Development of 2′-O-Methyl and LNA Antisense Oligonucleotides for SMN2 Splicing Correction in SMA Cells
Abstract
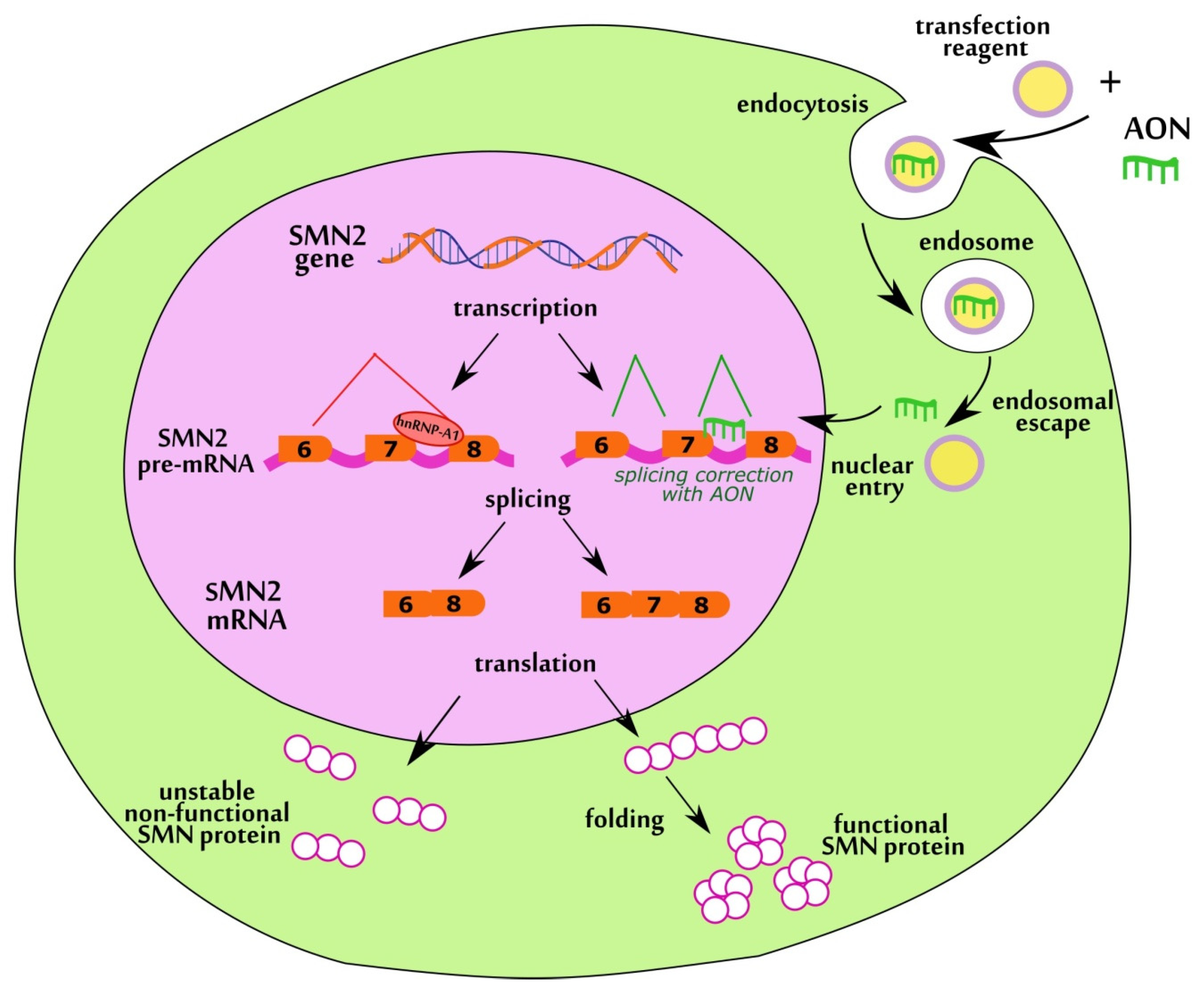
1. Introduction
2. Materials and Methods
2.1. Transfection
2.2. Analysis of Toxicity of RNA/Carrier Complexes
2.3. RNA Isolation and cDNA Synthesis
2.4. Semiquantitative and Quantitative Fluorescence RT-PCR
2.5. Immunocytochemistry
2.6. Statistical Methods
3. Results
3.1. Design of Antisense RNA Oligonucleotides Complementary to Negative Splicing Elements of SMN2 Gene
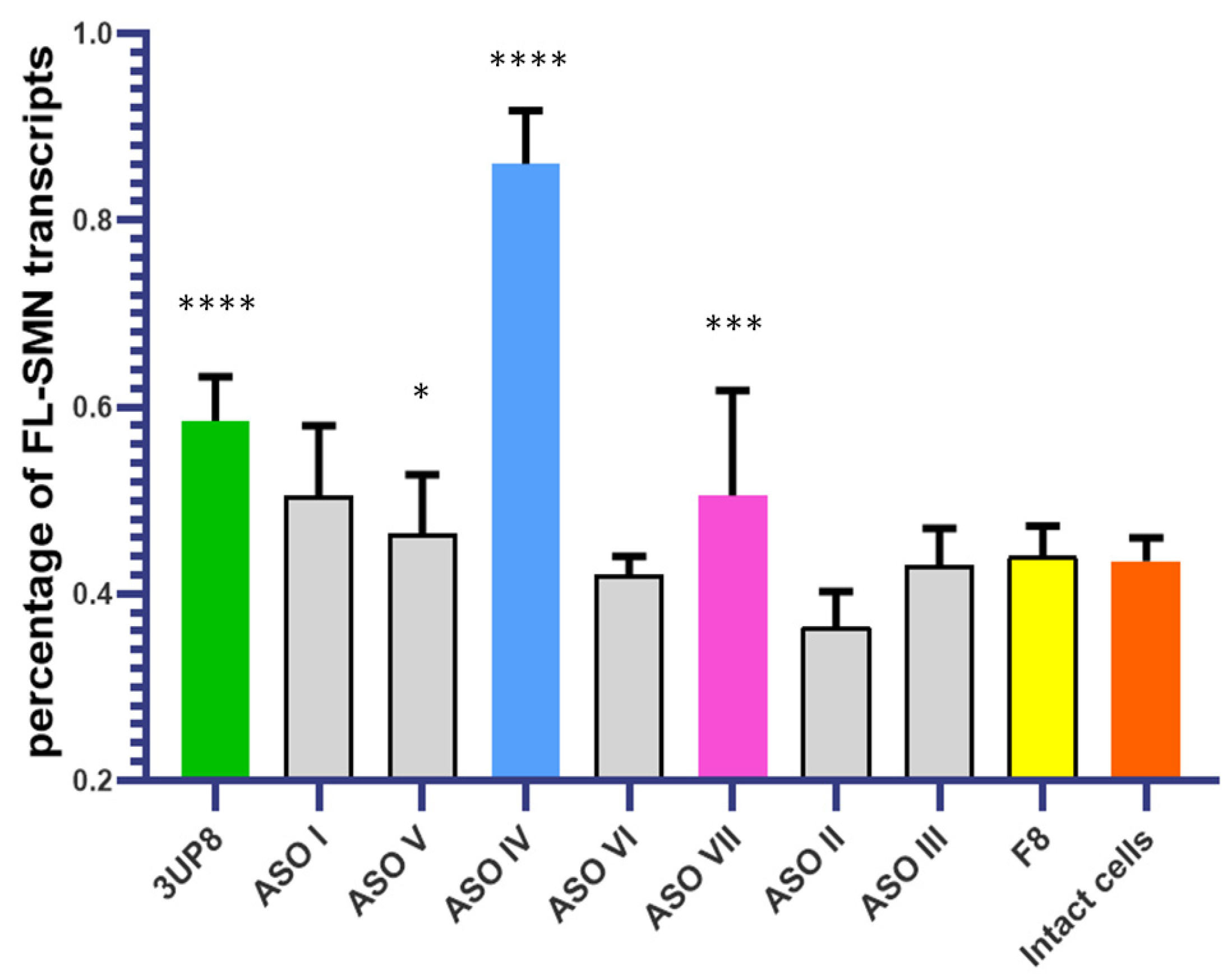
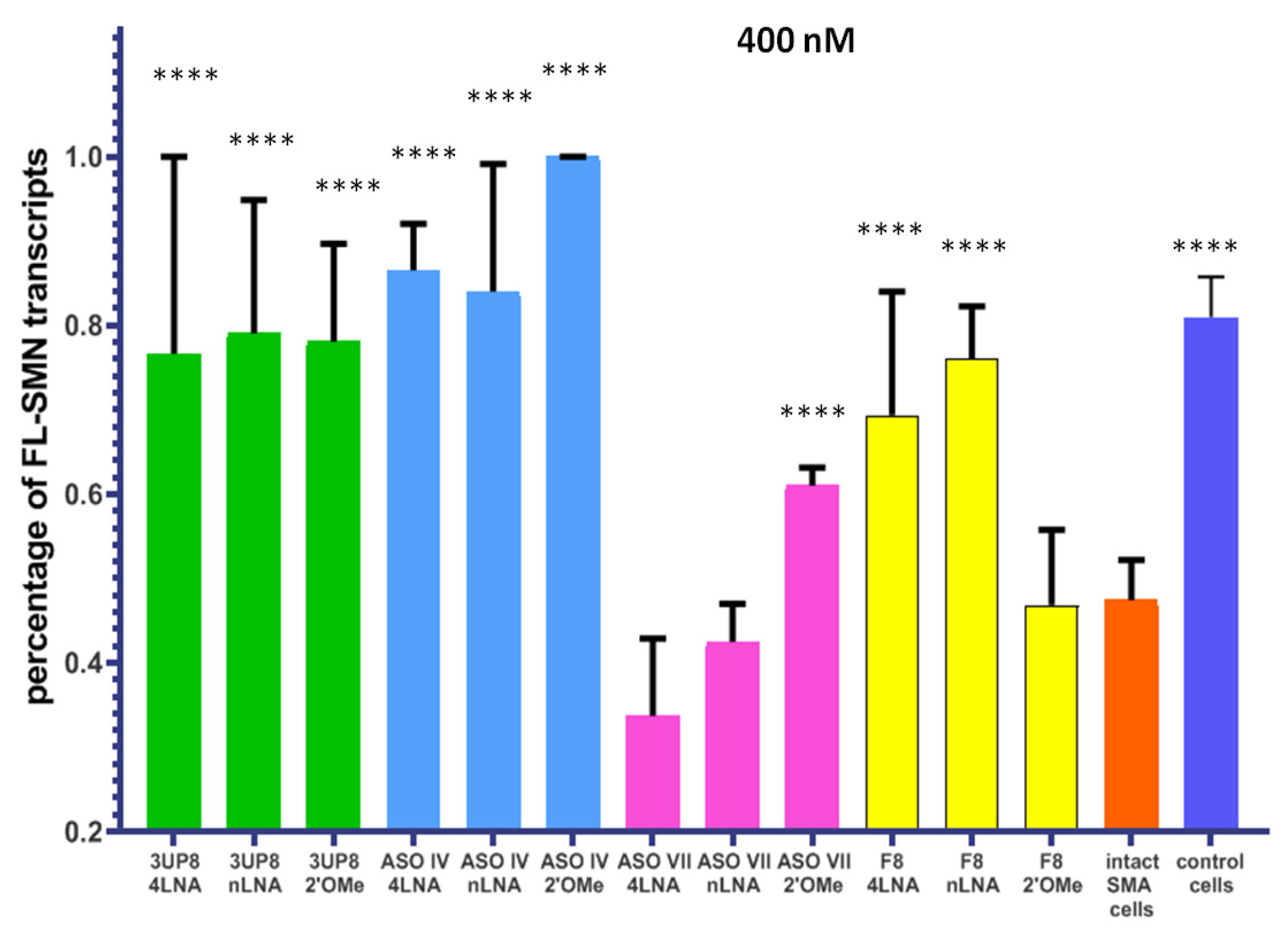
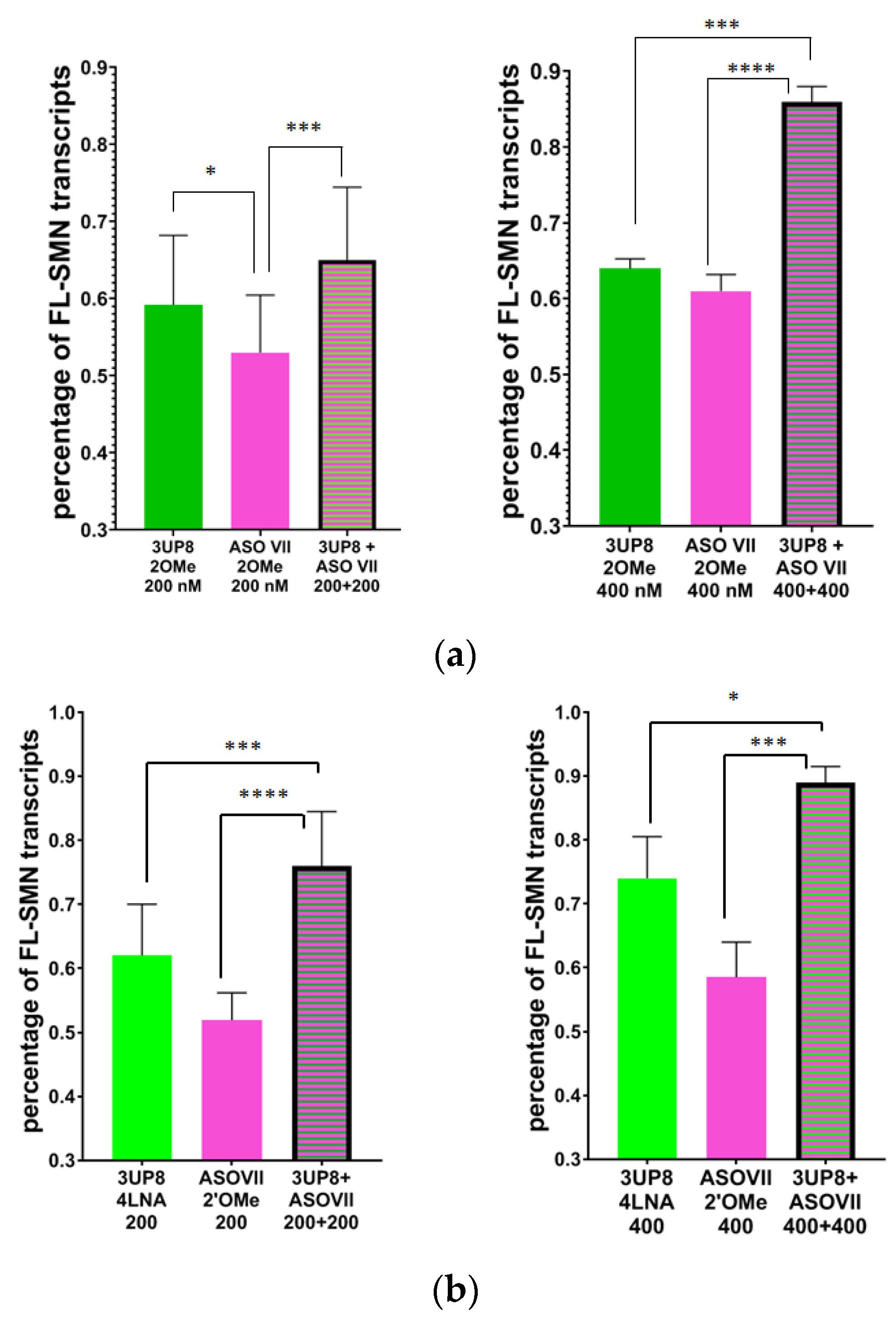
3.2. Evaluation of the AONs Efficiency at Different Concentrations
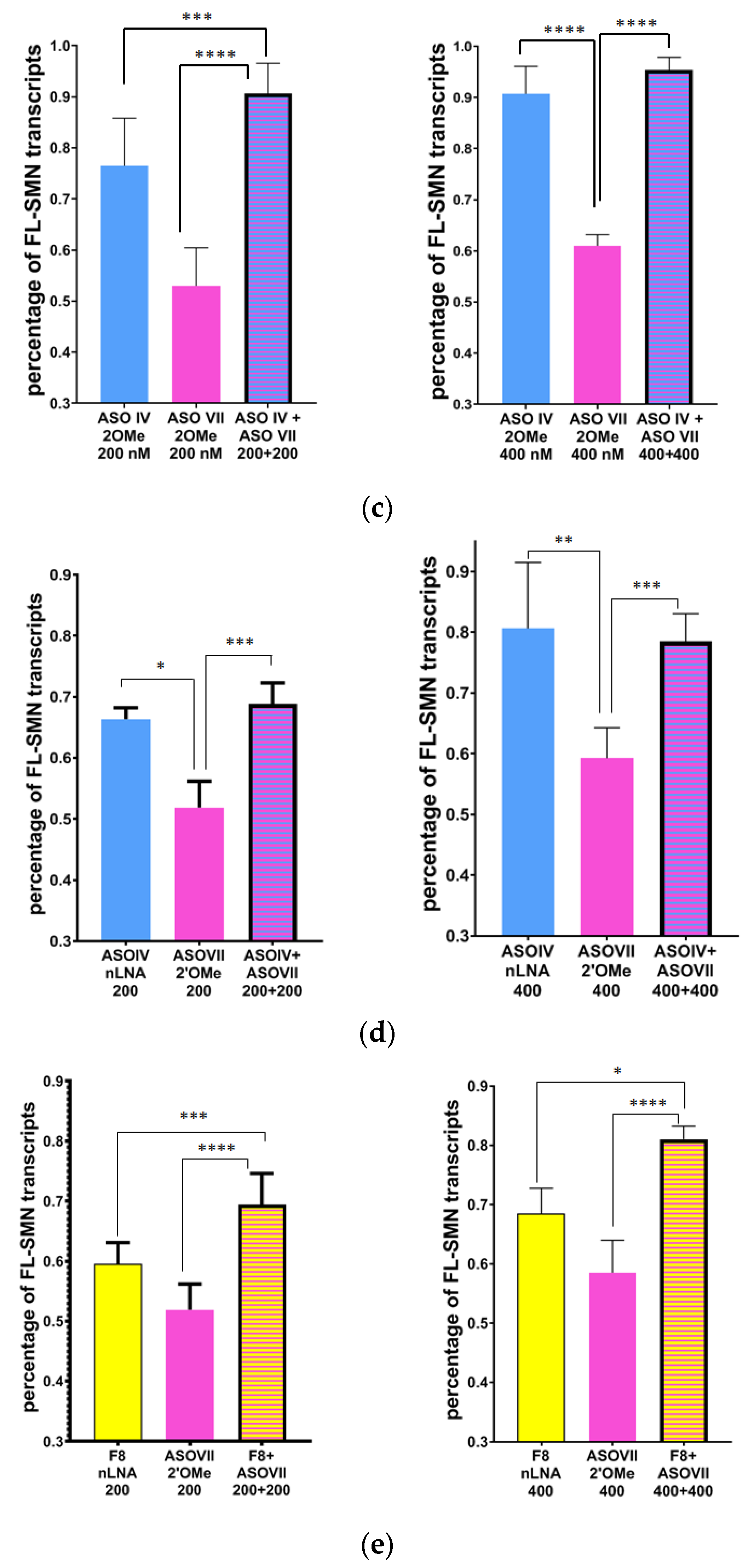
3.3. Simultaneous Targeting Two Nonadjacent SRE
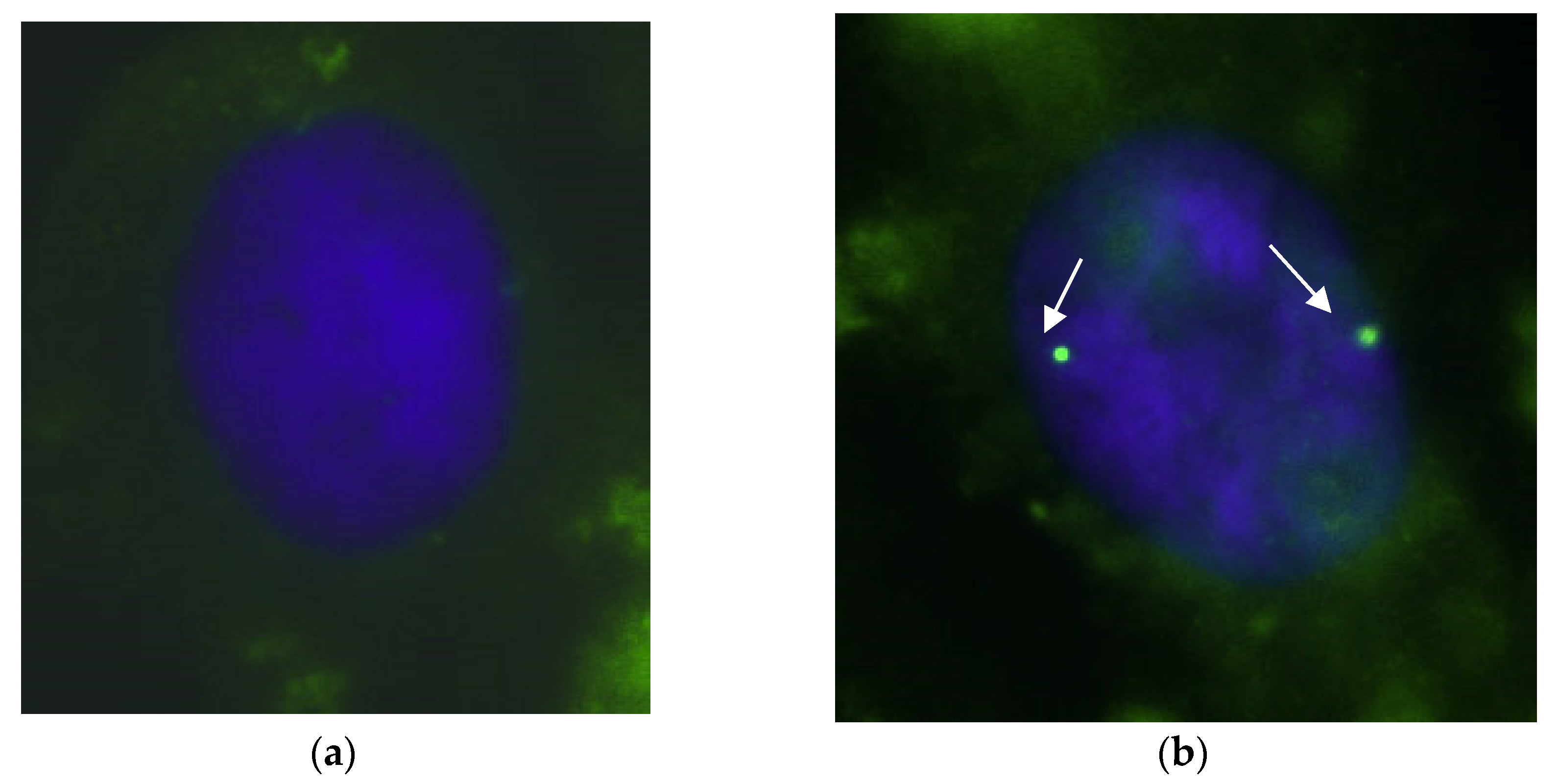
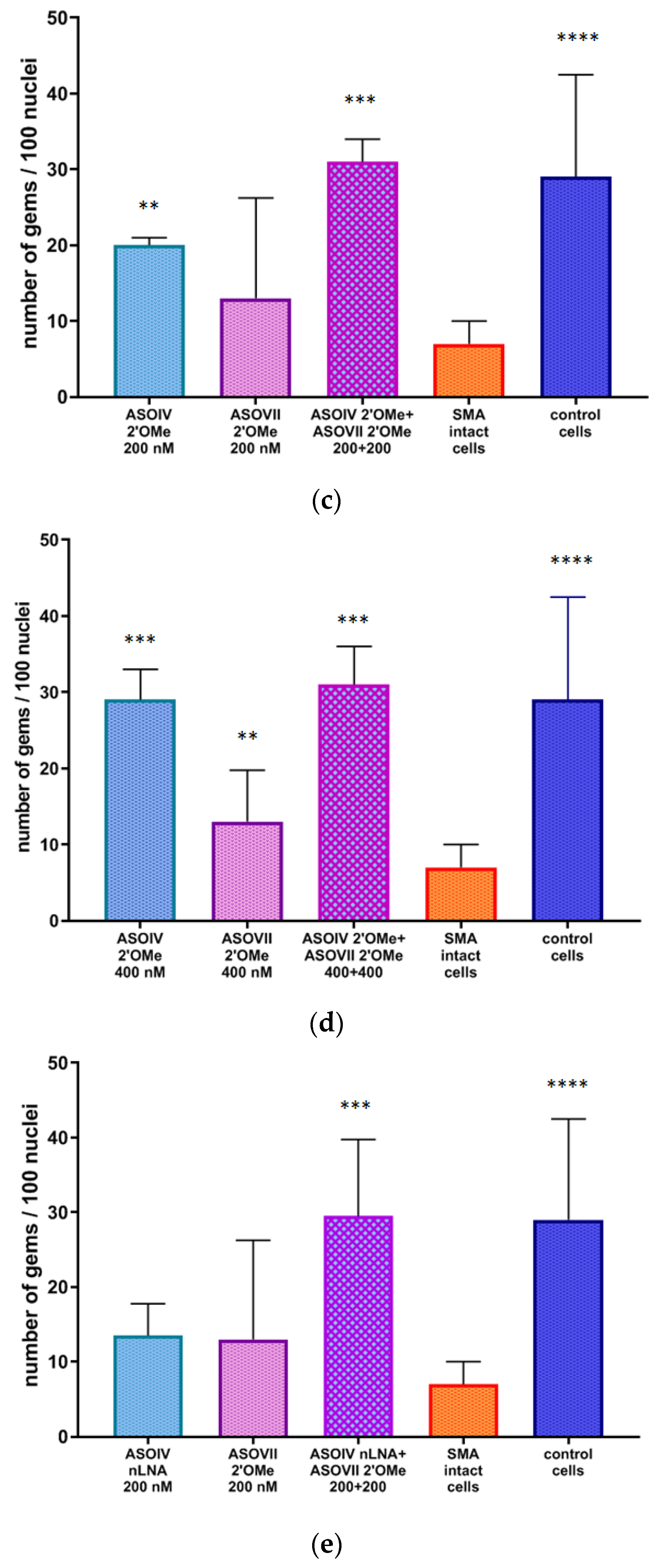
3.4. Nuclear Gems Detection in Transfected SMA Fibroblasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogino, S.; Wilson, R.B. Genetic testing and risk assessment for spinal muscular atrophy (SMA). Hum. Genet. 2002, 111, 477–500. [Google Scholar] [CrossRef] [PubMed]
- Russman, B.S. Spinal muscular atrophy: Clinical classification and disease heterogeneity. J. Child Neurol. 2007, 22, 946–951. [Google Scholar] [CrossRef] [PubMed]
- Rudnik-Schöneborn, S.; Heller, R.; Berg, C.; Betzler, C.; Grimm, T.; Eggermann, T.; Eggermann, K.; Wirth, R.; Wirth, B.; Zerres, K. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet. 2008, 45, 635–638. [Google Scholar] [CrossRef]
