Abstract
(1) Background: Sepsis is present in nearly 90% of critically ill patients with community-acquired pneumonia (CAP). This systematic review updates the information on studies that have assessed gene expression profiles in critically ill septic patients with CAP. (2) Methods: We searched for studies that satisfied the following criteria: (a) expression profile in critically ill patients with sepsis due to CAP, (b) presence of a control group, and (c) adult patients. Over-representation analysis was performed with clusterProfiler using the Hallmark and Reactome collections. (3) Results: A total of 4312 differentially expressed genes (DEGs) and sRNAs were included in the enrichment analysis. In the Hallmark collection, genes regulated by nuclear factor kappa B in response to tumor necrosis factor, genes upregulated by signal transducer and activator of transcription 5 in response to interleukin 2 stimulation, genes upregulated in response to interferon-gamma, genes defining the inflammatory response, a subgroup of genes regulated by MYC—version 1 (v1), and genes upregulated during transplant rejection were significantly enriched in critically ill septic patients with CAP. Moreover, 88 pathways were identified in the Reactome database. (4) Conclusions: This study summarizes the reported DEGs in critically ill septic patients with CAP and investigates their functional implications. The results highlight the complexity of immune responses during CAP.
1. Introduction
Community-acquired pneumonia (CAP) remains one of the most common infectious diseases and a major cause of mortality worldwide [1]. CAP is defined as an acute infection of the lungs (alveoli and distal airways) acquired outside hospitals or long-term care facilities. Typical clinical manifestations included fever, cough, mucopurulent sputum, pleuritic chest pain, and dyspnea [2,3]. Acute respiratory distress syndrome, sepsis, pleural effusion, and worsened preexisting comorbidities are complications of CAP. Viruses and bacteria are the most frequent etiology. Studies have found that influenza is the most common cause of viral CAP, and Streptococcus pneumoniae, Klebsiella pneumoniae, Haemophilus influenzae, anaerobes, and atypical bacteria (i.e., Mycoplasma pneumoniae and Legionella species) represent common causes of bacterial CAP [2]. A considerable number of factors related to sociodemographic characteristics, clinical manifestations or complications, and causative pathogens can affect mortality in CAP patients [4]. Similarly, studies have shown that an inappropriate inflammatory response is a major cause of treatment failure and mortality in patients with CAP [5,6,7]. Kellum et al. [7] found that high levels of both proinflammatory and anti-inflammatory cytokines were related to higher risk of mortality. The overall mortality rate of CAP has changed little in recent decades, and in patients admitted to intensive care units (ICU), it continues to exceed 20% [8,9].
Moreover, sepsis is a medical emergency characterized by a dysregulated host inflammatory and immune response to an infectious process that results in organ injury and death [10]. Studies have documented that the incidence of sepsis has increased continuously in recent decades and can be attributed to different factors, such as the aging of the population, resistance to antibiotics, the greater number of immunosuppressed patients, and the increased need for invasive procedures [10,11]. Despite an increase in knowledge of the pathophysiological characteristics of sepsis, the ability to intervene and modify the evolution of the disease has not been entirely satisfactory. Patients who develop sepsis during an infectious process have a high risk of mortality and represent about 20% of all global deaths [10,12].
Sepsis is common in patients with CAP; it is present in more than 50% of those admitted to the ward and in nearly 90% of patients treated in the ICU [13]. Sepsis and organ dysfunction in patients with CAP are risk factors for poor outcomes, especially in patients who present with septic shock or require mechanical ventilation [9,13].
Timely initiation of appropriate broad-spectrum antibiotic therapy is the cornerstone of treatment for CAP and sepsis. Current guidelines recommend a β-lactam plus a macrolide or fluoroquinolone monotherapy as empiric antibiotic therapy for hospitalized patients with CAP. Other recommendations to take into account in the management of CAP and sepsis include the appropriate use of fluids, intensive support of organ dysfunction, control of the infectious focus, nutritional support, glucose management, serial analytical monitoring, and corticosteroids [3,14]. In recent years, new therapeutic interventions for CAP and sepsis have been evaluated. In this regard, studies have documented that mesenchymal stem cells have immunomodulatory properties in various diseases. These cells also been described as having angiogenic, antiapoptotic, antibacterial and tissue repair activity [15,16]. The above characteristics have made mesenchymal stem cells candidates for the treatment of CAP and sepsis [15]. A meta-analysis of preclinical studies found that mesenchymal stem cell therapy was associated with reduced mortality in animal models of sepsis [16]. Recently, several clinical phase 1/2 trials have evaluated mesenchymal stem cell treatment in critically ill patients with acute respiratory distress syndrome (ARDS) and sepsis. These studies found that mesenchymal stem cell therapy is safe, although the number of patients who have received therapy to date is small [17].
Information derived from gene expression studies may improve our understanding of the immune response during sepsis and CAP and may help to identify new pathways or candidate biomarkers and targets for possible interventions. Recently, interesting studies on the blood transcriptome of patients with sepsis and CAP have been published [18,19,20]. Gene expression profiling in septic patients has revealed differential regulation compared to healthy volunteers or non-septic patients with CAP. Genes related to host defenses and inflammatory responses have been found to be upregulated during sepsis. Furthermore, gene expression studies have allowed the stratification of CAP as a heterogeneous disease into subtypes with possible diagnostic and/or prognostic impacts [21].
This systematic review updates the information on studies that have evaluated the expression profiles in critically ill septic patients with CAP, focusing on identifying which genes are differentially expressed in these patients compared to controls (i.e., critically ill adult patients without sepsis, healthy adults, or others).
2. Materials and Methods
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [22]. We searched the PubMed/MEDLINE, Cochrane Central Register of Controlled Trials (CENTRAL), and LILACS (Literatura Latino-Americana e do Caribe de Informação em Ciências da Saúde) databases from inception to December 2021 for studies that satisfied the following eligibility criteria: (a) expression profile in critically ill patients with sepsis due to CAP, (b) presence of a control group, and (c) adult age. Our search also identified relevant publications from the references of the articles included. The search terms used were “community-acquired pneumonia”, “sepsis”, “intensive care unit”, “gene expression”, and “transcriptome”. We included observational studies (cohort and case-control studies) in English and Spanish, excluding studies on animals and pediatric populations, abstracts, and case reports (<5 cases). Likewise, patients with SARS-CoV-2 disease (COVID-19) were also excluded. The study was registered with the International Registry of Systematic Reviews/Meta-Analyses (PROSPERO CRD42022342685).
Two pairs of reviewers (DV, CR, FF, and CD) assessed titles and abstracts for inclusion. Eligibility of these article was determined by independent review of the full text. If any disagreement occurred, it was resolved by another reviewer (JC). The following information was extracted from the included articles and collected on a standardized form created for the review: author, year, journal, country in which the study was conducted, original study design, sample size, sample size of the subgroup, tissue investigated, methods for analyzing gene expression changes, and differentially expressed genes comparing patients with sepsis due to CAP and a comparator group. A modified Quality of Genetic Association Studies (Q-Genie) tool was used for critical appraisal of the included studies [23]. Two reviewers (CR and DV) independently performed these quality assessments. If any disagreement occurred, it was resolved by consensus between the reviewers.
Statistical Analysis
A description was made on the basis of the data extracted and main findings of each study. Datasets were assessed for differentially expressed genes (DEGs) and sRNAs between critically ill patients with sepsis due to CAP and controls. All DEGs were filtered to have an absolute fold change (FC) > 1.7 and p value < 0.05, based on the maximum FC threshold reported by Severino et al. [24]. Over-representation analysis (ORA) was performed with clusterProfiler [25] using Hallmark and Reactome (C2.Reactome) collections from the MSigDB (v. 2023). Gene sets from a collection were considered significative if adj.p.val < 0.05. Analyses were performed in R [26]. Table S1 shows the codes used in R to analyze the data.
