Histone H3 Acetylation Is Involved in Retinoid Acid-Induced Neural Differentiation through Increasing Mitochondrial Function
Abstract
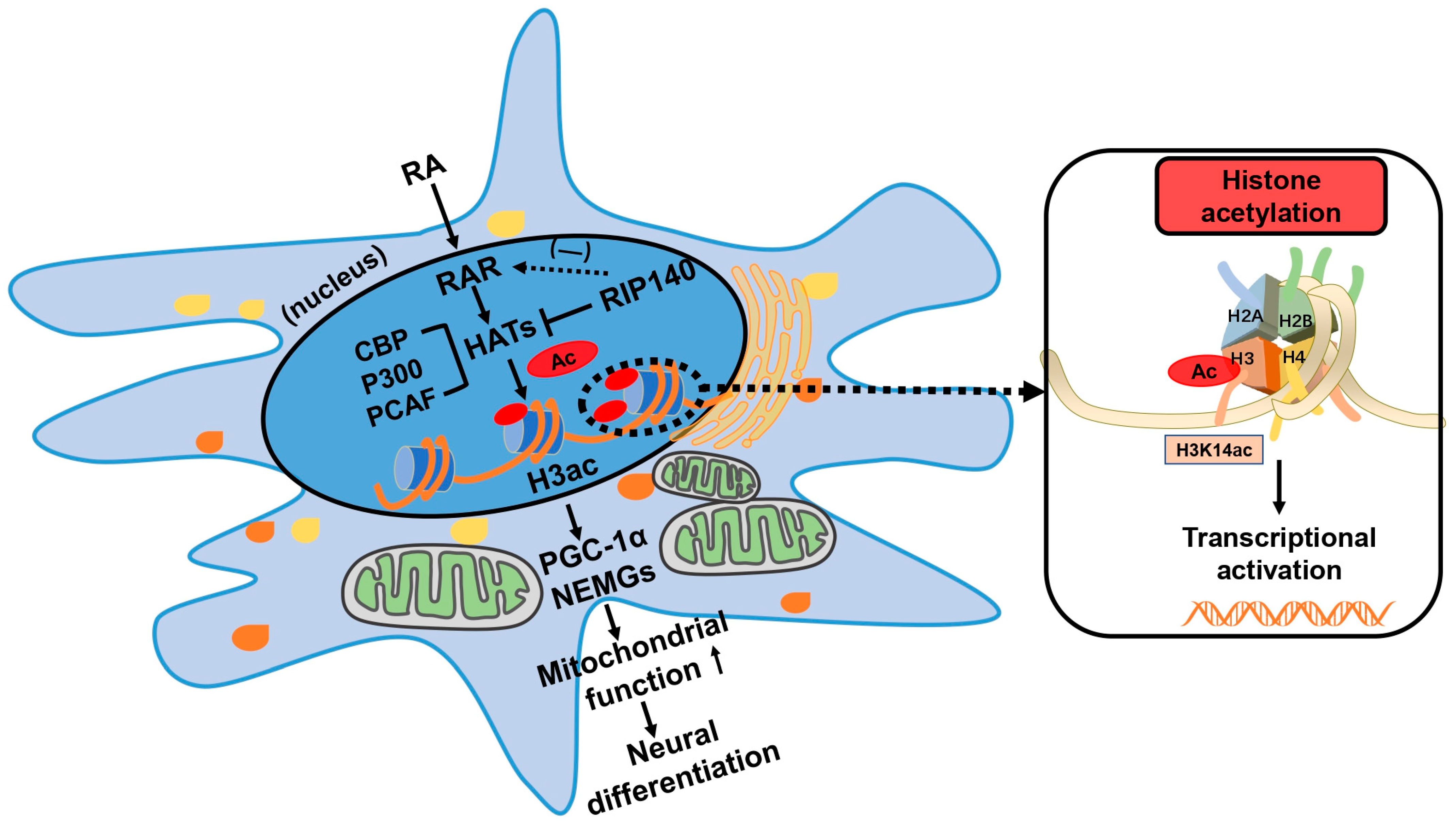
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Generation of a Stable RIP140-Overexpressing Cell Line
2.3. Hematoxylin and Eosin Staining (H&E Staining)
2.4. Immunofluorescence
2.5. Transmission Electron Microscopy (TEM)
2.6. Detection of Reactive Oxygen Species (ROS)
2.7. RNA Preparation and Real-Time Quantitative PCR (RT-qPCR)
2.8. Protein Preparation and Western Blot
2.9. Co-Immunoprecipitation
2.10. HAT Activity Assay
2.11. Statistical Analysis
3. Results
3.1. The Neural Differentiation Model Is Established with SH-SY5Y Cells Induced by RA
3.2. RA Activates PCAF/CBP/P300 and Increases the Acetylation of Lysine 14 in Histone H3 during Neural Differentiation
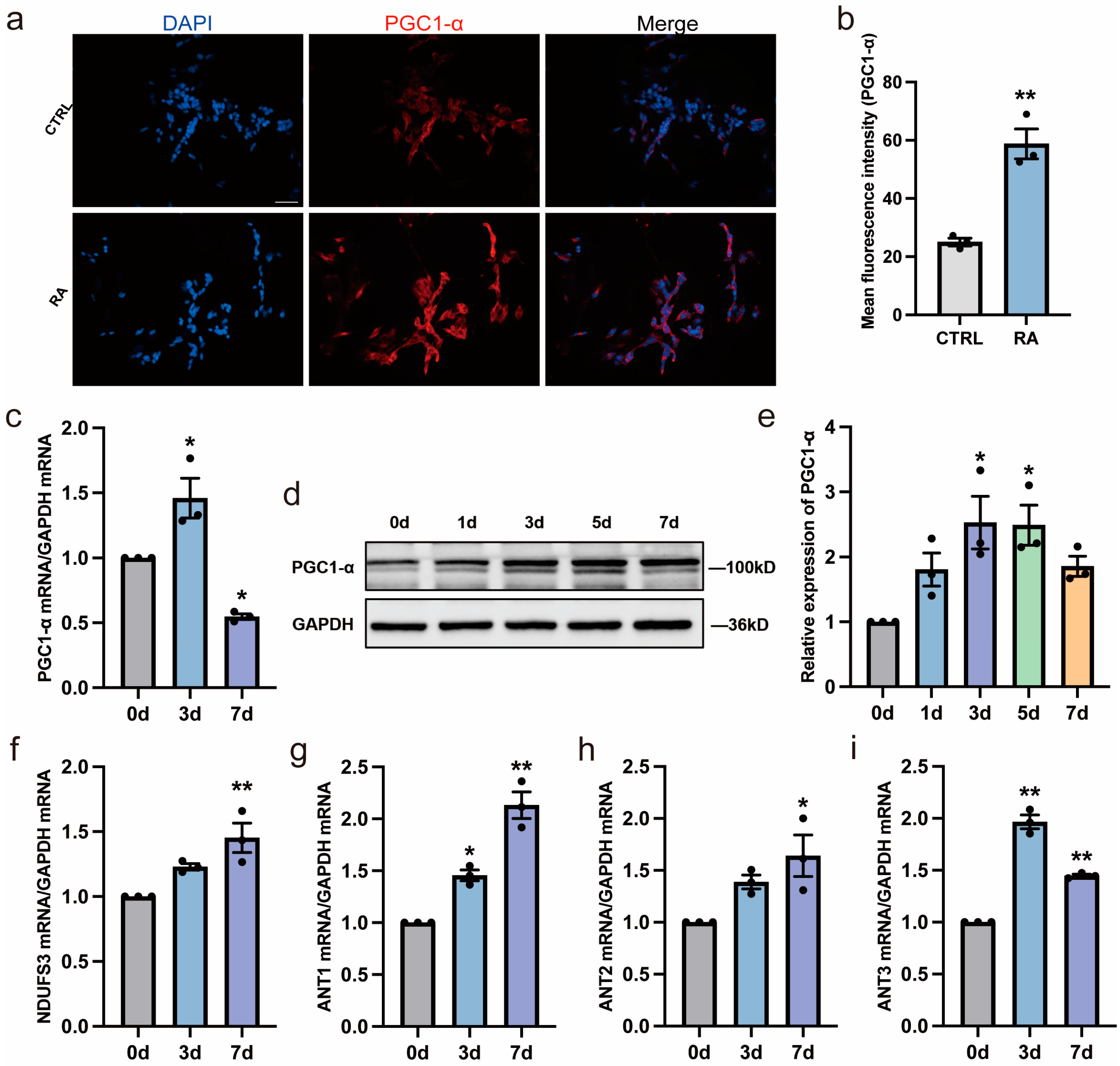
3.3. RA Increases Mitochondrial Biogenesis and Function during Neural Differentiation
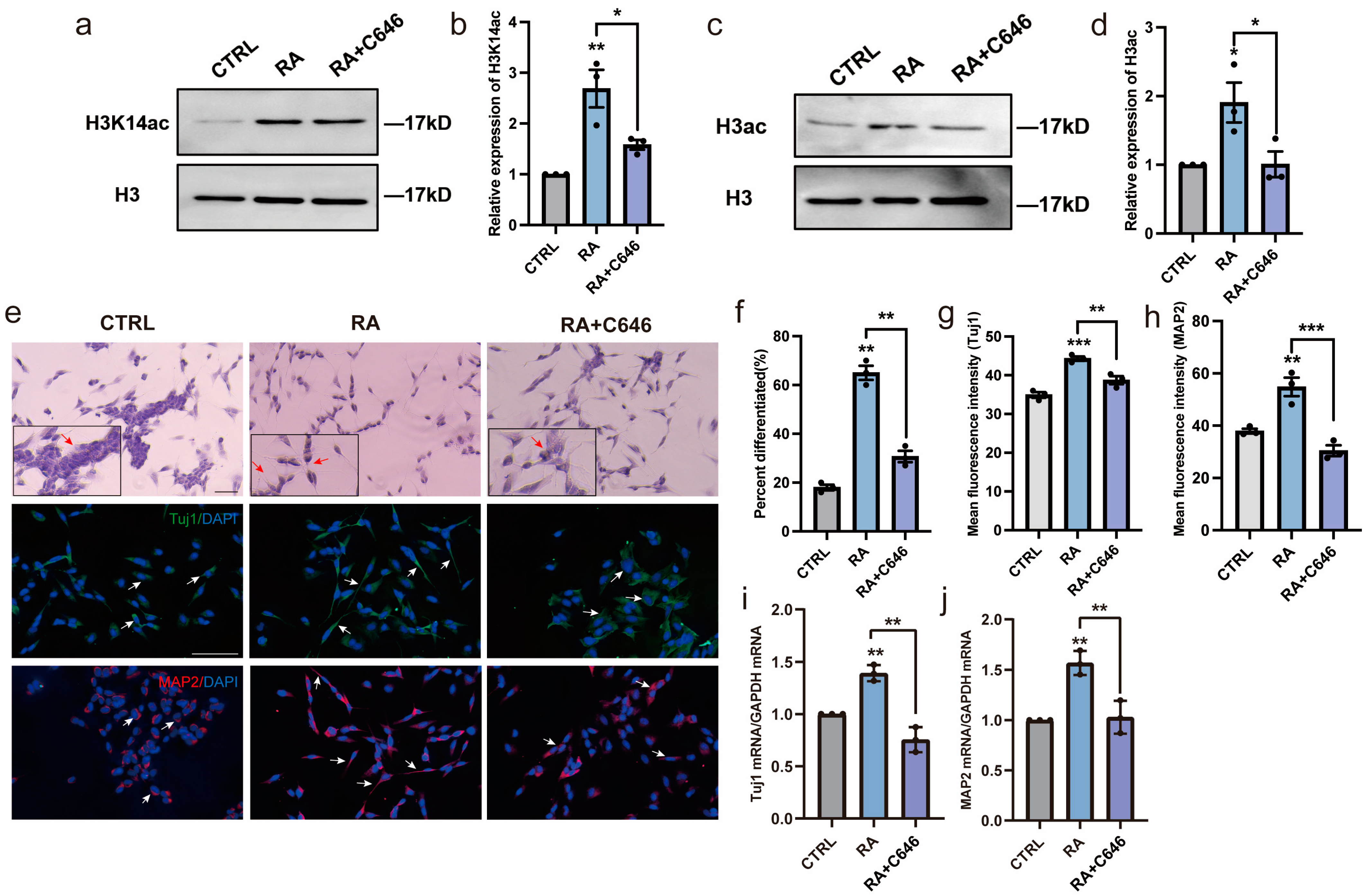
3.4. HAT Activity Inhibitor Inhibits Neural Differentiation of SH-SY5Y Cells
3.5. Histone Acetylation Is Involved in Regulating Mitochondrial Morphology and Function
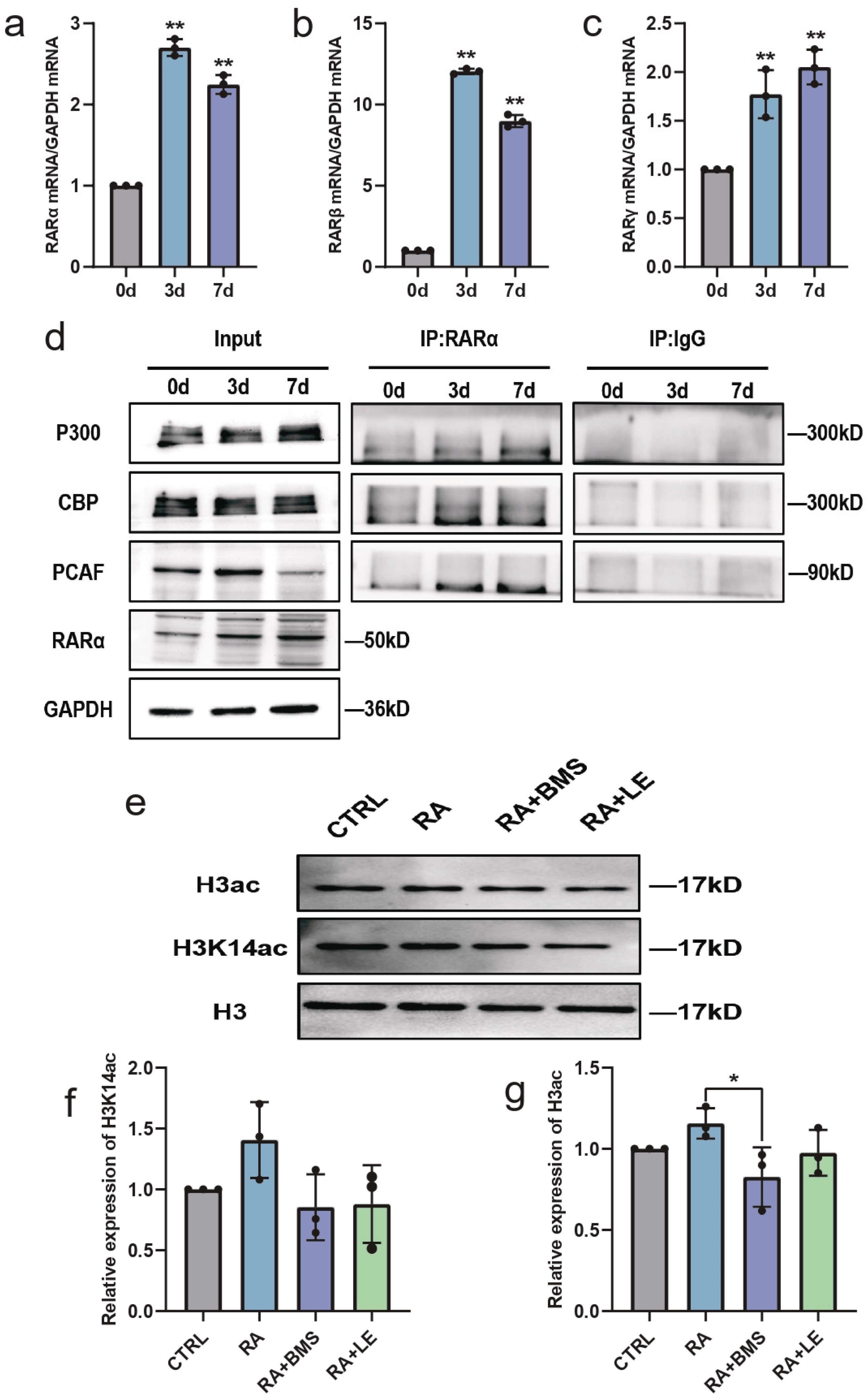
3.6. RA Promotes HATs and Increases Histone Acetylation via RA Signaling during Neural Differentiation
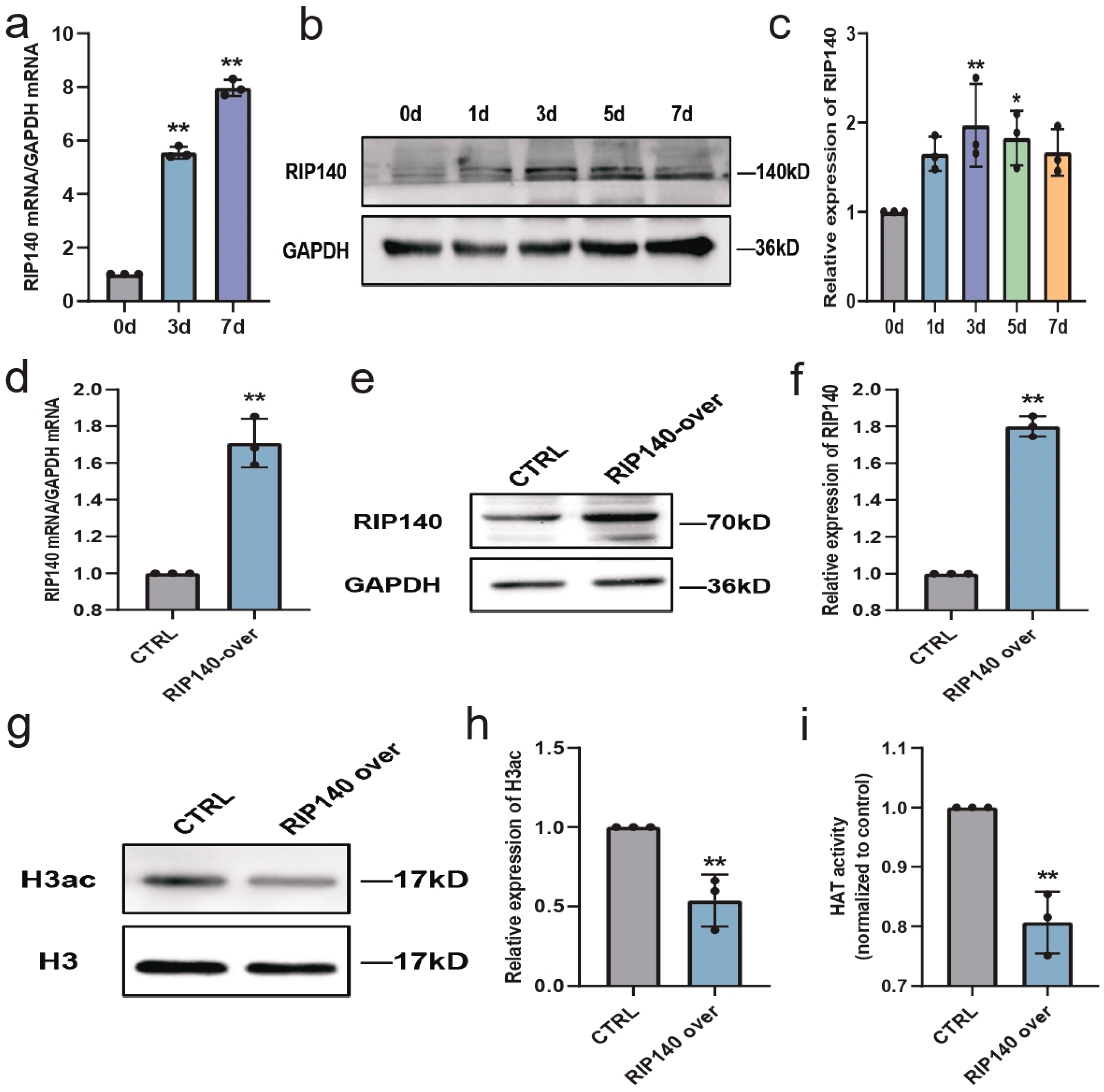
3.7. RIP140, a RAR Co-Repressor, Is Involved in Regulating Histone Acetylation during Neural Differentiation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fregeac, J.; Moriceau, S.; Poli, A.; Nguyen, L.S.; Oury, F.; Colleaux, L. Loss of the neurodevelopmental disease-associated gene miR-146a impairs neural progenitor differentiation and causes learning and memory deficits. Mol. Autism 2020, 11, 22. [Google Scholar] [CrossRef]