- Tizzano, E.F.; Finkel, R.S. Spinal muscular atrophy: A changing phenotype beyond the clinical trials. Neuromuscul. Disord. 2017, 27, 883–889. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Butchbach, M.E.R. Genomic Variability in the Survival Motor Neuron Genes (SMN1 and SMN2): Implications for Spinal Muscular Atrophy Phenotype and Therapeutics Development. Int. J. Mol. Sci. 2021, 22, 7896. [Google Scholar] [CrossRef] [PubMed]
- Monani, U.R.; Lorson, C.L.; Parsons, D.W.; Prior, T.W.; Androphy, E.J.; Burghes, A.H.M.; McPherson, J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999, 8, 1177–1183. [Google Scholar] [CrossRef]
- Wirth, B.; Brichta, L.; Hahnen, E. Spinal Muscular Atrophy: From Gene to Therapy. Semin. Pediatr. Neurol. 2006, 13, 121–131. [Google Scholar] [CrossRef]
- Maretina, M.A.; Zheleznyakova, G.Y.; Lanko, K.M.; Egorova, A.A.; Baranov, V.S.; Kiselev, A.V. Molecular Factors Involved in Spinal Muscular Atrophy Pathways as Possible Disease-modifying Candidates. Curr. Genom. 2018, 19, 339–355. [Google Scholar] [CrossRef]
- Zheleznyakova, G.Y.; Nilsson, E.K.; Kiselev, A.V.; Maretina, M.A.; Tishchenko, L.I.; Fredriksson, R.; Baranov, V.S.; Schiöth, H.B. Methylation levels of SLC23A2 and NCOR2 genes correlate with spinal muscular atrophy severity. PLoS ONE 2015, 10, e0121964. [Google Scholar] [CrossRef]
- Liu, Q.; Fischer, U.; Wang, F.; Dreyfuss, G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 1997, 90, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Crawford, T.O.; Paushkin, S.V.; Kobayashi, D.T.; Forrest, S.J.; Joyce, C.L.; Finkel, R.S.; Kaufmann, P.; Swoboda, K.J.; Tiziano, D.; Lomastro, R.; et al. Evaluation of SMN Protein, Transcript, and Copy Number in the Biomarkers for Spinal Muscular Atrophy (BforSMA) Clinical Study. PLoS ONE 2012, 7, e33572. [Google Scholar] [CrossRef] [PubMed]