3. Results
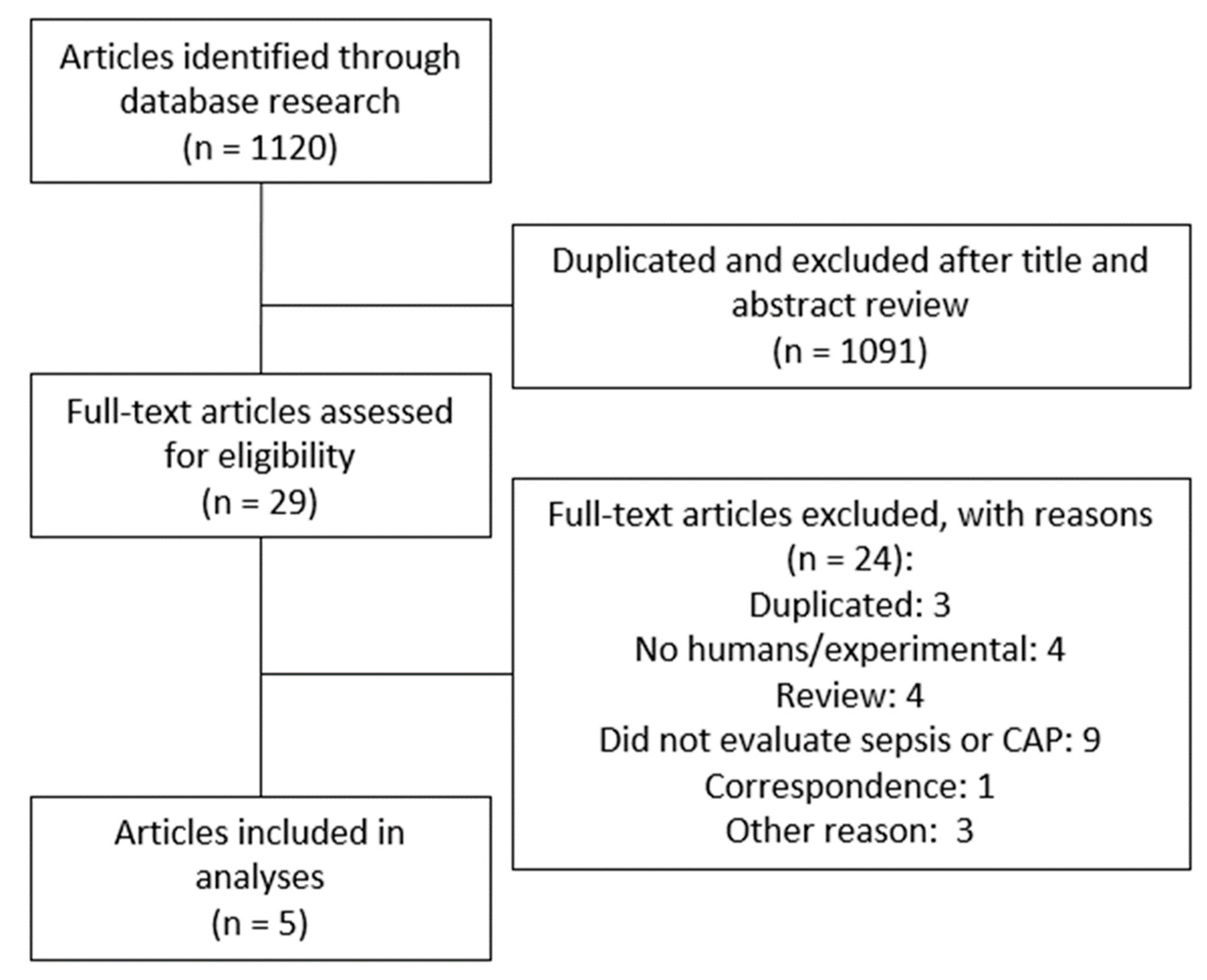
Figure 1 shows the PRISMA flowchart describing the process of the systematic literature search to identify all eligible studies. We identified and screened 1120 papers. A total of 1091 articles were excluded because they did not meet the inclusion criteria based on title and abstract or duplication. Therefore, 29 articles were considered relevant for full-text review and eligibility, of which five were included in this systematic review [24,27,28,29,30].
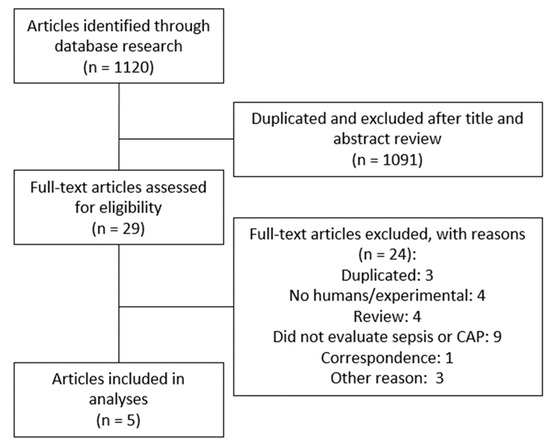
Figure 1.
Flow diagram of the study selection process.
The main characteristics of the studies included are summarized in Table 1. In all, there were 231 critically ill patients with sepsis due to CAP and 71 healthy controls. Studies investigated expression in whole or peripheral blood leukocytes and monocytes. Three of the studies used gene expression microarrays, one using next-generation RNA sequencing (RNA-Seq) studied small RNA, and the other estimated the expression of 35 genes involved in NLR inflammasome pathways using real-time polymerase chain reaction (RT-PCR). The quality assessment scores of the included studies are found in Table S2. Total quality assessment scores ranged between 52 and 56 (median = 53).

Table 1.
Main characteristics of the studies.
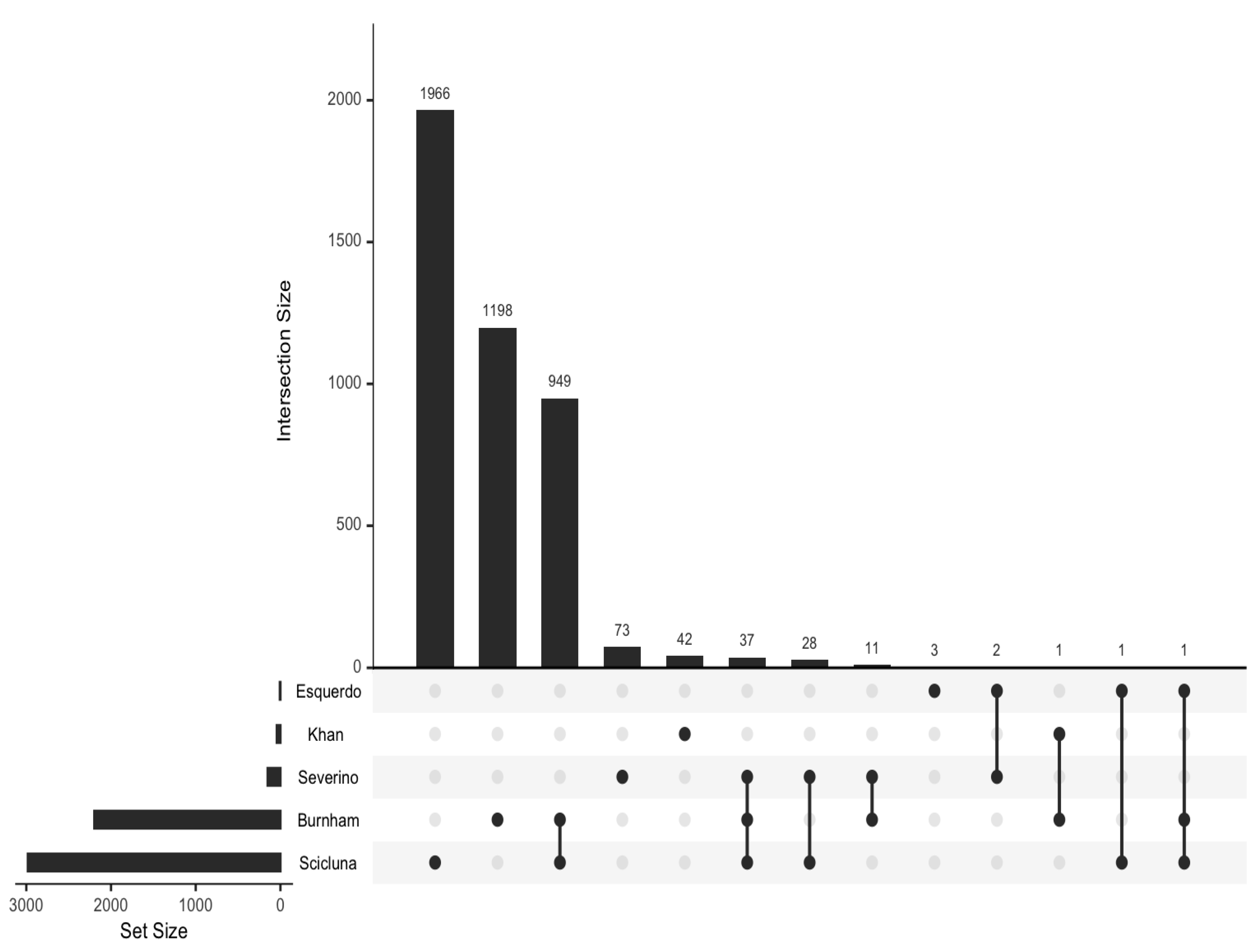