- Kurabayashi, N.; Sanada, K. Molecular Mechanism Underlying Abnormal Differentiation of Neural Progenitor Cells in the Developing Down Syndrome Brain. Yakugaku Zasshi 2017, 137, 795–800. [Google Scholar] [CrossRef][Green Version]
- Zhou, W.; Zhao, T.; Du, J.; Ji, G.; Li, X.; Ji, S.; Tian, W.; Wang, X.; Hao, A. TIGAR promotes neural stem cell differentiation through acetyl-CoA-mediated histone acetylation. Cell Death Dis. 2019, 10, 198. [Google Scholar] [CrossRef]
- Uittenbogaard, M.; Brantner, C.A.; Chiaramello, A. Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells. Cell Death Dis. 2018, 9, 360. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Zhou, W.; Ji, S.; Li, X.; Li, G.; Liu, G.; Wang, F.; Hao, A. Effect of melatonin on neuronal differentiation requires CBP/p300-mediated acetylation of histone H3 lysine 14. Neuroscience 2017, 364, 45–59. [Google Scholar] [CrossRef]
- Nieto-Estevez, V.; Changarathil, G.; Adeyeye, A.O.; Coppin, M.O.; Kassim, R.S.; Zhu, J.; Hsieh, J. HDAC1 Regulates Neuronal Differentiation. Front. Mol. Neurosci. 2021, 14, 815808. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R.; Zhou, M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Mizar, P.; Cassel, R.; Neidl, R.; Selvi, B.R.; Mohankrishna, D.V.; Vedamurthy, B.M.; Schneider, A.; Bousiges, O.; Mathis, C.; et al. A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J. Neurosci. 2013, 33, 10698–10712. [Google Scholar] [CrossRef] [PubMed]