- Maretina, M.; Egorova, A.; Lanko, K.; Baranov, V.; Kiselev, A. Evaluation of Mean Percentage of Full-Length SMN Transcripts as a Molecular Biomarker of Spinal Muscular Atrophy. Genes 2022, 13, 1911. [Google Scholar] [CrossRef]
- Al-hilal, H.; Maretina, M.; Egorova, A.; Glotov, A.; Kiselev, A. Quantification of the Number of Nuclear Gems as a Potential Biomarker for Spinal Muscular Atrophy. Preprints 2023, 2023091527. [Google Scholar] [CrossRef]
- Haché, M.; Swoboda, K.J.; Sethna, N.; Farrow-Gillespie, A.; Khandji, A.; Xia, S.; Bishop, K.M. Intrathecal Injections in Children with Spinal Muscular Atrophy: Nusinersen Clinical Trial Experience. J. Child Neurol. 2016, 31, 899–906. [Google Scholar] [CrossRef]
- Feldman, A.G.; Parsons, J.A.; Dutmer, C.M.; Veerapandiyan, A.; Hafberg, E.; Maloney, N.; Mack, C.L. Subacute Liver Failure Following Gene Replacement Therapy for Spinal Muscular Atrophy Type 1. J. Pediatr. 2020, 225, 252–258.e1. [Google Scholar] [CrossRef]
- Chand, D.; Mohr, F.; McMillan, H.; Tukov, F.F.; Montgomery, K.; Kleyn, A.; Sun, R.; Tauscher-Wisniewski, S.; Kaufmann, P.; Kullak-Ublick, G. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J. Hepatol. 2021, 74, 560–566. [Google Scholar] [CrossRef]
- Singh, R.N.; Ottesen, E.W.; Singh, N.N. The First Orally Deliverable Small Molecule for the Treatment of Spinal Muscular Atrophy. Neurosci. Insights 2020, 15, 263310552097398. [Google Scholar] [CrossRef]
