of effective therapies for oral cancer. It is crucial to establish PDTX models that accurately represent the phenotypic and genotypic characteristics of oral cancer. PDTX, an innovative preclinical animal model, consistently maintains the tumor’s morphology and genetic stability and is one of the most promising platforms for simulating human cancer and its complexity. The histopathology of PDTX tumors is very similar to the histopathology of donor lesions. A large amount of evidence, including mutation status, transcriptome, histology, polymorphism, and copy number variation with high fidelity, also supports the view that the PDTX model is very similar to human tumors’ pathophysiology than the traditional cancer-derived xenograft model [44,45].
AITC has been shown to inhibit several common cancer types both in vitro and in vivo [46,47,48], but there are limited studies on its inhibitory effects on oral cancer cells [29,49]. As a result, this study utilizes a preclinical PDTX model and an oral cancer cell line to explore whether AITC’s anticancer effects on cell proliferation in oral cancer are modulated through KDM8 and CCNA1.
2. Materials and Methods
2.1. Chemicals and Reagents
The following chemicals and reagents were procured from Sigma-Aldrich (St. Louis, MO, USA): AITC, methylene blue, propidium iodide (PI), ethanol, DMSO, and isopropanol. A concentration of 10 mM AITC was prepared by dissolving it in DMSO and storing it at 4 °C. RPMI 1640 and fetal bovine serum (FBS) were obtained from Biological Industries (Beit-Haemek, Israel), while RIPA buffer was purchased from Millipore (Burlington, MA, USA). The present study utilized primary antibodies against KDM8 (AVIVA SYSTEMS BIOLOGY; ARP58120_P050), H3K36me2 (GeneTex; GTX54108), Cyclin A1 (GeneTex; GTX02524), and GAPDH (GeneTex; GTX100118), as well as secondary antibodies (GeneTex; GTX213110-01, GTX213111-01).
2.2. Tumor Specimens
Samples of tumors and adjacent normal oral mucosa were collected from 27 patients with OSCC at the Tri-Service General Hospital of National Defense Medical Center (Taipei, Taiwan) with their consent and in compliance with Institutional Review Board protocols (TSGH-2-105-05-004). Tumor tissue samples were obtained from archived formalin-fixed, paraffin-embedded (FFPE) specimens collected during diagnosis and stored at hospital pathology departments. The tissue microarray (TMA) procedure was carried out using a method previously described [27].
2.3. Immunohistochemistry
The immunohistochemical staining protocol was utilized to evaluate KDM8 antibody binding in all tissue specimens, and the resulting staining scores were determined according to previously published methods [27]. Specifically, the immunostaining score was calculated by multiplying the score for stained tumor cells by the intensity score. The intensity of immune reactivity towards tumor cells was graded on a 0–3 scale, with 0 indicating no staining, 1 indicating weak intensity, 2 indicating moderate intensity, and 3 indicating strong intensity. For each intensity score, the percentage of tumor cells exhibiting nuclear or cytosolic staining was graded on a 5-point scale, with 0 indicating no staining and 4 indicating staining in 75–100% of cells.
2.4. Cell Lines and Cell Culture
The cell lines were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mmol/L L-glutamine, following established protocols [27]. The SCC25 tongue cancer cell line was obtained from the American Type Culture Collection (ATCC), while the SAS tongue cancer cell line was kindly provided by Dr. Lo from the Institute of Oral Biology, Department of Dentistry, National Yang-Ming University, Taipei, Taiwan.
2.5. In Vitro Cell Proliferation Assay
The antitumor effect of AITC on oral cancer cell growth was performed by the methylene blue dye assay, as described previously [50].
2.6. Cell Cycle Analysis
The study utilized a cell cycle assay method previously described in the literature [51]. Briefly, SAS oral cancer cells were seeded in tissue culture plates at a density of 1 × 106 cells/dish and subjected to flow cytometry analysis using the FACSCalibur instrument (Becton Dickinson, Franklin Lakes, NJ, USA). The cells were treated with different concentrations of AITC (0, 25, and 50 μM) for 24 and 48 h. Subsequently, the cells were harvested, fixed in chilled ethanol overnight at 4 °C, washed with PBS, and resuspended in PBS. The cells were then incubated with 0.5 mL of PI/RNase for 15 min at room temperature.
2.7. Western Blot Analysis
A previously published protocol [50] was followed to perform Western blot analysis. In summary, cell pellets were lysed directly in RIPA buffer with added protease and phosphatase inhibitors (Calbiochem, San Diego, CA, USA). The protein concentration of the resulting supernatants was measured using a BCA protein assay kit (Thermo Scientific, Waltham, MA, USA). Afterward, 30 μg of cell lysate protein was loaded onto each lane of a 10% SDS-PAGE gel, separated, and then transferred onto a polyvinyldifluoride membrane (Amersham, Freiburg im Breisgau, Germany). Finally, specific antibodies were used to probe the membranes for target proteins.
2.8. Establishment of PDTX Models and Treatment Protocol
The methods used to establish PDTX were outlined in a previous study [51]. In brief, tumor fragments were repeatedly washed in penicillin/streptomycin culture medium. All surgical instruments were sterilized before use. The procedure involved subcutaneous transplantation by the operating personnel, also known as a survival surgery. The surgical process included wiping the Biose-cure operating table and all tools with Virkon and injecting 100 μL of ketamine mixed with Rompun into the abdominal cavity. Subcutaneous transplantation was performed after anesthesia took effect. After implantation, the wound was sutured with stitches. Postoperatively, the wound site was continuously monitored until the experiment’s completion (see Figure S1 for the workflow), and the histological patterns of the tumor tissue were verified using H&E staining. The establishment of PDTX models and treatment protocols was approved by ethical guidelines and the Institutional Review Board (TSGH-2-102-05-111). Tumor specimens were obtained from patients with OSCC during their initial surgical treatment and were classified as T4aN2b based on World Health Organization criteria. These tumors were then maintained in RPMI 1640 medium and implanted subcutaneously into nonobese diabetic/severe combined immunodeficiency/gamma (NSG) mice aged 6–10 weeks. The xenograft models were monitored at least twice a week, and the tumor volume was calculated using the formula V = 1/2 × (length × width2). Tumor tissues were removed and transplanted serially if the tumor volume reached approximately 3000 mm3. When the tumor volume reached approximately 500 mm3, mice with seventh-generation SC179-PDTXs were randomly assigned to three groups: one receiving AITC (50 mg/Kg/daily, n = 4), one receiving cisplatin (10 mg/Kg/daily, n = 4), and a third group receiving Phosphate buffered saline (PBS, n = 4) as a control. Treatment was administered by intraperitoneal injection for 24 days, and body weight and tumor volume were measured at least twice a week. At the end of the treatment, the mice were sacrificed, and tumors were removed, weighed, and observed. The animal experiments were approved by the National Defense Medical Center Institutional Animal Care and Use Committee, Taipei, Taiwan (IACUC; 16-244 and 18-027).
2.9. Mouse SAS Xenograft Model
Previously described methods [50] were used to perform SAS xenograft animal models. Approval for all experiments was obtained from the National Defense Medical Center Institutional Animal Care and Use Committee, Taipei, Taiwan (IACUC; 16-244). Nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice that were eight weeks old were housed in microisolators under specific pathogen-free conditions. The mice were divided into three groups: the AITC treatment group (n = 4), which received 50 mg/Kg body weight/daily intraperitoneal (i.p.) treatment; the positive control group (n = 4), which received 15 mg/Kg body weight/daily i.p. treatment of 5-Flurouracil (5-FU); and the vehicle control group (n = 4), which received PBS treatment. Each group of mice was subcutaneously injected with 2 × 106 SAS oral cancer cells. Drug treatments began on the third day after the tumor injection and continued until day 21. Tumor size was monitored at least twice a week, and tumor volume was calculated using the formula V = 1/2 × (length × width2). At the end of the experiment, the mice were sacrificed, and the tumors were excised, weighed, and examined.