- LoPresti, P. HDAC6 in Diseases of Cognition and of Neurons. Cells 2020, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Yu, W.; Zheng, S.; Shi, H.; Li, M.; Sun, J.; Wang, D.; Hou, X.; Liu, L.; Wang, X.; et al. RIP140/PGC-1α axis involved in vitamin A-induced neural differentiation by increasing mitochondrial function. Artif. Cells Nanomed. Biotechnol. 2018, 46 (Suppl. 1), 806–816. [Google Scholar] [CrossRef]
- Mendivil-Perez, M.; Soto-Mercado, V.; Guerra-Librero, A.; Fernandez-Gil, B.I.; Florido, J.; Shen, Y.Q.; Tejada, M.A.; Capilla-Gonzalez, V.; Rusanova, I.; Garcia-Verdugo, J.M.; et al. Melatonin enhances neural stem cell differentiation and engraftment by increasing mitochondrial function. J. Pineal Res. 2017, 63, e12415. [Google Scholar] [CrossRef]
- Cabello-Rivera, D.; Sarmiento-Soto, H.; López-Barneo, J.; Muñoz-Cabello, A.M. Mitochondrial Complex I Function Is Essential for Neural Stem/Progenitor Cells Proliferation and Differentiation. Front. Neurosci. 2019, 13, 664. [Google Scholar] [CrossRef] [PubMed]
- Khacho, M.; Harris, R.; Slack, R.S. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat. Rev. Neurosci. 2019, 20, 34–48. [Google Scholar] [CrossRef]
- Izzo, A.; Mollo, N.; Nitti, M.; Paladino, S.; Calì, G.; Genesio, R.; Bonfiglio, F.; Cicatiello, R.; Barbato, M.; Sarnataro, V.; et al. Mitochondrial dysfunction in down syndrome: Molecular mechanisms and therapeutic targets. Mol. Med. (Camb. Mass.) 2018, 24, 2. [Google Scholar] [CrossRef]
- Kong, L.; Zhao, Y.; Zhou, W.J.; Yu, H.; Teng, S.W.; Guo, Q.; Chen, Z.; Wang, Y. Direct Neuronal Glucose Uptake Is Required for Contextual Fear Acquisition in the Dorsal Hippocampus. Front. Mol. Neurosci. 2017, 10, 388. [Google Scholar] [CrossRef] [PubMed]