- Kray, K.M.; McGovern, V.L.; Chugh, D.; Arnold, W.D.; Burghes, A.H.M. Dual SMN inducing therapies can rescue survival and motor unit function in symptomatic ∆7SMA mice. Neurobiol. Dis. 2021, 159, 105488. [Google Scholar] [CrossRef]
- Mirea, A.; Shelby, E.-S.; Axente, M.; Badina, M.; Padure, L.; Leanca, M.; Dima, V.; Sporea, C. Combination Therapy with Nusinersen and Onasemnogene Abeparvovec-xioi in Spinal Muscular Atrophy Type I. J. Clin. Med. 2021, 10, 5540. [Google Scholar] [CrossRef]
- Singh, N.N.; Shishimorova, M.; Cao, L.C.; Gangwani, L.; Singh, R.N. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009, 6, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Vickers, T.A.; Okunola, H.L.; Bennett, C.F.; Krainer, A.R. Antisense Masking of an hnRNP A1/A2 Intronic Splicing Silencer Corrects SMN2 Splicing in Transgenic Mice. Am. J. Hum. Genet. 2008, 82, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Porensky, P.N.; Mitrpant, C.; McGovern, V.L.; Bevan, A.K.; Foust, K.D.; Kaspar, B.K.; Wilton, S.D.; Burghes, A.H.M. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012, 21, 1625–1638. [Google Scholar] [CrossRef]
- Singh, N.K.; Singh, N.N.; Androphy, E.J.; Singh, R.N. Splicing of a Critical Exon of Human Survival Motor Neuron Is Regulated by a Unique Silencer Element Located in the Last Intron. Mol. Cell. Biol. 2006, 26, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, H.; Miyaso, H.; Okumura, M.; Kurisu, J.; Imaizumi, K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002, 277, 23271–23277. [Google Scholar] [CrossRef]