2.10. UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html, URL (accessed on 17 July 2023)) is a web-based tool that provides a thorough examination of gene expression data derived from The Cancer Genome Atlas (TCGA) database [52,53]. The tool’s “TCGA Gene analysis” module was employed to investigate the mRNA levels of KDM8/JMJD5 in head and neck squamous carcinoma (HNSC) patients and healthy individuals. The analysis also explored the correlation of these levels with clinicopathological parameters. The TCGA HNSC dataset, comprising genetic data from 520 individuals, was used for the analysis. The significance of the findings was determined at p < 0.05.
2.11. Statistical Analysis
The statistical analysis of the data was carried out using the GraphPad Prism software (GraphPad Software 8.0, San Diego, CA, USA). The statistical significance of the results was determined by performing unpaired, two-tailed Student’s t-tests. p < 0.05 was considered statistically significant.
3. Results
3.1. KDM8 and CCNA1 Expression Exhibited a Significant Increase in Both Human OSCC and Cell Lines
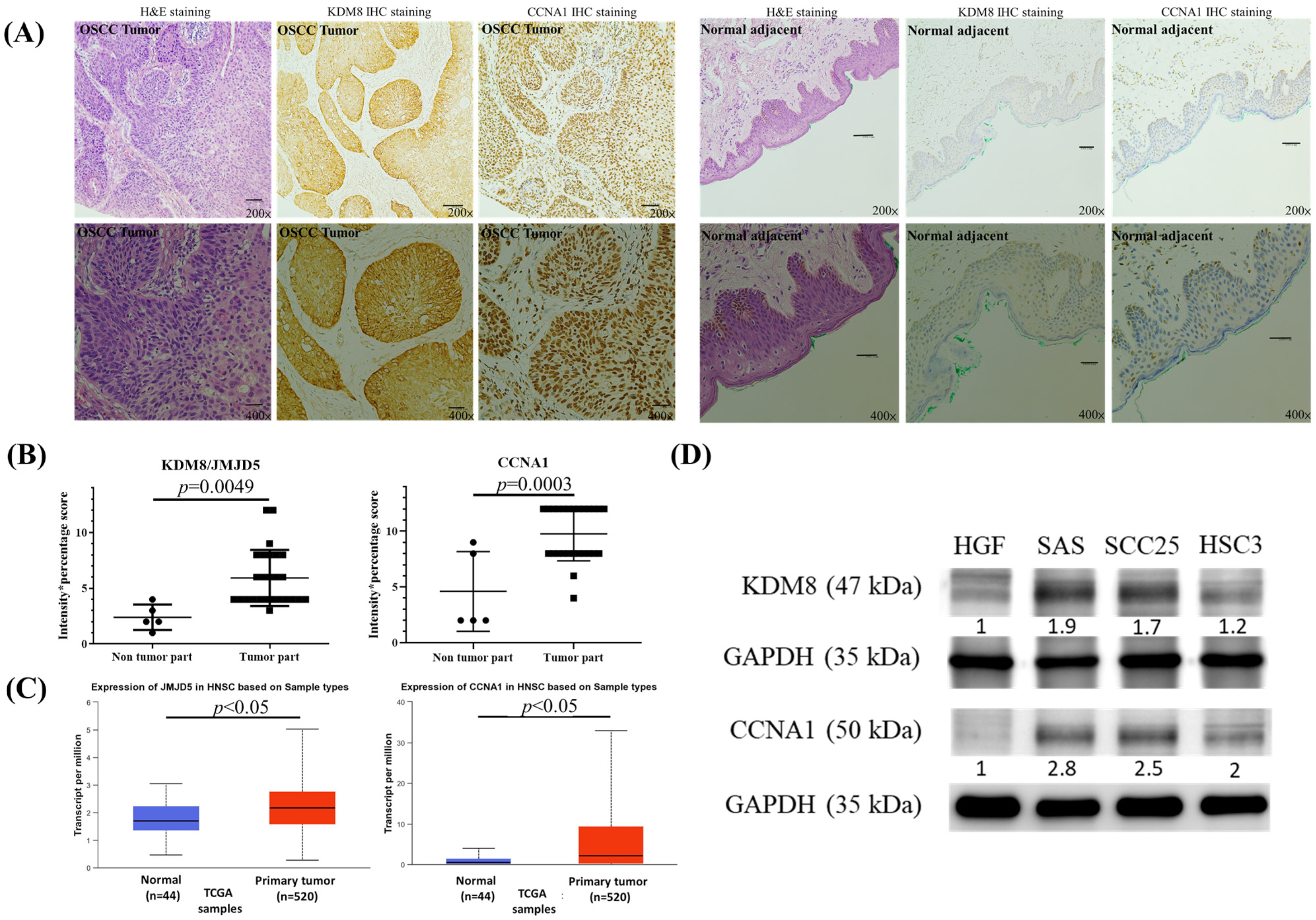
At the beginning, we used immunohistochemical stain analysis to determine the expression status of the KDM8 and CCNA1 protein in 27 OSCC samples TMA (Table 1) and 5 normal oral mucosa samples adjacent to the tumor. Our findings, presented in Figure 1A,B, revealed a substantial increase in the levels of KDM8 and CCNA1 protein expression in the OSCC tumor samples relative to the normal mucosa samples. To validate our results, we utilized the UALCAN web tool, which utilizes the TCGA database, to analyze the mRNA levels of KDM8 and CCNA1 in HNSC tissues and normal tissues. Our analysis, presented in Figure 1C, showed a significant elevation in KDM8 and CCNA1 expression levels in HNSC tissues compared to normal tissues (p < 0.05). To further support our observations, we performed Western blotting analysis on various oral cancer cell lines (SAS, SCC25, and HSC3) and human normal gingival fibroblast cells (HGF) to investigate KDM8 and CCNA1 expression. Our results, illustrated in Figure 1D, demonstrated significantly higher levels of KDM8 and CCNA1 expression in the oral cancer cell lines relative to HGF cells.

Table 1.
The clinical parameters of OSCC patients in TMA (n = 27).

Figure 1.
High expression of KDM8 existed in oral cancer. (A) H&E and KDM8 IHC stain for normal tissues adjacent to the tumor (magnification 200× and 400×). (B) Representative samples scored for KDM8 IHC results in normal mucosal and oral cancer tissues are shown. The score was defined as intensity score x percentage score. Data are expressed as mean ± standard deviation. p < 0.05 (Student’s t-test) represents a significant difference. (C) Expression levels of the KDM8 in HNSC patients based on the UALCAN database. (D) KDM8 and CCNA1 protein levels in oral cancer cell lines were determined through Western blot analysis. Normal human gingival fibroblast HGF-1 cells were used as the negative control.