- Anwer, S.T.; Mobashir, M.; Fantoukh, O.I.; Khan, B.; Imtiyaz, K.; Naqvi, I.H.; Rizvi, M.M.A. Synthesis of Silver Nano Particles Using Myricetin and the In-Vitro Assessment of Anti-Colorectal Cancer Activity: In-Silico Integration. Int. J. Mol. Sci. 2022, 23, 11024. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lv, H.; Zhang, B.; Duan, R.; Zhang, X.; Lin, P.; Song, C.; Liu, Y. Growth Differentiation Factor 15 Protects SH-SY5Y Cells From Rotenone-Induced Toxicity by Suppressing Mitochondrial Apoptosis. Front. Aging Neurosci. 2022, 14, 869558. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Du, Z.; Li, X.; Wang, L.; Wang, F.; Shi, W.; Hao, A. Protein Palmitoylation Regulates Neural Stem Cell Differentiation by Modulation of EID1 Activity. Mol. Neurobiol. 2016, 53, 5722–5736. [Google Scholar] [CrossRef]
- Kovalevich, J.; Santerre, M.; Langford, D. Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. Methods Mol. Biol. 2021, 2311, 9–23. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, R.; Yang, X.; Tang, K.; Jing, N. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J. Biol. Chem. 2015, 290, 2508–2520. [Google Scholar] [CrossRef]
- Hou, N.; Ren, L.; Gong, M.; Bi, Y.; Gu, Y.; Dong, Z.; Liu, Y.; Chen, J.; Li, T. Vitamin A deficiency impairs spatial learning and memory: The mechanism of abnormal CBP-dependent histone acetylation regulated by retinoic acid receptor alpha. Mol. Neurobiol. 2015, 51, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Park, S.W.; Farooqui, M.; Wei, L.N. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 2007, 35, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Izzo, A.; Manco, R.; Bonfiglio, F.; Calì, G.; De Cristofaro, T.; Patergnani, S.; Cicatiello, R.; Scrima, R.; Zannini, M.; Pinton, P.; et al. NRIP1/RIP140 siRNA-mediated attenuation counteracts mitochondrial dysfunction in Down syndrome. Hum. Mol. Genet. 2014, 23, 4406–4419. [Google Scholar] [CrossRef] [PubMed]