- Kashima, T.; Rao, N.; Manley, J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 3426–3431. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Lawler, M.N.; Ottesen, E.W.; Upreti, D.; Kaczynski, J.R.; Singh, R.N. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013, 41, 8144–8165. [Google Scholar] [CrossRef]
- Lim, S.R.; Hertel, K.J. Modulation of Survival Motor Neuron Pre-mRNA Splicing by Inhibition of Alternative 3′ Splice Site Pairing. J. Biol. Chem. 2001, 276, 45476–45483. [Google Scholar] [CrossRef]
- Pao, P.W.; Wee, K.B.; Yee, W.C.; Dwipramono, Z.A. Dual masking of specific negative splicing regulatory elements resulted in maximal exon 7 inclusion of SMN2 gene. Mol. Ther. 2014, 22, 854–861. [Google Scholar] [CrossRef]
- Brad Wan, W.; Seth, P.P. The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem. 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Testa, C.M. Antisense Oligonucleotide Therapeutics for Neurodegenerative Disorders. Curr. Geriatr. Rep. 2022, 11, 19–32. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef]
- Touznik, A.; Maruyama, R.; Hosoki, K.; Echigoya, Y.; Yokota, T. LNA/DNA mixmer-based antisense oligonucleotides correct alternative splicing of the SMN2 gene and restore SMN protein expression in type 1 SMA fibroblasts. Sci. Rep. 2017, 7, 3672. [Google Scholar] [CrossRef] [PubMed]
- Kingwell, K. Small activating RNAs lead the charge to turn up gene expression. Nat. Rev. Drug Discov. 2021, 20, 573–574. [Google Scholar] [CrossRef]
- Grigor’eva, E.V.; Valetdinova, K.R.; Ustyantseva, E.I.; Shevchenko, A.I.; Medvedev, S.P.; Mazurok, N.A.; Maretina, M.A.; Kuranova, M.L.; Kiselev, A.V.; Baranov, V.S.; et al. Neural differentiation of patient-specific induced pluripotent stem cells from patients with a hereditary form of spinal muscular atrophy. Genes Cells 2016, 11, 70–81. [Google Scholar]