3.2. AITC Exerts Cytotoxic Effects in Oral Cancer Cells
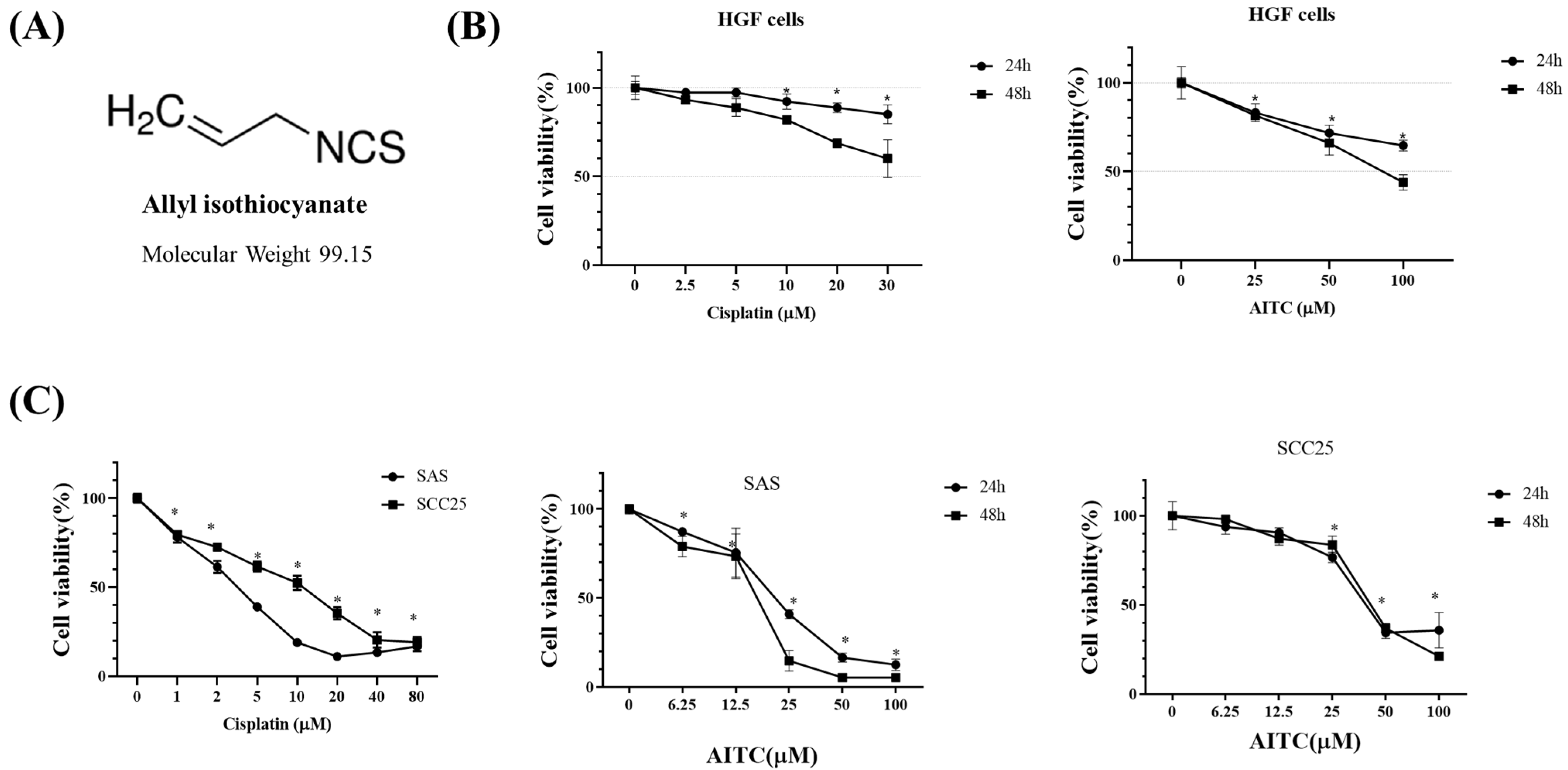
We investigated the impact of AITC (Figure 2A) on the cytotoxicity of normal human gingival fibroblast HGF (Figure 2B) and oral cancer cells (Figure 2C). The positive control chemotherapy cisplatin was also tested. To assess this, SAS and SCC25 oral cancer cells were exposed to varying concentrations of AITC for 24 and 48 h (as shown in Figure 2C). Our findings revealed that AITC exhibited a dose- and time-dependent inhibitory effect on the proliferation of tongue cancer cells (as depicted in Figure 2C). After 24 h of treatment, the IC50 of AITC for SAS was approximately 25 μM.

Figure 2.
Effects of AITC on proliferation in SAS and SCC25 cells. The number of cells with methylene blue dye assay correspond to the mean ± SD of three independent experiments carried out in duplicate. (*) p < 0.05 compared to the negative control. (A) Structure of AITC. (B) Normal human gingival fibroblast HGF cells as the control. HGF cells were treated with various concentrations of cisplatin and AITC for 24 and 48 h, and then cell viability was determined by the methylene blue assay. (C) Cells were treated with various concentrations of cisplatin and AITC for 24 and 48 h, and then cell viability was determined by the methylene blue assay.
3.3. AITC Inhibited the Growth of Oral Cancer in Preclinical Models
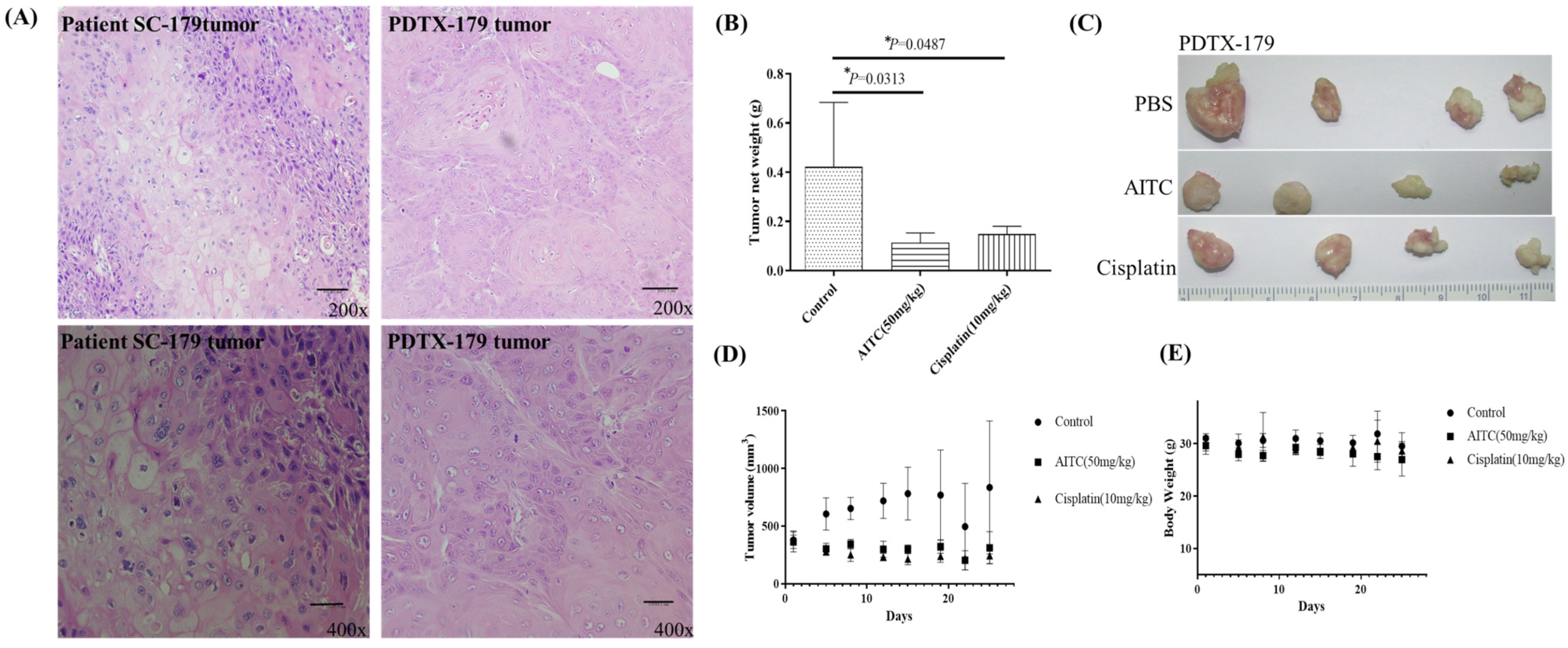
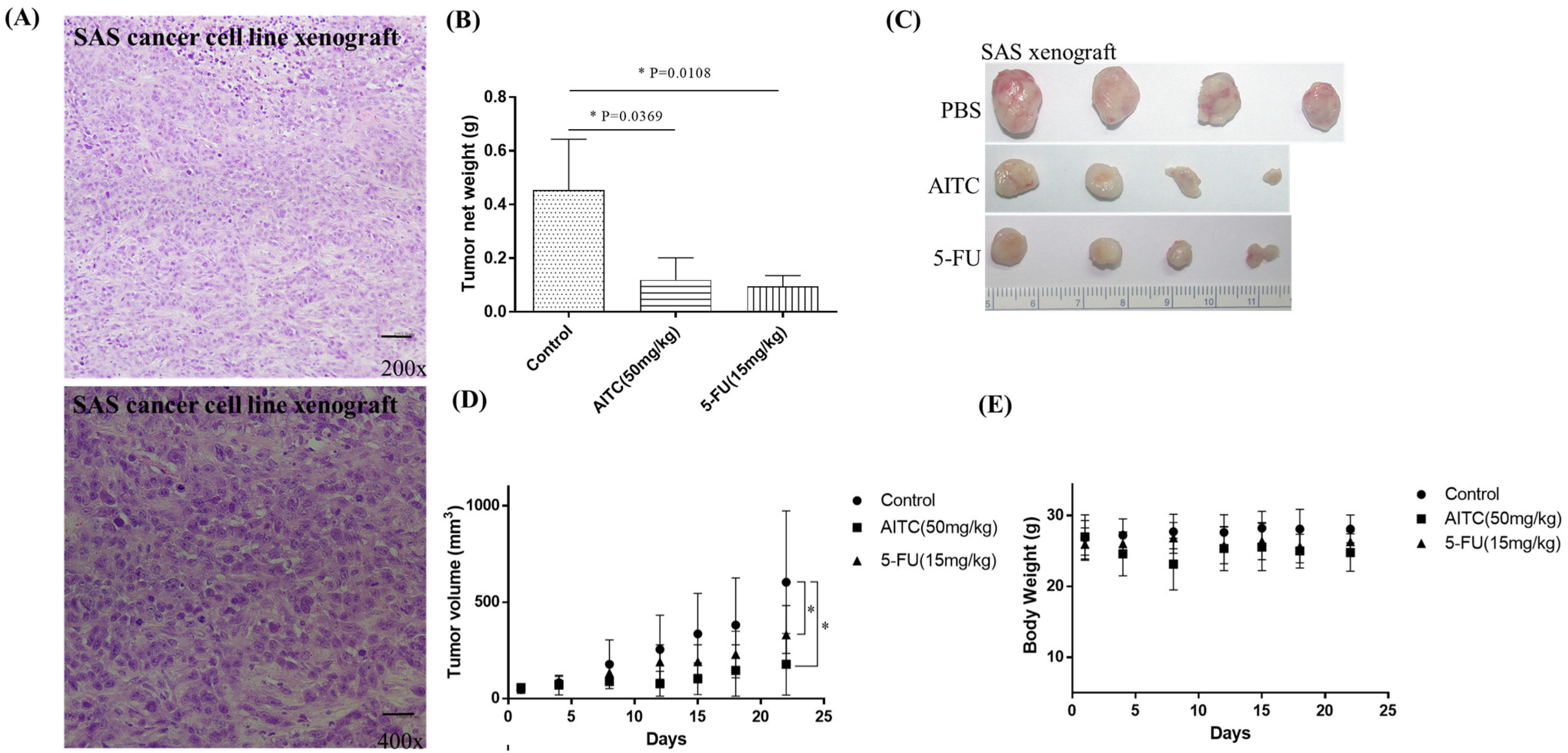
The initial chemotherapy approach for oral cancer can vary based on several factors, such as the cancer’s location and stage. Nonetheless, the most frequently employed chemotherapy method for oral cancer is a combination of cisplatin and 5-fluorouracil (5-FU). To determine the potential impact of allyl isothiocyanate (AITC) on PDTX proliferation in NOD SCID mice, we conducted further research. Our findings demonstrated that AITC, administered at a dosage of 50 mg/Kg, exhibited significant antitumor activity, similar to that of cisplatin administered at a dosage of 10 mg/Kg, compared to the PBS-treated control group (as depicted in Figure 3B). Additionally, the outcomes showed no discernible toxic effects or considerable loss of body weight among the different treatment groups (as illustrated in Figure 