- Bifari, F.; Dolci, S.; Bottani, E.; Pino, A.; Di Chio, M.; Zorzin, S.; Ragni, M.; Zamfir, R.G.; Brunetti, D.; Bardelli, D.; et al. Complete neural stem cell (NSC) neuronal differentiation requires a branched chain amino acids-induced persistent metabolic shift towards energy metabolism. Pharmacol. Res. 2020, 158, 104863. [Google Scholar] [CrossRef]
- Ranea-Robles, P.; Galino, J.; Espinosa, L.; Schlüter, A.; Ruiz, M.; Calingasan, N.Y.; Villarroya, F.; Naudí, A.; Pamplona, R.; Ferrer, I.; et al. Modulation of mitochondrial and inflammatory homeostasis through RIP140 is neuroprotective in an adrenoleukodystrophy mouse model. Neuropathol. Appl. Neurobiol. 2022, 48, e12747. [Google Scholar] [CrossRef] [PubMed]
- Janesick, A.; Wu, S.C.; Blumberg, B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 2015, 72, 1559–1576. [Google Scholar] [CrossRef] [PubMed]
- White, K.A.; Yore, M.M.; Warburton, S.L.; Vaseva, A.V.; Rieder, E.; Freemantle, S.J.; Spinella, M.J. Negative feedback at the level of nuclear receptor coregulation. Self-limitation of retinoid signaling by RIP140. J. Biol. Chem. 2003, 278, 43889–43892. [Google Scholar] [CrossRef]
- Wei, L.N. Retinoids and receptor interacting protein 140 (RIP140) in gene regulation. Curr. Med. Chem. 2004, 11, 1527–1532. [Google Scholar] [CrossRef]
- Wei, L.N.; Hu, X.; Chandra, D.; Seto, E.; Farooqui, M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 2000, 275, 40782–40787. [Google Scholar] [CrossRef]
- Castet, A.; Boulahtouf, A.; Versini, G.; Bonnet, S.; Augereau, P.; Vignon, F.; Khochbin, S.; Jalaguier, S.; Cavaillès, V. Multiple domains of the Receptor-Interacting Protein 140 contribute to transcription inhibition. Nucleic Acids Res. 2004, 32, 1957–1966. [Google Scholar] [CrossRef]