- Zheleznyakova, G.Y.; Kiselev, A.V.; Vakharlovsky, V.G.; Rask-Andersen, M.; Chavan, R.; Egorova, A.A.; Schiöth, H.B.; Baranov, V.S. Genetic and expression studies of SMN2 gene in Russian patients with spinal muscular atrophy type II and III. BMC Med. Genet. 2011, 12, 96. [Google Scholar] [CrossRef]
- Osman, E.Y.; Washington, C.W.; Kaifer, K.A.; Mazzasette, C.; Patitucci, T.N.; Florea, K.M.; Simon, M.E.; Ko, C.P.; Ebert, A.D.; Lorson, C.L. Optimization of morpholino antisense oligonucleotides targeting the intronic repressor element1 in spinal muscular atrophy. Mol. Ther. 2016, 24, 1592–1601. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.H.; Sun, J.; Krainer, A.R.; Hua, Y.; Prior, T.W. A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum. Mol. Genet. 2017, 26, 2768–2780. [Google Scholar] [CrossRef]
- Stein, C.A.; Subasinghe, C.; Shinozuka, K.; Cohen, J.S. Physicochemical properties of phospborothioate oligodeoxynucleotides. Nucleic Acids Res. 1988, 16, 3209–3221. [Google Scholar] [CrossRef]
- Yoo, B.H.; Bochkareva, E.; Bochakarev, A.; Mou, T.C.; Gray, D.M. 2′-O-methyl-modified phoshorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004, 32, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Heemskerk, H.A.; de Winter, C.L.; de Kimpe, S.J.; van Kuik-Romeijn, P.; Heuvelmans, N.; Platenburg, G.J.; van Ommen, G.-J.B.; van Deutekom, J.C.T.; Aartsma-Rus, A. In vivo comparison of 2′- O -methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. J. Gene Med. 2009, 11, 257–266. [Google Scholar] [CrossRef]