3C,D). Moreover, we assessed AITC’s in vivo cytotoxic potential by analyzing its capacity to suppress the growth of SAS xenograft oral cancer cells. The results revealed that AITC, administered at a dosage of 50 mg/Kg, demonstrated a substantial inhibition of tumor growth, which was akin to that of 5-FU (administered at a dosage of 15 mg/Kg) in the oral cancer xenograft (as demonstrated in Figure 4).

Figure 3.
AITC inhibited tumor growth in oral cancer PDTX models. (A) H&E staining of the SC-179 clinical sample and PDTX-179. (B) In PDTX, NOD/SCID mice bearing patient-derived tumor tissue xenograft were treated with PBS (n = 4), AITC (n = 4; 50 mg/Kg/d), and cisplatin (n = 4; 10 mg/Kg/d). The average tumor weight of each group was compared with that of the control (p < 0.05 by Student’s t-test). (C) Tumor images for each group. (D) Changes in tumor volume in oral cancer PDTX models treated for 24 days. Diameters were measured twice a week for 24 days by using Vernier calipers, and the tumor volume was calculated as 1/2 × L × W2, where W and L are the shortest and longest diameters, respectively. Tumor volumes were compared with those of controls. All data are expressed as mean ± SD. p < 0.05 (Student t-test). (E) No significant change was observed in the mice’s body weight compared with that of the vehicle control. *: statistically significant difference.

Figure 4.
AITC inhibited oral cancer growth in SAS xenograft. (A) H&E staining of the SAS cancer cell linr xenograft. (B) In vivo, NOD/SCID mice bearing subcutaneous SAS cells were treated with PBS (n = 4), AITC (n = 4; 50 mg/Kg/d), and 5-FU (n = 4; 15 mg/Kg/d). The average tumor weight of each group was compared with that of the control (* p < 0.05 by Student’s t-test). (C) Tumor images for each group. (D) Changes in tumor volume in oral cancer SAS xenograft model, which was treated for 21 days. Diameters were measured twice a week for 21 days by using Vernier calipers, and the tumor volume was calculated as 1/2 × L × W2, where W and L are the shortest and longest diameters, respectively. Tumor volumes were compared with those of controls. All data are expressed as mean ± SD. * p < 0.05 (Student t-test). (E) No significant change was observed in the mice’s body weight compared with that of the vehicle control.