- Vissers, C.; Sinha, A.; Ming, G.L.; Song, H. The epitranscriptome in stem cell biology and neural development. Neurobiol. Dis. 2020, 146, 105139. [Google Scholar] [CrossRef]
- Haran, V.; Lenka, N. Deciphering the Epitranscriptomic Signatures in Cell Fate Determination and Development. Stem Cell Rev. Rep. 2019, 15, 474–496. [Google Scholar] [CrossRef] [PubMed]
- Sunami, Y.; Araki, M.; Kan, S.; Ito, A.; Hironaka, Y.; Imai, M.; Morishita, S.; Ohsaka, A.; Komatsu, N. Histone Acetyltransferase p300/CREB-binding Protein-associated Factor (PCAF) Is Required for All-trans-retinoic Acid-induced Granulocytic Differentiation in Leukemia Cells. J. Biol. Chem. 2017, 292, 2815–2829. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef]
- Luebben, W.R.; Sharma, N.; Nyborg, J.K. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc. Natl. Acad. Sci. USA 2010, 107, 19254–19259. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, Y.; Wu, N.H.; Shen, Y.F. Histone marks and chromatin remodelers on the regulation of neurogenin1 gene in RA induced neuronal differentiation of P19 cells. J. Cell. Biochem. 2009, 107, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.G.; Choi, B.; Kim, S.; Lee, J.Y.; Kim, M.J. Trichostatin A and Sirtinol Regulate the Expression and Nucleocytoplasmic Shuttling of Histone Deacetylases in All-Trans Retinoic Acid-Induced Differentiation of Neuroblastoma Cells. J. Mol. Neurosci. 2018, 64, 501–511. [Google Scholar] [CrossRef]