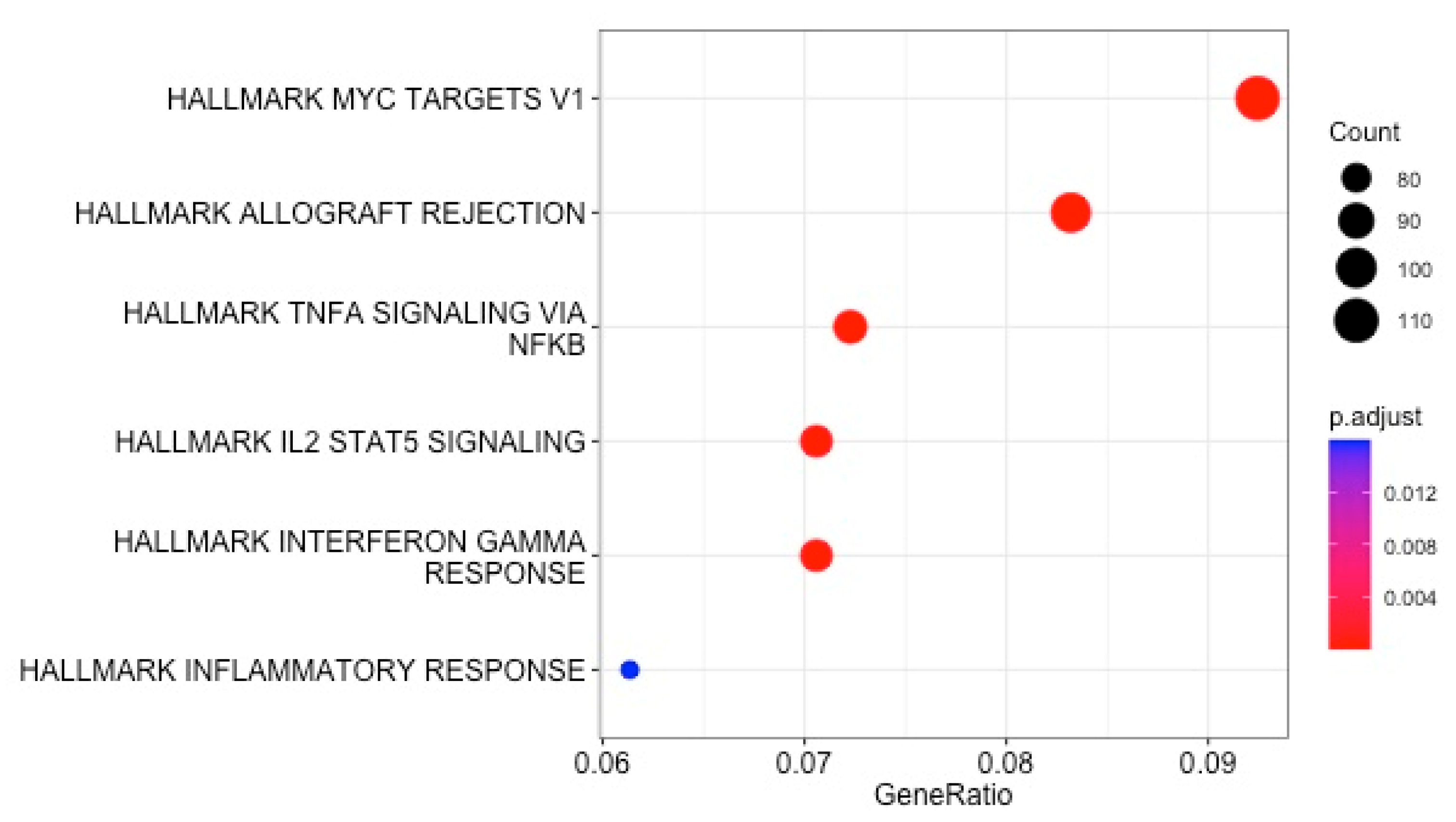
All 4312 reported DEGs and small RNAs (FC > 1.7 and p value < 0.05) (Table S3) were included in an enrichment analysis using the Hallmark and Reactome gene set collections from the Molecular Signatures Database (mSigDB) (v. 2023). Minimal overlap of the DEGs from the included studies was noted (Figure 2). Regarding the Hallmark gene set collection, the following DEGs were significantly over-represented in critically ill septic patients with CAP: genes regulated by nuclear factor kappa B (NF-kB) in response to tumor necrosis factor (TNF), genes upregulated by signal transducer and activator of transcription 5 (STAT5) in response to interleukin 2 (IL2) stimulation, genes upregulated in response to interferon-gamma (IFN-γ), genes defining an inflammatory response, a subgroup of genes regulated by MYC—version 1 (v1), and genes upregulated during transplant rejection (Table 2 and Figure 3).
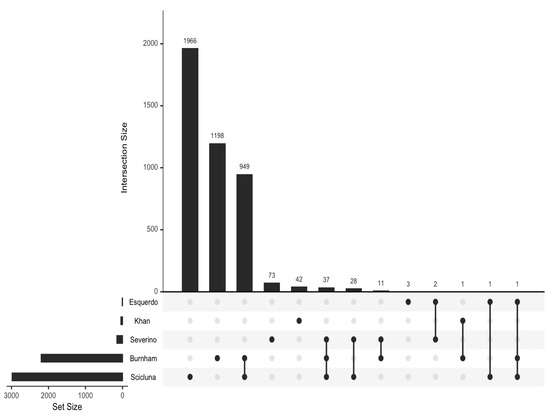
Figure 2.
UpSet plot to visualize the overlap between sets of differentially expressed genes.

Table 2.
Gene set enrichment Hallmark analysis of differentially expressed genes in critically ill septic patients with community-acquired pneumonia.
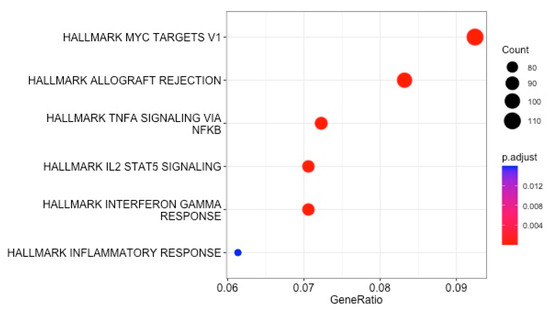
Figure 3.
Enrichment analysis using the Hallmark collection from the MSigDB: dot plot of enriched gene sets.