3.4. AITC Induced G2/M Cell Cycle Arrest and Apoptosis through the Modulation of G2/M-Associated Proteins KDM8 and CCNA1 in Oral Cancer Cells
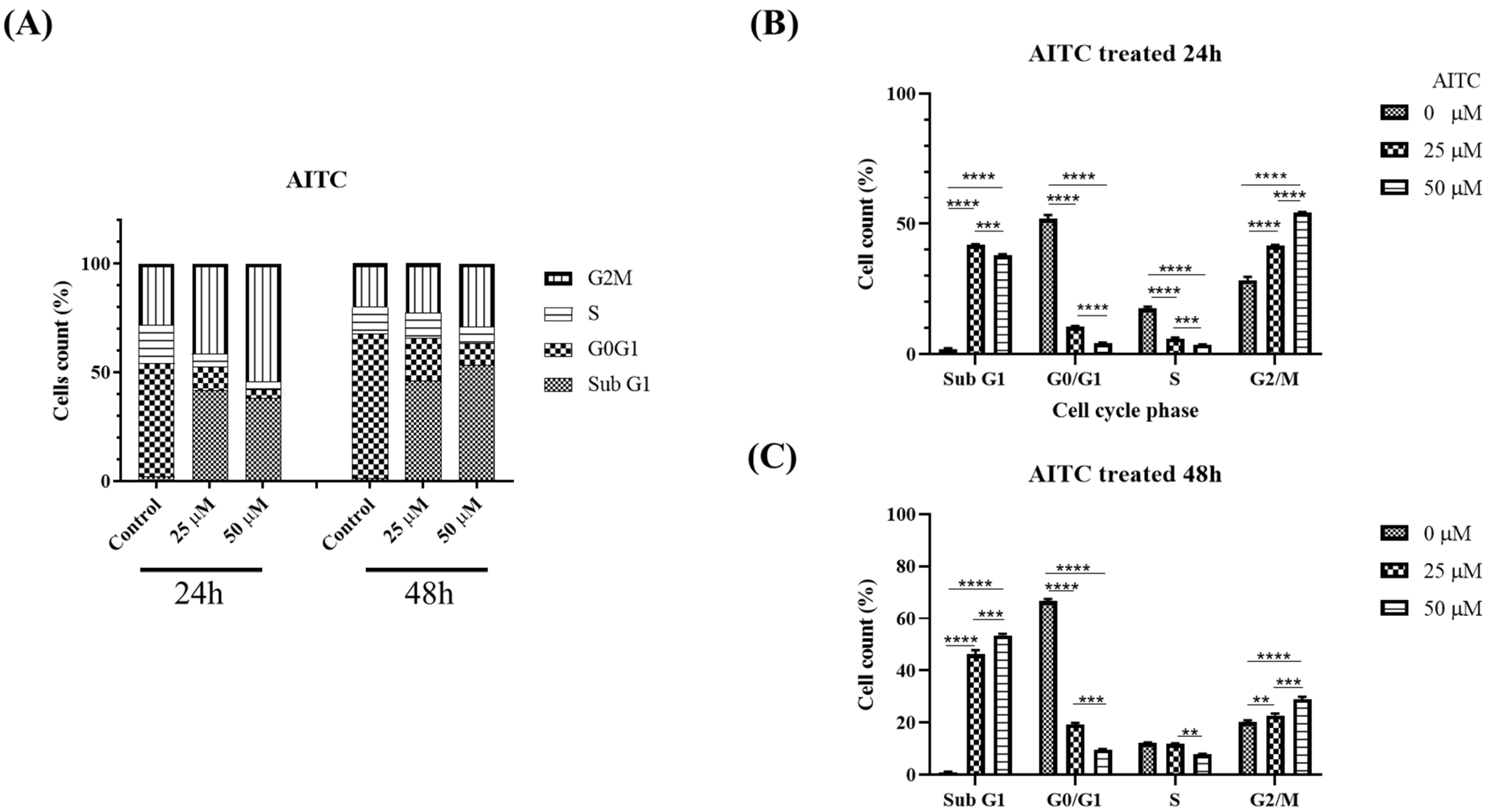
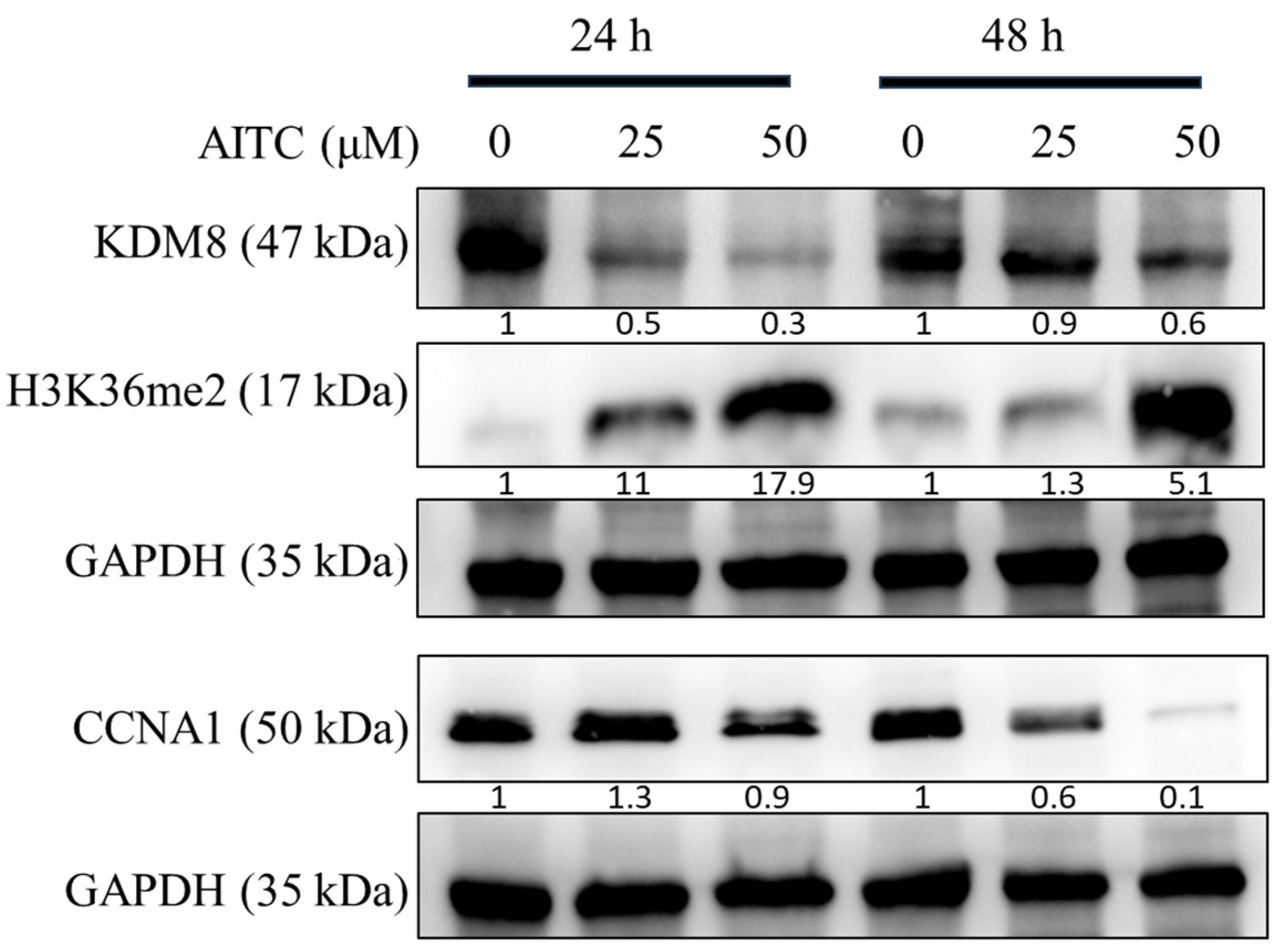
The AITC compound was found to induce cell cycle arrest at the G2/M phase, leading to reduced cell proliferation, potentially due to apoptosis, as evidenced by the sub-G1 population. This was determined through the examination of cell cycle progression profiles and the percentage of cells in each phase, as presented in Figure 5. A significant arrest at the G2/M phase and an increase in apoptotic cells in the sub-G1 groups were observed at a concentration of 25 μM of AITC. Additionally, AITC was observed to affect the levels of proteins linked to the G2/M phase. To confirm the buildup of the G2/M population in SAS oral cancer cells prompted by AITC, the levels of regulated proteins were evaluated. The results demonstrated a significant decline in KDM8 and CCNA1, and an increase in H3K36me2 in SAS cells exposed to AITC, as shown in Figure 6.

Figure 5.
AITC induced oral cancer G2/M arrest. Flow cytometric cell cycle analysis of SAS cells treated with varying concentrations of AITC (25–50 μM) or DMSO (1 μL/mL) for 24 and 48 h. Graphs show cell cycle distribution (A) and distribution quantification percentage (B) for AITC treated 24 h and (C) AITC treated 48 h. **: statistically significant as p < 0.01; ***: statistically significant as p < 0.001; ****: statistically significant as p < 0.0001.

Figure 6.
AITC downregulated KDM8 and CCNA1 expression in oral cancer cells. Western blot analysis and densitometry for KDM8, H3K36me2, and CCNA1 after SAS cells treated with AITC for 24 h and 48 h.