- Ho, C.F.; Bon, C.P.; Ng, Y.K.; Herr, D.R.; Wu, J.S.; Lin, T.N.; Ong, W.Y. Expression of DHA-Metabolizing Enzyme Alox15 is Regulated by Selective Histone Acetylation in Neuroblastoma Cells. Neurochem. Res. 2018, 43, 540–555. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.G.; Zhu, X. Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim. Biophys. Acta 2014, 1842, 1240–1247. [Google Scholar] [CrossRef]
- Xavier, J.M.; Rodrigues, C.M.; Solá, S. Mitochondria: Major Regulators of Neural Development. Neuroscientist 2016, 22, 346–358. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, X.; Wei, L.N. Molecular interaction of retinoic acid receptors with coregulators PCAF and RIP140. Mol. Cell. Endocrinol. 2004, 226, 43–50. [Google Scholar] [CrossRef]
- Dilworth, F.J.; Fromental-Ramain, C.; Yamamoto, K.; Chambon, P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell 2000, 6, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Katsanis, N.; Ives, J.H.; Groet, J.; Nizetic, D.; Fisher, E.M. Localisation of receptor interacting protein 140 (RIP140) within 100 kb of D21S13 on 21q11, a gene-poor region of the human genome. Hum. Genet. 1998, 102, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Kerley, J.S.; Olsen, S.L.; Freemantle, S.J.; Spinella, M.J. Transcriptional activation of the nuclear receptor corepressor RIP140 by retinoic acid: A potential negative-feedback regulatory mechanism. Biochem. Biophys. Res. Commun. 2001, 285, 969–975. [Google Scholar] [CrossRef] [PubMed]





| Genes | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| TUJ1 | CAAGGTGCGTGAGGAGTAT | GTCTGACACCTTGGGTGAGG |
| Tuj1 (M) | ACCCCGTGGGCTCAAAAT | CCGGAACATGGCTGTGAACT |
| MAP2 | CTCAGCACCGCTAACAGAGG | CATTGGCGCTTCGGACAAG |
| PGC1-α | TGGTGCCACCACCATCAAAGA | TCACCAAACAGCCGCAGACTG |
| NDUFS3F | ATCATATGGCGGCGGCGGC | TGCTCGAGCTACTTGGCATCAGGCTTC |