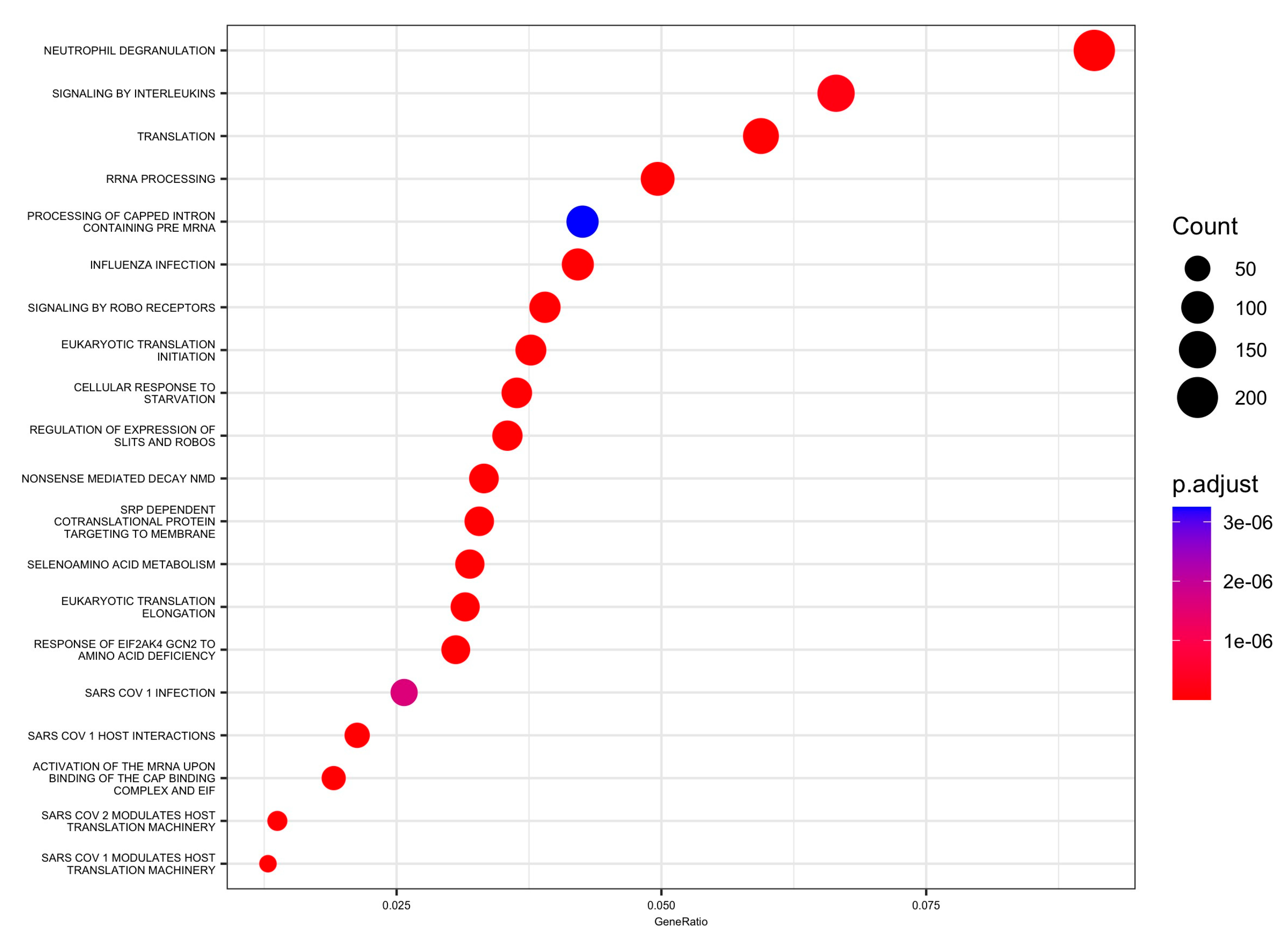
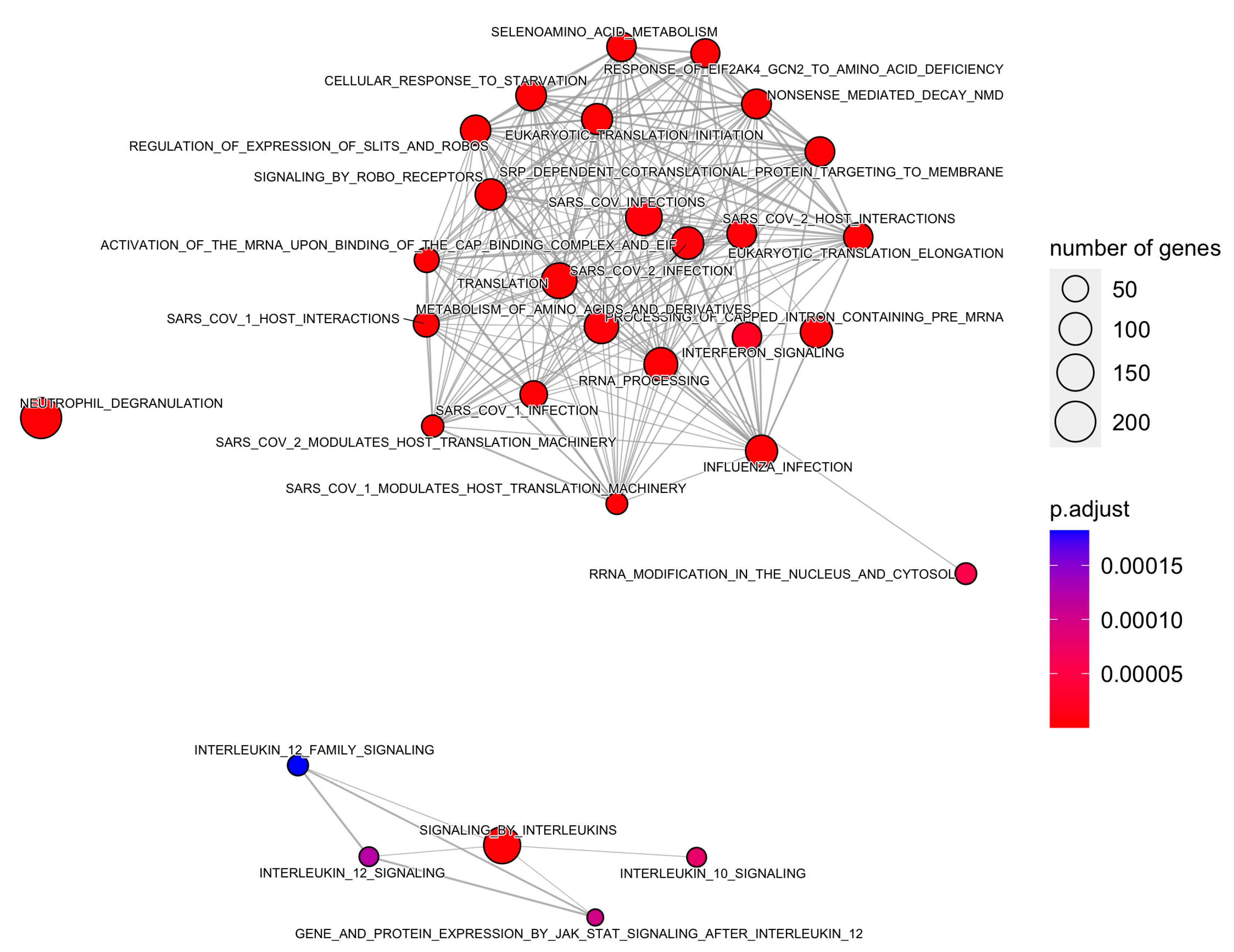
Moreover, 88 pathways in the Reactome database were identified with DEGs and small RNAs. The top enriched Reactome pathways were annotated as related to translation, neutrophil degranulation, influenza infection, rRNA processing, SRP-dependent cotranslational protein targeting to the membrane, nonsense-mediated decay (NMD), response of EIF2AK4 (GCN2) to amino acid deficiency, selenoamino acid metabolism, and others (Table 3 and Figure 4 and Figure 5).

Table 3.