4. Discussion
KDM8, a member of an evolutionarily conserved protein family with a Jumonji domain, is currently recognized as an oncogene in colon and breast cancer [21,22,25,26]. In this study, we confirmed previous findings that KDM8 is overexpressed in OSCC [20,27]. KDM8 promotes breast cancer cell proliferation by activating CCNA1, a regulator of the G1/S and G2/M transition [13,54,55]. Additionally, KDM8 inhibits p21, a potent inhibitor of a cyclin-dependent kinase, which leads to cell growth retardation, the loss of pluripotency in embryonic stem cells [56], and embryonic lethality when KDM8 is deficient in mouse embryonic cells [57]. These results provide strong evidence that KDM8 is crucial in regulating the G2/M cell cycle. Furthermore, knocking down KDM8 using siRNA inhibited the growth of oral cancer cells (Figure S2). The results showed a similarity with a previous study [20]. Previously conducted studies have indicated that allyl isothiocyanate (AITC) can prompt G2/M cell cycle arrest in various types of cancer, such as brain glioma, colorectal adenocarcinoma, and breast adenocarcinoma[40,58,59]. CCNA1, which is a member of the highly conserved cyclin family, may be involved in regulating the cell cycle at both G1/S and G2/M transition points [6]. Based on immunohistochemistry-based tests, several studies have suggested that CCNA1 may be linked to poor prognosis in OSCC [12,60]. In the current study, we discovered that AITC could cause G2/M arrest in OSCC cells and reduce CCNA1 expression. Thus, these results imply that the potential anti-oral cancer effects of AITC may be associated with the suppression of CCNA1.
Histone modification is a crucial mechanism of epigenetic regulation. During the post-translational modification (PTM) of histones, histone methylation marks have various effects on transcriptional activation [61]. Several studies have shown that the global regulation of the H3K36me2 histone mark is responsible for epithelial plasticity and metastatic progression [62]. Furthermore, Jumonji domain 2 (JMJD2/KDM4) cluster members are capable of demethylating H3K36me3/2 [61,63]. These JMJD2 family members are overexpressed in various human cancers [7,28,29], indicating that H3K36me3 may serve as a tumor suppressor marker. In this study, our findings suggest that the downregulation of KDM8 expression by AITC treatment could induce H3K36me2 expression.
Although AITC has shown promise in its ability to combat cancer, there are still obstacles to its clinical application [29,64]. PDTX is a type of preclinical cancer model where the tumor tissue from a patient is implanted into immunodeficient mice to create a tumor that closely retains the tumor heterogeneity present in the original patient sample, which enables them to replicate the complexity of the tumor, including the tumor microenvironment. In this study, we created a PDTX model which is actually similar to the original tumor by histopathological analysis (H&E stain) (Figure S1). This feature is critical for drug development [65]. PDTX models have several advantages for cancer research and drug development, including having good biological relevance, being more predictive of patient response to treatment than cell line models, being suitable for personalized medicine, and having high fidelity. PDTX models can be propagated indefinitely, allowing for long-term studies and drug development. Additionally, PDX models can be used to study tumor evolution and drug resistance, which is difficult to do in patients due to ethical and practical limitations. However, there are also many disadvantages for PDTX models which are costly and time-consuming, have limited engraftment success, and show variability in tumor growth rates. PDTX models may exhibit variations in tumor growth rates, which can complicate drug testing standardization. In this study, our results in Figure 3 have shown variations in tumor growth volume and tumor mass. Although different cancer types of preclinical PDTX models have been established, research on PDTX models for oral cancer is limited [66]. To address this gap, this study aimed to establish PDTX models and verify the anticancer activity of AITC in oral cancer.
5. Conclusions
AITC inhibited oral cancer growth in vitro, in vivo, and in PDTX models. KDM8 and CCNA1 repression by AITC markedly suppressed oral cancer’s proliferation or growth. Thus, AITC might be the candidate adjuvant of OSCC. However, further study should still reveal the mechanism of the AITC-regulated anticancer effect.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines11102669/s1, Figure S1: The PDTX establishment process. Figure S2: KDM8 suppression alleviates OSCC proliferation.
Author Contributions
Conceptualization, C.-C.H., W.-T.C. and Y.-W.C.; Data curation, C.-Y.Y.; Funding acquisition, Y.-W.C.; Investigation, C.-C.H., C.-Y.Y., B.P., S.-L.H., C.-H.T., C.-K.L., C.-S.L. and H.-Y.L.; Project administration, W.-T.C. and Y.-W.C.; Resources, G.-J.L., H.-C.H. and S.-C.C.; Supervision, H.-K.S., W.-T.C. and Y.-W.C.; Writing—original draft, C.-C.H.; Writing—review and editing, W.-T.C. and Y.-W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by research grants from the Tri-Service General Hospital, Taiwan, Republic of China (grants No. TSGH-C01-109017, TSGH-C05-110035, TSGH-C04-111037, TSGH-D-110148, TSGH-D-110149, TSGH-D-110151, TSGH-D-110152, TSGH-D-110154, TSGH-C02-112032), Ministry of National Defense, Taiwan, Republic of China (grants No. MAB-E-109003, MAB-D-110003, MND-MAB-110-043, MND-MAB-110-076, MND-MAB-C-111036, MAB-E-111002, MND-MAB-D-111149, MND-MAB-D-112176, MND-MAB-C08-112033), Ministry of Science and Technology, Taiwan, Republic of China (grants No. MOST 108-2314-B-016-005), Kaohsiung Armed Forces General Hospital, Taiwan, Republic of China (grant No. KAFGH-E-111047, KAFGH_E_112061), Hualien Armed Forces General Hospital, Taiwan, Republic of China (grant No. HAFGH_E_112018), Kaohsiung Veterans General Hospital, Taiwan, Republic of China (grant No. KSVGH112-135), and Cardinal Tien Hospital, Taipei, Taiwan, Republic of China (grant No. CTH107A-2C01).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Tri-Service General Hospital of National Defense Medical Center (Taipei, Taiwan) (TSGH-2-105-05-004 and TSGH-2-102-05-111)” for studies involving humans. The animal study protocol was approved by the National Defense Medical Center Institutional Animal Care and Use Committee, Taipei, Taiwan (IACUC; 16-244 and 18-027) for studies involving animals.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data created or analyzed during this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors acknowledge the technical services provided by the Instrument Center of the National Defense Medical Center and the laboratory animal center of the National Defense Medical Center. This research was supported by the Cancer Registry Group, Tri-Service General Hospital.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sehgal, S.; Mishra, M.K.; Gupta, S. Shivangi Assessing long non-coding RNAs in tobacco-associated oral cancer. Curr. Cancer Drug Targets 2022, 22, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Vallina, C.; López-Pintor, R.M.; González-Serrano, J.; de Vicente, J.C.; Hernández, G.; Lorz, C. Genes involved in the epithelial-mesenchymal transition in oral cancer: A systematic review. Oral Oncol. 2021, 117, 105310. [Google Scholar] [CrossRef] [PubMed]