- Le, B.T.; Adams, A.M.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Rational Design of Short Locked Nucleic Acid-Modified 2′-O-Methyl Antisense Oligonucleotides for Efficient Exon-Skipping In Vitro. Mol. Ther. Nucleic Acids 2017, 9, 155–161. [Google Scholar] [CrossRef]
- Kurreck, J.; Wyszko, E.; Gillen, C.; Erdmann, V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Vester, B.; Wengel, J. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Wancewicz, E.V.; Witchell, D.; Swayze, E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009, 52, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Shimo, T.; Tachibana, K.; Saito, K.; Yoshida, T.; Tomita, E.; Waki, R.; Yamamoto, T.; Doi, T.; Inoue, T.; Kawakami, J.; et al. Design and evaluation of locked nucleic acid-based splice-switching oligonucleotides in vitro. Nucleic Acids Res. 2014, 42, 8174–8187. [Google Scholar] [CrossRef]
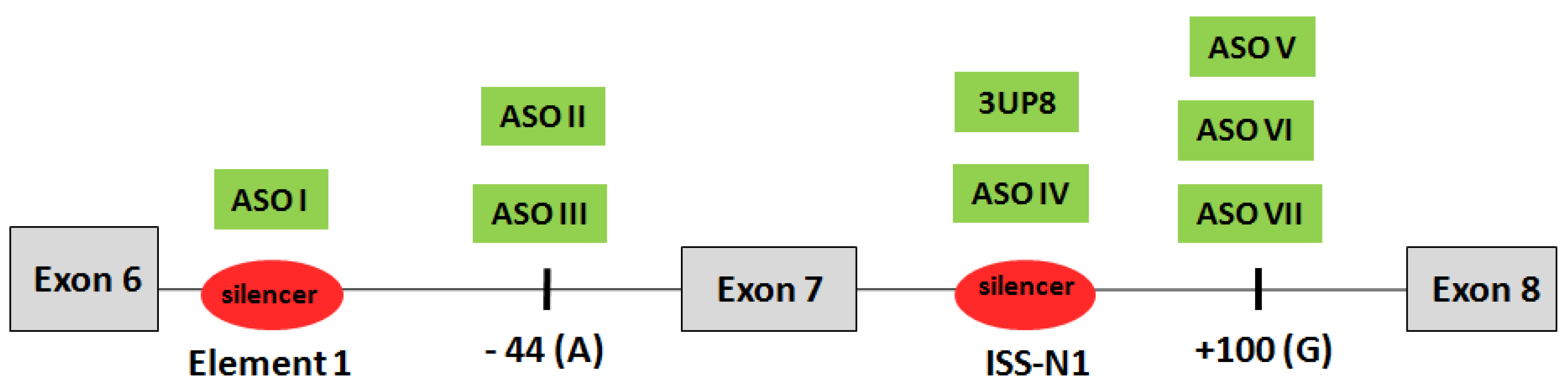



| Name | Length, nt | Sequence | Reference |
|---|---|---|---|
| Phosphorothioate RNA AONs modified at each nucleotide by 2′-O-methyl moiety | |||
| ASO I | 20 | 5′-CUAUAUAUAGUUAUUCAACA-3′ | [38] |
| ASO II | 20 | 5′-CUAUAUAGAUAUAGAUAGCU-3′ | this study |
| ASO III | 20 | 5′-AAUAGCUAUAGAUAGCUUUA-3′ | this study |
| ASO IV | 18 | 5′-UCACUUUCAUAAUGCUGG-3′ | [23] |
| ASO V | 19 | 5′-ACCUUUCAACUUUCUAACA-3′ | [30] |
| ASO VI | 14 | 5′- UCAACUUUCUAACA-3′ | this study |
| ASO VII | 20 | 5′-UUCAACUUUCUAACAUCUGA-3′ | this study |
| 3UP8 | 8 | 5′-GCUGGCAG-3′ | [22] |