Gene set enrichment Reactome analysis of differentially expressed genes in critically ill septic patients with community-acquired pneumonia.
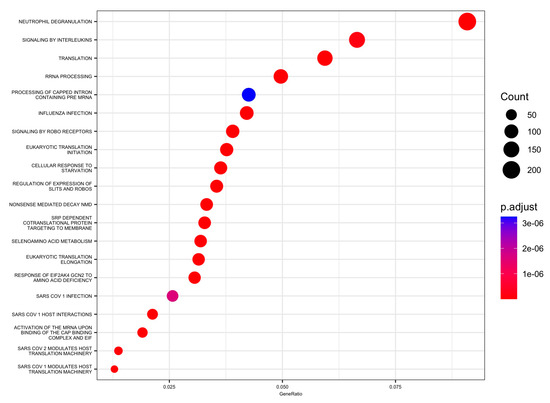
Figure 4.
Enrichment using the Reactome collection from the MSigDB: top 20 significant pathways dot plot.
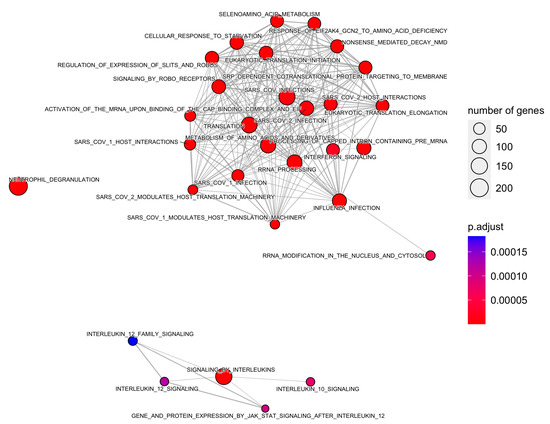
Figure 5.
Enrichment map of Reactome pathways using a minimum similarity score of 0.1.
4. Discussion
This is a systematic review of gene expression studies that have investigated critically ill septic patients with CAP compared to control groups (healthy adults or others). We have listed all reported DEGs in critically ill septic patients with CAP and have investigated the functional implications of these genes. The functional analyses summarize the molecular mechanisms of the inflammatory response in CAP found in those studies. Our functional analyses of DEGs were related to genes regulated by NF-kB in response to TNF, genes upregulated in response to IFN-gamma, genes defining the inflammatory response, genes upregulated by STAT5 in response to IL2 stimulation, a subgroup of regulated genes with MYC—version 1 (v1), and genes upregulated during transplant rejection.
Sepsis, a life-threatening condition, is one of the main complications of CAP. Mortality associated with CAP remains high, particularly in cases requiring ICU admission [31], even when adequate antibiotic therapy has been provided. The host immune response to sepsis can be altered in several ways, and previous studies have highlighted the complexity of this response during acute inflammation and subsequent resolution of pneumonia [32,33]. A variety of biomarkers have also been investigated, alone or in combination, for the purposes of diagnosis, etiology, mortality, and guiding antibiotic therapy, but none is ideal. Transcriptomic, proteomic, and metabolomic studies of host immune response or biomarker signatures have shown encouraging preliminary results [32].
Regarding the gene sets found in the functional analysis of the present review, studies have documented that the transcription factor MYC may be an important regulatory gene in the dysfunction of sepsis or acute respiratory distress syndrome (ARDS) secondary to sepsis [34]. On the other hand, NF-kB is an important transcriptional regulator during the process of inflammation and injury and plays a role in inflammatory disorders since it modulates the response of immunoregulatory genes, such as cytokines and chemokines, cell adhesion molecules, and antimicrobial peptides [35]. Experimental studies in animals have shown that regulation of the NF-kB signaling pathway reduces lung inflammation during pneumonia [36,37]. STAT5 signaling plays a critical role in the maintenance of lung homeostasis [38], and IFN-γ produced during pneumonia induces the transcription of target genes in the lungs, which are critical for host defenses. Studies have shown that the production of IFN-γ early during pneumonia regulates bacterial clearance [39].
In addition, 88 pathways were identified in the Reactome database. These pathways were related to translation, neutrophil degranulation, influenza infection, SARS-CoV infections, signaling by interleukins and related cytokines, IFN signaling, gene and protein expression by JAK-STAT signaling, regulation of expression of SLITs and ROBOs, immunoregulatory interactions between lymphoid and non-lymphoid cells, and others. In this regard, neutrophils are essential in the defense against invading microorganisms. Neutrophils have different tools to kill invading microbes, including distinct granule subsets that contain a variety of antimicrobial peptides and enzymes [40]. However, neutrophil granule proteins also are directly responsible for lung damage. In mice, the Streptococcus pyogenes M1 protein causes degranulation of all neutrophil granule subsets, leading to the accumulation of neutrophils in lung tissue, and has also been linked to increased vascular permeability and acute lung damage [41]. Moreover, cytokines are also important mediators of the innate and acquired immune response. The main functions of cytokines during an infection are cell differentiation, chemotaxis, and the regulation of inflammatory and anti-inflammatory processes. During CAP, a systemic and local inflammatory response occurs, and the number of cytokines identified and the knowledge of their functions have increased over the years. Cytokines are major players in cases of severe lung infection [42]. In addition, the Slit-induced signaling pathway is a modulator of vascular stability. Activation of this pathway reduces capillary leakage, multiorgan edema, and death in multiple animal models of infections. One experimental study found that administration of the Slit2N ligand strengthens the endothelial barrier and attenuates vascular leakage in response to the cytokine storm [43]. Furthermore, studies have suggested that HSF1 is required to initiate the host defense against bacterial infection through early activation of TLR2 signaling. HSF1(−/−) mice had a higher lung bacterial load and delayed associated inflammation compared to HSF1(+/+) mice [44].
Gene expression studies in patients with CAP have not only allowed a better understanding of the immune response but could also have applications in clinical practice. These studies have evaluated the utility of gene expression in stratifying patients into subtypes according to the prognosis of CAP, determining the fingerprints of the host response to viruses and bacteria that might be used for etiological diagnosis, and proposing new biomarkers that might be suitable for diagnosis or the development of therapies. Studies with these objectives were not included in the present systematic review. Davenport et al. [21] found that transcriptomic profiling of circulating peripheral blood leukocytes from CAP patients defined two individual sepsis response signatures (SRS1 and SRS2) related with outcomes. SRS1 was related to a higher risk of early mortality than SRS2 (14-day mortality 22% vs. 10%). The investigators documented that SRS1 was associated with an immunosuppressed phenotype. The features of this group of patients included T-cell exhaustion, endotoxin tolerance, and downregulation of human leukocyte antigen (HLA) class II. Similarly, Severino et al. [24] evaluated patterns of gene expression in blood mononuclear cells from patients with CAP and sepsis. They found that differences in oxidative phosphorylation were associated with prognosis. In addition, gene expression profiles also differed between patients who died and survived, with decreased expression of genes related to immune functions. In another study, comparison of gene expression profiles between 185 surviving and 13 non-surviving hospitalized CAP patients yielded 49 DEGs [18]. Gene set enrichment analysis found four positively enriched gene sets in survivors and seven positively enriched gene sets in the patients who died. These gen sets were related with the interferon-alpha response, apoptosis, sex hormone pathways, oxidative stress, endoplasmic reticulum stress, oxidative phosphorylation, and angiogenesis pathways.
In addition, another study reported that the whole-blood gene-expression profile of pneumonia caused by influenza A(H1N1) differed notably from those of bacterial pneumonia [45]. A total of 1416 genes were uniquely upregulated in influenza A (H1N1) pneumonia. Analysis of biological pathways revealed over-representation related to the cell cycle and its regulation, DNA damage response, apoptosis, and protein degradation. Similarly, a study evaluated gene expression using RNASeq and qPCR to discriminate bacterial from non-bacterial infectious agents in adults with lower respiratory tract infection [46]. For molecular analyses, 41 subjects were considered to have a bacterial infection, and 53 subjects were classified with a non-bacterial infection. Influenza A was the most common virus, and Streptococcus pneumoniae was the most common bacteria documented. The investigators found that lymphocyte, α-linoleic acid metabolism, and IGF regulation pathways including eleven genes were markers for distinguishing bacterial infection. Interestingly, Pereverzeva et al. [19] performed a study to evaluate host response biomarkers and transcriptomes between Gram-positive and Gram-negative bacteria in CAP patients in the ICU. There was no differences in blood leukocyte transcriptomes in patients with Gram-positive and Gram-negative causative pathogens.