- Myoung, H.; Kim, M.-J.; Lee, J.-H.; Ok, Y.-J.; Paeng, J.-Y.; Yun, P.-Y. Correlation of proliferative markers (Ki-67 and PCNA) with survival and lymph node metastasis in oral squamous cell carcinoma: A clinical and histopathological analysis of 113 patients. Int. J. Oral Maxillofac. Surg. 2006, 35, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Crooker, K.; Aliani, R.; Ananth, M.; Arnold, L.; Anant, S.; Thomas, S.M. A Review of Promising Natural Chemopreventive Agents for Head and Neck Cancer. Cancer Prev. Res. 2018, 11, 441–450. [Google Scholar] [CrossRef]
- Rivera, A.; Mavila, A.; Bayless, K.J.; Davis, G.E.; Maxwell, S.A. Cyclin A1 is a p53-induced gene that mediates apoptosis, G2/ M arrest, and mitotic catastrophe in renal, ovarian, and lung carcinoma cells. Cell. Mol. Life Sci. 2006, 63, 1425–1439. [Google Scholar] [CrossRef]
- Ji, P.; Agrawal, S.; Diederichs, S.; Bäumer, N.; Becker, A.; Cauvet, T.; Kowski, S.; Beger, C.; Welte, K.; Berdel, W.E.; et al. Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell cycle progression in somatic cells. Oncogene 2005, 24, 2739–2744. [Google Scholar] [CrossRef]
- Yang, R.; Müller, C.; Huynh, V.; Fung, Y.K.; Yee, A.S.; Koeffler, H.P. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol. Cell. Biol. 1999, 19, 2400–2407. [Google Scholar] [CrossRef]
- Munari, E.; Chaux, A.; Maldonado, L.; Compérat, E.; Varinot, J.; Bivalacqua, T.J.; Hoque, M.O.; Netto, G.J. Cyclin A1 expression predicts progression in pT1 urothelial carcinoma of bladder: A tissue microarray study of 149 patients treated by transurethral resection. Histopathology 2015, 66, 262–269. [Google Scholar] [CrossRef]
- He, X.; Li, S.; Shi, W.; Lin, Q.; Ma, J.; Liu, Y.; Feng, T.; Cao, X. Cyclin A1 is associated with poor prognosis in oesophageal squamous cell carcinoma. Oncol. Lett. 2019, 18, 706–712. [Google Scholar] [CrossRef]
- Kannan, S.; Roshan, V.D.; Sinto, M.S.; Vargees, B. Loss of CDKN2A and CDKN2B expression is associated with disease recurrence in oral cancer. J. Oral Maxillofac. Pathol. 2019, 23, 82–89. [Google Scholar] [CrossRef]
- Juodzbalys, G.; Kasradze, D.; Cicciù, M.; Sudeikis, A.; Banys, L.; Galindo-Moreno, P.; Guobis, Z. Modern molecular biomarkers of head and neck cancer. Part I. Epigenetic diagnostics and prognostics: Systematic review. Cancer Biomark. 2016, 17, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Bourova-Flin, E.; Derakhshan, S.; Goudarzi, A.; Wang, T.; Vitte, A.L.; Chuffart, F.; Khochbin, S.; Rousseaux, S.; Aminishakib, P. The combined detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 expression predicts aggressive forms of oral squamous cell carcinoma. Br. J. Cancer 2021, 125, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- Hsia, D.A.; Tepper, C.G.; Pochampalli, M.R.; Hsia, E.Y.C.; Izumiya, C.; Huerta, S.B.; Wright, M.E.; Chen, H.-W.; Kung, H.-J.; Izumiya, Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. USA 2010, 107, 9671–9676. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.I.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Oh, S.; Janknecht, R. Histone demethylase JMJD5 is essential for embryonic development. Biochem. Biophys. Res. Commun. 2012, 420, 61–65. [Google Scholar] [CrossRef]
- Wang, H.-J.; Hsieh, Y.-J.; Cheng, W.-C.; Lin, C.-P.; Lin, Y.-S.; Yang, S.-F.; Chen, C.-C.; Izumiya, Y.; Yu, J.-S.; Kung, H.-J.; et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Youn, M.-Y.; Yokoyama, A.; Fujiyama-Nakamura, S.; Ohtake, F.; Minehata, K.-I.; Yasuda, H.; Suzuki, T.; Kato, S.; Imai, Y. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J. Biol. Chem. 2012, 287, 12994–13004. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A.; Covington, M.F.; DiTacchio, L.; Vollmers, C.; Panda, S.; Harmer, S.L. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 2010, 107, 21623–21628. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhou, W.Y.; He, R.X. Down-regulation of JMJD5 suppresses metastasis and induces apoptosis in oral squamous cell carcinoma by regulating p53/NF-kappaB pathway. Biomed. Pharmacother. 2019, 109, 1994–2004. [Google Scholar] [CrossRef]
- Zhao, Z.; Sun, C.; Li, F.; Han, J.; Li, X.; Song, Z. Overexpression of histone demethylase JMJD5 promotes metastasis and indicates a poor prognosis in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10325–10334. [Google Scholar] [PubMed]
- Li, H.; Wang, Y.; Liu, L.; Ji, F.; Jiang, J.; Yu, Y.; Sheng, S. Soybean (Glycine max) prevents the progression of breast cancer cells by downregulating the level of histone demethylase JMJD5. J. Cancer Res. Ther. 2018, 14, S609–S615. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Ma, X.; Fang, M.; Zhong, L.; Liu, H.; Liu, H.; Tong, Y. Jumonji domain containing 5 is a potential prognostic indicator in non-small cell lung cancer patients who received platinum-based chemotherapy. Transl. Cancer Res. 2019, 8, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Q.; Jing, H.; Zhao, J.; Zhang, H.; Ma, X.; Wei, L.; Dai, R.; Sun, W.; Suo, Z. Expression and prognosis analysis of JMJD5 in human cancers. Front. Biosci. (Landmark Ed.) 2021, 26, 707–716. [Google Scholar] [PubMed]
- Wang, H.J.; Pochampalli, M.; Wang, L.Y.; Zou, J.X.; Li, P.S.; Hsu, S.C.; Wang, B.-J.; Huang, S.-H.; Yang, P.; Yang, J.C.; et al. KDM8/JMJD5 as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in CRPC. Oncogene 2019, 38, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Huang, Q.; Li, Y.; Song, Y.; Li, Y. JMJD5 is a potential oncogene for colon carcinogenesis. Int. J. Clin. Exp. Pathol. 2015, 8, 6482–6489. [Google Scholar]
- Yang, C.-Y.; Tsao, C.-H.; Hsieh, C.-C.; Lin, C.-K.; Lin, C.-S.; Li, Y.-H.; Chang, W.-C.; Cheng, J.-C.; Lin, G.-J.; Sytwu, H.-K.; et al. Downregulation of Jumonji-C domain-containing protein 5 inhibits proliferation by silibinin in the oral cancer PDTX model. PLoS ONE 2020, 15, e0236101. [Google Scholar] [CrossRef]
- Haque, A.; Brazeau, D.; Amin, A.R. Perspectives on natural compounds in chemoprevention and treatment of cancer: An update with new promising compounds. Eur. J. Cancer 2021, 149, 165–183. [Google Scholar] [CrossRef]
- Chang, P.-Y.; Tsai, F.-J.; Bau, D.-T.; Hsu, Y.-M.; Yang, J.-S.; Tu, M.-G.; Chiang, S.-L. Potential effects of allyl isothiocyanate on inhibiting cellular proliferation and inducing apoptotic pathway in human cisplatin-resistant oral cancer cells. J. Formos. Med. Assoc. 2021, 120, 515–523. [Google Scholar] [CrossRef]
- Ling, X.; Westover, D.; Cao, F.; Cao, S.; He, X.; Kim, H.-R.; Zhang, Y.; Chan, D.C.; Li, F. Synergistic effect of allyl isothiocyanate (AITC) on cisplatin efficacy in vitro and in vivo. Am. J. Cancer Res. 2015, 5, 2516–2530. [Google Scholar]
- Zhang, Y. Allyl isothiocyanate as a cancer chemopreventive phytochemical. Mol. Nutr. Food Res. 2010, 54, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J. The Glucosinolates: A Sulphur Glucoside Family of Mustard Anti-Tumour and Antimicrobial Phytochemicals of Potential Therapeutic Application. Biomedicines 2019, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, M.K.; Preetha, R.; Haque, S.; Akhter, N.; Khan, S.; Ahmad, S.; Hussain, A. Dietary isothiocyanates inhibit cancer progression by modulation of epigenome. Semin. Cancer Biol. 2022, 83, 353–376. [Google Scholar] [CrossRef]
- Mitsiogianni, M.; Mantso, T.; Trafalis, D.T.; Rupasinghe, H.P.V.; Zoumpourlis, V.; Franco, R.; Botaitis, S.; Pappa, A.; Panayiotidis, M.I. Allyl isothiocyanate regulates lysine acetylation and methylation marks in an experimental model of malignant melanoma. Eur. J. Nutr. 2020, 59, 557–569. [Google Scholar] [CrossRef]