| F8 | 8 | 5′-AAUGCUGG-3′ | [22] |
| Phosphorothioate RNA AONs modified by LNA (indicated as capital letters) | |||
| ASO IV 4LNA | 18 | 5′-UCacuuucauaaugcUGg-3′ | this study |
| ASO IV nLNA | 18 | 5′-UcAcUuUcAuAaUgCuGg-3′ | this study |
| ASO VII 4LNA | 20 | 5′-UUcaacuuucuaacaucUGa-3′ | this study |
| ASO VII nLNA | 20 | 5′-UuCaAcUuUcUaAcAuCuGa-3′ | this study |
| 3UP8 4LNA | 8 | 5′-GCuggCAg-3′ | this study |
| 3UP8 nLNA | 8 | 5′-GcUgGcAg-3′ | this study |
| F8 4LNA | 8 | 5′-AAugcUGg-3′ | this study |
| F8 nLNA | 8 | 5′-AaUgCuGg-3′ | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maretina, M.; Il’ina, A.; Egorova, A.; Glotov, A.; Kiselev, A. Development of 2′-O-Methyl and LNA Antisense Oligonucleotides for SMN2 Splicing Correction in SMA Cells. Biomedicines 2023, 11, 3071. https://doi.org/10.3390/biomedicines11113071
Maretina M, Il’ina A, Egorova A, Glotov A, Kiselev A. Development of 2′-O-Methyl and LNA Antisense Oligonucleotides for SMN2 Splicing Correction in SMA Cells. Biomedicines. 2023; 11(11):3071. https://doi.org/10.3390/biomedicines11113071
Chicago/Turabian StyleMaretina, Marianna, Arina Il’ina, Anna Egorova, Andrey Glotov, and Anton Kiselev. 2023. "Development of 2′-O-Methyl and LNA Antisense Oligonucleotides for SMN2 Splicing Correction in SMA Cells" Biomedicines 11, no. 11: 3071. https://doi.org/10.3390/biomedicines11113071
APA StyleMaretina, M., Il’ina, A., Egorova, A., Glotov, A., & Kiselev, A. (2023). Development of 2′-O-Methyl and LNA Antisense Oligonucleotides for SMN2 Splicing Correction in SMA Cells. Biomedicines, 11(11), 3071. https://doi.org/10.3390/biomedicines11113071