Transcriptional expression studies have also been used to propose new biomarkers in CAP. One study has proposed the FAIM3:PLAC8 ratio as a candidate biomarker to aid in the diagnosis of CAP in patients requiring ICU admission. The area under the curve (0.845, 95% confidence interval: 0.764–0.917) showed that the FAIM3:PLAC8 ratio was better in discriminating between patients with CAP and without CAP compared to procalcitonin, IL8, and IL6 [27]. Likewise, an experimental study carried out in murine models of lung infection identified in its transcriptional analysis an interferon signature associated with S. pneumoniae infection. Likewise, the chemokines CXCL9 and CXCL10 had the best sensitivity, specificity, and predictive power to differentiate between S. pneumoniae and Staphylococcus aureus pneumonia [47]. Other investigators analyzed both protein-coding mRNA and regulatory miRNAs in peripheral blood mononuclear cells in order to identify markers that may be suitable as diagnostic biomarkers between CAP and acute exacerbations of chronic obstructive pulmonary disease (COPD) [48]. A module of 120 genes was particularly suitable to discriminate acute exacerbations of COPD and CAP and identified HNF4A, MCC, and MUC1 as the most important discriminatory markers.
COVID-19 is a recently recognized illness, characterized as a predominantly respiratory disease that can lead to pneumonia and ARDS. Although patients with COVID-19 were excluded from the present review, several studies have evaluated gene expression profiles of this disease [49,50,51]. The gene expression profiles have been obtained from various types of samples, including whole blood; blood components, such as peripheral blood mononuclear cells, monocytes or T cells; bronchoalveolar lavage fluid; and autopsy samples. A recent review performed an analysis of the enriched pathways in COVID-19, and the main ones found were those related to regulatory aspects of the immune system. The most common pathways were associated with cytokines, especially interferons. Other pathways found were related to the immune response against bacteria, cell division and the cell cycle, organization of the extracellular matrix, exocytosis and hemostasis [49]. Moreover, a study determined the transcriptomic profile of patients with COVID-19 who required hospital admission and described those patients who developed severe disease [52]. The researchers found that a dysregulated inflammatory response was the most important factor related to severe pneumonia in SARS-CoV-2 infection. There was increased gene expression related to the pro-inflammatory state and activation of neutrophils and macrophages, in addition to a loss of immune regulation. Additionally, increased expression of genes linked to reactive oxygen species, metalloproteinases, and protein polyubiquitination was documented.
Other studies have evaluated circulating microRNAs (miRNAs) in patients with CAP or sepsis. miRNAs can regulate gene expression and are widely involved in the inflammatory response and immune regulation in infectious processes. However, miRNA research still faces several challenges, such as low sensitivity, specificity, and silencing efficiency; off-target effects; and toxic reactions [53]. Galván-Román et al. [54] assessed the usefulness of miRNA expression as a prognostic biomarker in patients with CAP who required hospital admission. High levels of miR-146a-5p and miR-16-5p were markers of good prognosis and associated with lower 30-day mortality. Likewise, another study documented that free miRNAs could be useful for diagnosis in patients with CAP or sepsis and predicting the severity of the disease [55]. miR-1246 levels were higher in patients with severe disease, while miR-193a-5p and miR-542-3p distinguished between infection (CAP or sepsis) and healthy volunteers. Interestingly, other researchers compared differential miRNA profiles between COVID-19 and CAP [56]. A signature of 15 dysregulated miRNAs was found between patients with COVID-19 and CAP. Furthermore, 4 miRNAs (miR-106b-5p, miR-221-3p, miR-25-3p, and miR-30a-5p) significantly discriminated between both diseases, with a sensitivity of 93.7% and a specificity of 89%. Regarding the 15 dysregulated miRNAs, the enriched pathways were significantly associated with angiogenesis, regulation of endothelial cells or vasodilatation, cardiac muscle cell differentiation and proliferation, leukocyte adhesion, IL6-mediated signaling, Th1 response, macrophage differentiation and MyD88-dependent Toll-like receptor signaling.
The strength of this systematic review is that only studies evaluating expression in a specific population (critical adults) and infection (CAP) were included. This means, however, that the findings are unlikely to be valid for septic patients with other etiologies or CAP patients without sepsis. The limitations of this systematic review include the exclusion of studies that were not published in English or Spanish and studies that investigated animal models or cell lines. The studies included were small and did not report sample size estimation or power analysis; the studies did not state whether statistical corrections were used for multiple testing. Some of the studies were performed in a subgroup of patients from a larger cohort but used a different methodology to determine transcriptomic analysis. For example, Khan et al. [30] assessed small RNAs, whereas Esquerdo et al. [29] studied just 35 genes using RT-PCR. However, an analysis of studies included with the cohorts using microarrays showed similar results (Figures S1 and S2).
5. Conclusions
This systematic review summarizes all reported DEGs in critically ill septic patients with CAP and investigates their functional implications. The results highlight the complexity of the immune response during CAP. Gene expression studies in patients with CAP not only allow a better understanding of the immune response but may also have applications in clinical practice. In this regard, future trials should evaluate the role of gene expression for diagnosis and personalized treatment approaches in CAP.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines11102755/s1. Table S1: Codes used in R to analyze the data; Table S2: Quality assessment of included studies using the modified ‘Q-Genie’ assessment tool; Table S3: Differentially expressed genes in the studies; Figure S1: Enrichment analysis using the Hallmark collection from the MSigDB: dot plot of enriched gene sets from studies included with the cohorts using microarrays; Figure S2: Enrichment using the Reactome collection from the MSigDB: top 20 significant pathways dot plot of studies included with the cohorts using microarrays.