- Gerhauser, C. Cancer chemoprevention and nutri-epigenetics: State of the art and future challenges. Nat. Prod. Cancer Prev. Ther. 2013, 329, 73–132. [Google Scholar] [CrossRef]
- Ong, T.P.; Moreno, F.S.; Ross, S.A. Targeting the epigenome with bioactive food components for cancer prevention. Lifestyle Genom. 2011, 4, 275–292. [Google Scholar] [CrossRef]
- Huang, J.; Plass, C.; Gerhauser, C. Cancer chemoprevention by targeting the epigenome. Curr. Drug Targets 2011, 12, 1925–1956. [Google Scholar] [CrossRef]
- Nian, H.; Delage, B.; Ho, E.; Dashwood, R.H. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: Studies with sulforaphane and garlic organosulfur compounds. Environ. Mol. Mutagen. 2009, 50, 213–221. [Google Scholar] [CrossRef]
- Myzak, M.C.; Dashwood, R.H. Histone deacetylases as targets for dietary cancer preventive agents: Lessons learned with butyrate, diallyl disulfide, and sulforaphane. Curr. Drug Targets 2006, 7, 443–452. [Google Scholar] [CrossRef]
- Chen, N.G.; Chen, K.T.; Lu, C.C.; Lan, Y.H.; Lai, C.H.; Chung, Y.T.; Yang, J.S.; Lin, Y.C. Allyl isothiocyanate triggers G2/M phase arrest and apoptosis in human brain malignant glioma GBM 8401 cells through a mitochondria-dependent pathway. Oncol. Rep. 2010, 24, 449–455. [Google Scholar]
- Bo, P.; Lien, J.-C.; Chen, Y.-Y.; Yu, F.-S.; Lu, H.-F.; Yu, C.-S.; Chou, Y.-C.; Yu, C.-C.; Chung, J.-G. Allyl Isothiocyanate Induces Cell Toxicity by Multiple Pathways in Human Breast Cancer Cells. Am. J. Chin. Med. 2016, 44, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Rakariyatham, K.; Yang, X.; Gao, Z.; Song, M.; Han, Y.; Chen, X.; Xiao, H. Synergistic chemopreventive effect of allyl isothiocyanate and sulforaphane on non-small cell lung carcinoma cells. Food Funct. 2019, 10, 893–902. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Xiao, D.; Lew, K.L.; Hershberger, P.; Kokkinakis, D.M.; Johnson, C.S.; Trump, D.L.; Singh, S.V. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits growth of PC-3 human prostate cancer xenografts in vivo. Carcinogenesis 2003, 24, 1665–1670. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, S.; Hidalgo, M.; Kung, A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 2015, 15, 311–316. [Google Scholar] [CrossRef]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef]
- Lu, K.-W.; Lu, T.-J.; Chueh, F.-S.; Lai, K.-C.; Hsia, T.-C.; Peng, S.-F.; Cheng, C.-C.; Chou, Y.-C.; Hsu, F.-T. Allyl Isothiocyanate (AITC) Induces Apoptotic Cell Death In Vitro and Exhibits Anti-Tumor Activity in a Human Glioblastoma GBM8401/luc2 Model. Int. J. Mol. Sci. 2022, 23, 10411. [Google Scholar] [CrossRef]
- Rajakumar, T.; Pugalendhi, P.; Thilagavathi, S.; Ananthakrishnan, D.; Gunasekaran, K. Allyl isothiocyanate, a potent chemopreventive agent targets AhR/Nrf2 signaling pathway in chemically induced mammary carcinogenesis. Mol. Cell. Biochem. 2018, 437, 1–12. [Google Scholar] [CrossRef]
- Rajakumar, T.; Pugalendhi, P. Allyl isothiocyanate inhibits invasion and angiogenesis in breast cancer via EGFR-mediated JAK-1/STAT-3 signaling pathway. Amino Acids 2023, 55, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Kiss, F.; Kormos, V.; Szőke, É.; Kecskés, A.; Tóth, N.; Steib, A.; Szállási, Á.; Scheich, B.; Gaszner, B.; Kun, J.; et al. Functional Transient Receptor Potential Ankyrin 1 and Vanilloid 1 Ion Channels Are Overexpressed in Human Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 1921. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Lin, G.-J.; Chia, W.-T.; Lin, C.-K.; Chuang, Y.-P.; Sytwu, H.-K. Triptolide exerts anti-tumor effect on oral cancer and KB cells in vitro and in vivo. Oral Oncol. 2009, 45, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-Y.; Lin, C.-K.; Tsao, C.-H.; Hsieh, C.-C.; Lin, G.-J.; Ma, K.-H.; Shieh, Y.-S.; Sytwu, H.-K.; Chen, Y.-W. Melatonin exerts anti-oral cancer effect via suppressing LSD1 in patient-derived tumor xenograft models. Oncotarget 2017, 8, 33756–33769. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal For Facilitating Tumor Subgroup Gene Expression And Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-H.; Chen, H.; Cai, C.-M.; Fang, J.-Z.; Wu, C.-C.; Huang, L.-Y.; Wang, L.; Han, Z.-G. Epigenetic silencing of JMJD5 promotes the proliferation of hepatocellular carcinoma cells by down-regulating the transcription of CDKN1A 686. Oncotarget 2016, 7, 6847–6863. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, L.; Qi, H.; Shao, J.; Shen, J. Identification and functional implication of nuclear localization signals in the N-terminal domain of JMJD5. Biochimie 2013, 95, 2114–2122. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, A.; Minehata, K.I.; Terashima, M.; Kondoh, G.; Hara, T.; Suzuki, T. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 2012, 139, 749–759. [Google Scholar] [CrossRef]
- Ishimura, A.; Terashima, M.; Tange, S.; Suzuki, T. Jmjd5 functions as a regulator of p53 signaling during mouse embryogenesis. Cell Tissue Res. 2016, 363, 723–733. [Google Scholar] [CrossRef]
- Lau, W.S.; Chen, T.; Wong, Y.S. Allyl isothiocyanate induces G2/M arrest in human colorectal adenocarcinoma SW620 cells through down-regulation of Cdc25B and Cdc25C. Mol. Med. Rep. 2010, 3, 1023–1030. [Google Scholar]
- Tsai, S.C.; Huang, W.W.; Huang, W.C.; Lu, C.C.; Chiang, J.H.; Peng, S.F.; Chung, J.-G.; Lin, Y.-H.; Hsu, Y.-M.; Amagaya, S.; et al. ERK-modulated intrinsic signaling and G(2)/M phase arrest contribute to the induction of apoptotic death by allyl isothiocyanate in MDA-MB-468 human breast adenocarcinoma cells. Int. J. Oncol. 2012, 41, 2065–2072. [Google Scholar] [CrossRef]
- Nagata, M.; Kurita, H.; Uematsu, K.; Ogawa, S.; Takahashi, K.; Hoshina, H.; Takagi, R. Diagnostic value of cyclin-dependent kinase/cyclin-dependent kinase inhibitor expression ratios as biomarkers of locoregional and hematogenous dissemination risks in oral squamous cell carcinoma. Mol. Clin. Oncol. 2015, 3, 1007–1013. [Google Scholar] [CrossRef]
- Xiao, C.; Fan, T.; Tian, H.; Zheng, Y.; Zhou, Z.; Li, S.; Li, C.; He, J. H3K36 trimethylation-mediated biological functions in cancer. Clin. Epigenetics 2021, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Natesan, R.; Sanchez-Rivera, F.J.; Li, J.; Bhanu, N.V.; Yamazoe, T.; Lin, J.H.; Merrell, A.J.; Sela, Y.; Thomas, S.K.; et al. Global Regulation of the Histone Mark H3K36me2 Underlies Epithelial Plasticity and Metastatic Progression. Cancer Discov. 2020, 10, 854–871. [Google Scholar] [CrossRef] [PubMed]
- Cloos, P.A.; Christensen, J.; Agger, K.; Helin, K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes. Dev. 2008, 22, 1115–1140. [Google Scholar] [CrossRef] [PubMed]
- Tarar, A.; Peng, S.; Cheema, S.; Peng, C.-A. Anticancer Activity, Mechanism, and Delivery of Allyl Isothiocyanate. Bioengineering 2022, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. From Human to Mouse and Back: ‘Tumorgraft’ Models Surge in Popularity. JNCI J. Natl. Cancer Inst. 2009, 101, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Swick, A.D.; Stein, A.P.; McCulloch, T.M.; Hartig, G.K.; Ong, I.M.; Sampene, E.; Prabakaran, P.J.; Liu, C.Z.; Kimple, R.J. Defining the boundaries and expanding the utility of head and neck cancer patient derived xenografts. Oral Oncol. 2017, 64, 65–72. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).