| ANT1 | CTCTCCTTCTGGAGGGGTAAC | GAACTGCTTATGCCGATCCAC |
| ANT2 | GGGTCAAGCTGCTGCTGCAGG | CGGAATTCCCTTTCAGCTCCAGC |
| ANT3 | CACCAAGTCCGACGGCATCCG | ACGGTTGAGGATTCTACGTGG |
| RIP140 | CCCATTTGCAGCAGTATTCTC | GTAACTGCCAACATCCTTCTG |
| Rip140 (M) | GAACCTGGGCTTTTGAATGG | GTTTTGGTCAGTCTTGGAGAGTCTT |
| RARα | CTCTCCACCAAGTGCATCATTAAG | CAGCCTTGAGGAGGGTGATC |
| RARβ | TGACAGCTGAGTTGGACGAT | AGCACTGGAATTCGTGGTGT |
| RARγ | CTGTGCGAAATGACCGGAAC | CTGCACTGGAGTTCGTGGTA |
| GAPDH | GTGAAGCAGGCGTCGGAG | GCGTCAAAGGTGGAGGAGTG |
| β-actin (M) | CTGTCCCTGTATGCCTCTG | ATGTCACGCACGATTTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, X.; Mu, Q.; Hou, X.; Yu, W.; Guo, J. Histone H3 Acetylation Is Involved in Retinoid Acid-Induced Neural Differentiation through Increasing Mitochondrial Function. Biomedicines 2023, 11, 3251. https://doi.org/10.3390/biomedicines11123251
Zhang Y, Wang X, Mu Q, Hou X, Yu W, Guo J. Histone H3 Acetylation Is Involved in Retinoid Acid-Induced Neural Differentiation through Increasing Mitochondrial Function. Biomedicines. 2023; 11(12):3251. https://doi.org/10.3390/biomedicines11123251
Chicago/Turabian StyleZhang, Yang, Xinjuan Wang, Qing Mu, Xueyu Hou, Weidong Yu, and Jingzhu Guo. 2023. "Histone H3 Acetylation Is Involved in Retinoid Acid-Induced Neural Differentiation through Increasing Mitochondrial Function" Biomedicines 11, no. 12: 3251. https://doi.org/10.3390/biomedicines11123251
APA StyleZhang, Y., Wang, X., Mu, Q., Hou, X., Yu, W., & Guo, J. (2023). Histone H3 Acetylation Is Involved in Retinoid Acid-Induced Neural Differentiation through Increasing Mitochondrial Function. Biomedicines, 11(12), 3251. https://doi.org/10.3390/biomedicines11123251