Author Contributions
Conceptualization, D.V. and J.C.; Data curation, D.V., C.R., F.F. and C.D.-M.; Formal analysis: L.N.; Funding acquisition: J.C.; Supervision: D.V., L.N., C.R. and J.C.; Writing—original draft: D.V. and C.R.; Writing—review & editing: L.N., F.F., C.D.-M. and J.C. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Instituto de Salud Carlos III (research grants 11/01106, 14/00580, and 17/01332). CIBERINFEC (CB21/13/00009), Instituto de Salud Carlos III, Madrid, Spain. We thank the CERCA Programme/Generalitat de Catalunya for institutional support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Metlay, J.P.; Waterer, G.W.; Long, A.C.; Anzueto, A.; Brozek, J.; Crothers, K.; Cooley, L.A.; Dean, N.C.; Fine, M.J.; Flanders, S.A.; et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care. Med. 2019, 200, e45–e67. [Google Scholar] [CrossRef] [PubMed]
- Lutfiyya, M.N.; Henley, E.; Chang, L.F.; Reyburn, S.W. Diagnosis and Treatment of Community-Acquired Pneumonia. Am. Fam. Physician 2006, 73, 442–450. [Google Scholar] [PubMed]
- Womack, J.; Kropa, J. Community-Acquired Pneumonia in Adults: Rapid Evidence Review. Am. Fam. Physician 2022, 105, 625–630. [Google Scholar] [PubMed]
- Viasus, D.; Cillóniz, C.; Cardozo, C.G.; Puerta, P.; Garavito, A.; Torres, A.; Garcia-Vida, C. Early, short and long-term mortality in community-acquired pneumonia. Ann. Res. Hosp. 2018, 2, 5. [Google Scholar] [CrossRef]
- Siljan, W.W.; Holter, J.C.; Nymo, S.H.; Husebye, E.; Ueland, T.; Aukrust, P.; Mollnes, T.E.; Heggelund, L. Cytokine responses, microbial aetiology and short-term outcome in community-acquired pneumonia. Eur. J. Clin. Investig. 2018, 48, e12865. [Google Scholar] [CrossRef]
- Van Vught, L.A.; Scicluna, B.P.; Wiewel, M.A.; Hoogendijk, A.J.; Klein Klouwenberg, P.M.; Franitza, M.; Toliat, M.R.; Nürnberg, P.; Cremer, O.L.; Horn, J.; et al. Comparative Analysis of the Host Response to Community-acquired and Hospital-acquired Pneumonia in Critically Ill Patients. Am. J. Respir. Crit. Care. Med. 2016, 194, 1366–1374. [Google Scholar] [CrossRef]
- Kellum, J.A.; Kong, L.; Fink, M.P.; Weissfeld, L.A.; Yealy, D.M.; Pinsky, M.R.; Fine, J.; Krichevsky, A.; Delude, R.L.; Angus, D.C.; et al. Understanding the inflammatory cytokine response in pneumonia and sepsis. Arch. Intern. Med. 2007, 167, 1655–1663. [Google Scholar] [CrossRef]
- Simonetti, A.F.; Garcia-Vidal, C.; Viasus, D.; García-Somoza, D.; Dorca, J.; Gudiol, F.; Carratalà, J. Declining mortality among hospitalized patients with community-acquired pneumonia. Clin. Microbiol. Infect. 2016, 22, 567.e1–567.e7. [Google Scholar] [CrossRef]
- Niederman, M.S.; Torres, A. Severe community-acquired pneumonia. Eur. Respir. Rev. 2022, 31, 220123. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Moldawer, L.L.; Opal, S.M.; Reinhart, K.; Turnbull, I.R.; Vincent, J.L. Sepsis and septic shock. Nat. Rev. Dis. Primers 2016, 2, 16045. [Google Scholar] [CrossRef]
- Guarino, M.; Perna, B.; Cesaro, A.E.; Maritati, M.; Spampinato, M.D.; Contini, C.; De Giorgio, R. 2023 Update on Sepsis and Septic Shock in Adult Patients: Management in the Emergency Department. J. Clin. Med. 2023, 12, 3188. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, B.; Ramakrishna, K.; Dhamoon, A.S. Sepsis: The evolution in definition, pathophysiology, and management. SAGE Open. Med. 2019, 7, 2050312119835043. [Google Scholar] [CrossRef] [PubMed]
- Ceccato, A.; Torres, A. Sepsis and community-acquired pneumonia. Ann. Res. Hosp. 2018, 2, 7. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Crit. Care Med. 2021, 47, 1181–1247. [Google Scholar]
- Wang, C.; Zhao, D.; Zheng, L.; Bao, X.; Yang, Q.; Jiang, S.; Zhou, X.; Tang, L.; Liu, Z. Safety and efficacy of human umbilical cord mesenchymal stem cells for the treatment of sepsis induced by pneumonia: Study protocol for a single-centre, randomised single-blind parallel group trial. BMJ Open. 2022, 12, e058444. [Google Scholar] [CrossRef]
- Sun, X.Y.; Ding, X.F.; Liang, H.Y.; Zhang, X.J.; Liu, S.H.; Han, B.; Duan, X.G.; Sun, T.W. Efficacy of mesenchymal stem cell therapy for sepsis: A meta-analysis of preclinical studies. Stem Cell Res. Ther. 2020, 11, 214. [Google Scholar] [CrossRef]
- Masterson, C.H.; Ceccato, A.; Artigas, A.; Dos Santos, C.; Rocco, P.R.; Rolandsson Enes, S.; Weiss, D.J.; McAuley, D.; Matthay, M.A.; English, K.; et al. Mesenchymal stem/stromal cell-based therapies for severe viral pneumonia: Therapeutic potential and challenges. Intensive Care Med. Exp. 2021, 9, 61. [Google Scholar] [CrossRef]
- Viasus, D.; Simonetti, A.F.; Nonell, L.; Vidal, O.; Meije, Y.; Ortega, L.; Arnal, M.; Bódalo-Torruella, M.; Sierra, M.; Rombauts, A.; et al. Whole-Blood Gene Expression Profiles Associated with Mortality in Community-Acquired Pneumonia. Biomedicines 2023, 11, 429. [Google Scholar] [CrossRef]
- Pereverzeva, L.; Uhel, F.; Peters Sengers, H.; Butler, J.; van Vught, L.A.; Burnham, K.L.; Davenport, E.E.; Knight, J.C.; Cremer, O.L.; Schultz, M.J.; et al. Blood leukocyte transcriptomes in Gram-positive and Gram-negative community-acquired pneumonia. Eur. Respir. J. 2022, 59, 2101856. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Burnham, K.L.; Goh, C.; Allcock, A.; Malick, Z.H.; Overend, L.; Kwok, A.; Smith, D.A.; Peters-Sengers, H.; Antcliffe, D.; et al. An immune dysfunction score for stratification of patients with acute infection based on whole-blood gene expression. Sci. Transl. Med. 2022, 14, eabq4433. [Google Scholar] [CrossRef]
- Davenport, E.E.; Burnham, K.L.; Radhakrishnan, J.; Humburg, P.; Hutton, P.; Mills, T.C.; Rautanen, A.; Gordon, A.C.; Garrard, C.; Hill, A.V.; et al. Genomic landscape of the individual host response and outcomes in sepsis: A prospective cohort study. Lancet Respir. Med. 2016, 4, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.; Rajkumar, A.P. Systematic review of gene expression studies in people with Lewy body dementia. Acta Neuropsychiatr. 2020, 32, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; Silva, E.; Baggio-Zappia, G.L.; Brunialti, M.K.; Nucci, L.A.; Rigato, O., Jr.; da Silva, I.D.; Machado, F.R.; Salomao, R. Patterns of gene expression in peripheral blood mononuclear cells and outcomes from patients with sepsis secondary to community acquired pneumonia. PLoS. ONE 2014, 9, e91886. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 30 May 2023).
- Scicluna, B.P.; Klein Klouwenberg, P.M.; van Vught, L.A.; Wiewel, M.A.; Ong, D.S.; Zwinderman, A.H.; Franitza, M.; Toliat, M.R.; Nürnberg, P.; Hoogendijk, A.J.; et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am. J. Respir. Crit. Care Med. 2015, 192, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Burnham, K.L.; Davenport, E.E.; Radhakrishnan, J.; Humburg, P.; Gordon, A.C.; Hutton, P.; Svoren-Jabalera, E.; Garrard, C.; Hill, A.V.S.; Hinds, C.J.; et al. Shared and Distinct Aspects of the Sepsis Transcriptomic Response to Fecal Peritonitis and Pneumonia. Am. J. Respir. Crit. Care Med. 2017, 196, 328–339. [Google Scholar] [CrossRef]
- Esquerdo, K.F.; Sharma, N.K.; Brunialti, M.K.C.; Baggio-Zappia, G.L.; Assunção, M.; Azevedo, L.C.P.; Bafi, A.T.; Salomao, R. Inflammasome gene profile is modulated in septic patients, with a greater magnitude in non-survivors. Clin. Exp. Immunol. 2017, 189, 232–240. [Google Scholar] [CrossRef]
- Khan, H.N.; Jongejan, A.; van Vught, L.A.; Horn, J.; Schultz, M.J.; Zwinderman, A.H.; Cremer, O.L.; Bonten, M.J.; van der Poll, T.; Scicluna, B.P. The circulatory small non-coding RNA landscape in community-acquired pneumonia on intensive care unit admission. J. Cell Mol. Med. 2021, 25, 7621–7630. [Google Scholar] [CrossRef]
- Walden, A.P.; Clarke, G.M.; McKechnie, S.; Hutton, P.; Gordon, A.C.; Rello, J.; Chiche, J.D.; Stueber, F.; Garrard, C.S.; Hinds, C.J.; et al. Patients with community acquired pneumonia admitted to European intensive care units: An epidemiological survey of the GenOSept cohort. Crit. Care 2014, 18, R58. [Google Scholar] [CrossRef]
- Rombauts, A.; Abelenda-Alonso, G.; Cuervo, G.; Gudiol, C.; Carratalà, J. Role of the inflammatory response in community-acquired pneumonia: Clinical implications. Expert Rev. Anti Infect. Ther. 2022, 20, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Torres, A. Host-targeted approaches to sepsis due to community-acquired pneumonia. EBioMedicine 2022, 86, 104335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, Y.; Wang, X.; Zhu, J.; Li, Q.; Feng, J.; He, D.; Zhong, Z.; Zheng, X.; Lu, J.; et al. Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS. Ther. Adv. Respir. Dis. 2019, 13, 1753466619879840. [Google Scholar] [CrossRef] [PubMed]
- Batra, S.; Balamayooran, G.; Sahoo, M.K. Nuclear factor-κB: A key regulator in health and disease of lungs. Arch. Immunol. Ther. Exp. 2011, 59, 335–351. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.; Zhou, J.; Wang, D.; Zhou, T. Regulation of NF-κB/MAPK signaling pathway attenuates the acute lung inflammation in Klebsiella pneumonia rats by mollugin treatment. Microb. Pathog. 2019, 132, 369–373. [Google Scholar] [CrossRef]
- Devaney, J.; Curley, G.F.; Hayes, M.; Masterson, C.; Ansari, B.; O’Brien, T.; O’Toole, D.; Laffey, J.G. Inhibition of pulmonary nuclear factor kappa-B decreases the severity of acute Escherichia coli pneumonia but worsens prolonged pneumonia. Crit. Care 2013, 17, R82. [Google Scholar] [CrossRef]
- Eddy, W.E.; Gong, K.Q.; Bell, B.; Parks, W.C.; Ziegler, S.F.; Manicone, A.M. Stat5 Is Required for CD103+ Dendritic Cell and Alveolar Macrophage Development and Protection from Lung Injury. J. Immunol. 2017, 198, 4813–4822. [Google Scholar] [CrossRef]
- Gomez, J.C.; Yamada, M.; Martin, J.R.; Dang, H.; Brickey, W.J.; Bergmeier, W.; Dinauer, M.C.; Doerschuk, C.M. Mechanisms of interferon-γ production by neutrophils and its function during Streptococcus pneumoniae pneumonia. Am. J. Respir. Cell Mol. Biol. 2015, 52, 349–364. [Google Scholar] [CrossRef]
- Grudzinska, F.S.; Brodlie, M.; Scholefield, B.R.; Jackson, T.; Scott, A.; Thickett, D.R.; Sapey, E. Neutrophils in community-acquired pneumonia: Parallels in dysfunction at the extremes of age. Thorax 2020, 75, 164–171. [Google Scholar] [CrossRef]
- Soehnlein, O.; Oehmcke, S.; Ma, X.; Rothfuchs, A.G.; Frithiof, R.; van Rooijen, N.; Mörgelin, M.; Herwald, H.; Lindbom, L. Neutrophil degranulation mediates severe lung damage triggered by streptococcal M1 protein. Eur. Respir. J. 2008, 32, 405–412. [Google Scholar] [CrossRef]
- Rendon, A.; Rendon-Ramirez, E.J.; Rosas-Taraco, A.G. Relevant Cytokines in the Management of Community-Acquired Pneumonia. Curr. Infect. Dis. Rep. 2016, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- London, N.R.; Zhu, W.; Bozza, F.A.; Smith, M.C.; Greif, D.M.; Sorensen, L.K.; Chen, L.; Kaminoh, Y.; Chan, A.C.; Passi, S.F.; et al. Targeting Robo4-dependent Slit signaling to survive the cytokine storm in sepsis and influenza. Sci. Transl. Med. 2010, 2, 23ra19. [Google Scholar] [CrossRef] [PubMed]
- Gally, F.; Minor, M.N.; Smith, S.K.; Case, S.R.; Chu, H.W. Heat shock factor 1 protects against lung Mycoplasma pneumoniae infection in mice. J. Innate Immun. 2012, 4, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Parnell, G.P.; McLean, A.S.; Booth, D.R.; Armstrong, N.J.; Nalos, M.; Huang, S.J.; Manak, J.; Tang, W.; Tam, O.Y.; Chan, S.; et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit. Care 2012, 16, R157. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Rosenberg, A.F.; Peterson, D.R.; Grzesik, K.; Baran, A.M.; Ashton, J.M.; Gill, S.R.; Corbett, A.M.; Holden-Wiltse, J.; Topham, D.J.; et al. Transcriptomic Biomarkers to Discriminate Bacterial from Nonbacterial Infection in Adults Hospitalized with Respiratory Illness. Sci. Rep. 2017, 7, 6548. [Google Scholar] [CrossRef]
- Strehlitz, A.; Goldmann, O.; Pils, M.C.; Pessler, F.; Medina, E. An Interferon Signature Discriminates Pneumococcal from Staphylococcal Pneumonia. Front. Immunol. 2018, 9, 1424. [Google Scholar] [CrossRef]
- Bertrams, W.; Griss, K.; Han, M.; Seidel, K.; Klemmer, A.; Sittka-Stark, A.; Hippenstiel, S.; Suttorp, N.; Finkernagel, F.; Wilhelm, J.; et al. Transcriptional analysis identifies potential biomarkers and molecular regulators in pneumonia and COPD exacerbation. Sci. Rep. 2020, 10, 241. [Google Scholar] [CrossRef]
- Alqutami, F.; Senok, A.; Hachim, M. COVID-19 Transcriptomic Atlas: A Comprehensive Analysis of COVID-19 Related Transcriptomics Datasets. Front. Genet. 2021, 12, 755222. [Google Scholar] [CrossRef]
- Ilieva, M.; Tschaikowski, M.; Vandin, A.; Uchida, S. The current status of gene expression profilings in COVID-19 patients. Clin. Transl. Discov. 2022, 2, e104. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes. Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef]
- Rombauts, A.; Bódalo Torruella, M.; Abelenda-Alonso, G.; Perera-Bel, J.; Ferrer-Salvador, A.; Acedo-Terrades, A.; Gabarrós-Subirà, M.; Oriol, I.; Gudiol, C.; Nonell, L.; et al. Dynamics of Gene Expression Profiling and Identification of High-Risk Patients for Severe COVID-19. Biomedicines 2023, 11, 1348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhou, Y.; Ding, J. The current landscape of microRNAs (miRNAs) in bacterial pneumonia: Opportunities and challenges. Cell. Mol. Biol. Lett. 2022, 27, 70. [Google Scholar] [CrossRef]
- Galván-Román, J.M.; Lancho-Sánchez, Á.; Luquero-Bueno, S.; Vega-Piris, L.; Curbelo, J.; Manzaneque-Pradales, M.; Gómez, M.; de la Fuente, H.; Ortega-Gómez, M.; Aspa, J. Usefulness of circulating microRNAs miR-146a and miR-16-5p as prognostic biomarkers in community-acquired pneumonia. PLoS ONE 2020, 15, e0240926. [Google Scholar] [CrossRef] [PubMed]
- Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Borrmann, M.; Klein, M.; Kotschote, S.; Bonin, M.; Reithmair, M.; Kaufmann, I. Diagnostic potential of circulating cell-free microRNAs for community-acquired pneumonia and pneumonia-related sepsis. J. Cell. Mol. Med. 2020, 24, 12054–12064. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Fleta, P.; Vera-Tomé, P.; Jiménez-Fernández, M.; Requena, S.; Roy-Vallejo, E.; Sanz-García, A.; Lozano-Prieto, M.; López-Sanz, C.; Vara, A.; Lancho-Sánchez, Á. A Differential Signature of Circulating miRNAs and Cytokines between COVID-19 and Community-Acquired Pneumonia Uncovers Novel Physiopathological Mechanisms of COVID-19. Front. Immunol. 2022, 12, 815651. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).