Clinical and Microbiological Impact of Implementing a Decision Support Algorithm through Microbiologic Rapid Diagnosis in Critically Ill Patients: An Epidemiological Retrospective Pre-/Post-Intervention Study
Abstract
:1. Introduction
1.1. Primary Objective
1.2. Primary Outcome
1.3. Secondary Outcomes
1.4. Endpoints
2. Material and Method
2.1. Study Design and Population
- (1)
- The clinical decision algorithm was dissemination to all ICU staff physicians from the PROA team through regular face-to-face meetings.
- (2)
- Educational lectures related to the methodology and impact of antimicrobial treatment optimisation were offered to all ICU medical staff.
- (3)
- Biofire® Panel Pneumonia results were communicated in real time to the requesting physician via phone and electronic medical records.
- (4)
- Prospective audits were performed by the PROA team with real-time intervention and feedback to ICU attending physicians during the intervention period for all patients with a suspected LRTI.
2.2. Laboratory Methods
2.3. Reporting Methods
Nosocomial Infection Prevention Measures
2.4. Data Collection
2.4.1. Clinical and Laboratory Data
2.4.2. Antimicrobial Consumption Data
2.4.3. Microbiological Data
2.5. Study Definitions
2.6. Statistical Analysis
3. Results
3.1. Overall Population
3.1.1. Microbiological Findings
3.1.2. Antibiotic Consumption
3.1.3. Pseudomonas aeruginosa Susceptibility Pattern
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin-Loeches, I.; Reyes, L.F.; Nseir, S.; Ranzani, O.; Povoa, P.; Diaz, E.; Schultz, M.J.; Rodríguez, A.H.; Serrano-Mayorga, C.C.; De Pascale, G.; et al. European Network for ICU-Related Respiratory Infections (ENIRRIs): A multinational, prospective, cohort study of nosocomial LRTI. Intensive Care Med. 2023, 49, 1212–1222. [Google Scholar] [CrossRef]
- Pichon, M.; Cremniter, J.; Burucoa, C.; Abdallah, S.; Alauzet, C.; Alix, T.; Allouche, K.; Amara, M.; Anglade, F.; Anguel, N.; et al. French national epidemiology of bacterial superinfections in ventilator-associated pneumonia in patients infected with COVID-19: The COVAP study. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 50. [Google Scholar] [CrossRef]
- Freire, M.P.; de Assis, D.B.; Tavares, B.d.M.; Brito, V.O.; Marinho, I.; Lapchik, M.; Guedes, A.R.; Madalosso, G.; Oliveira, M.S.; de Lima, A.C.P.; et al. Impact of COVID-19 on healthcare-associated infections: Antimicrobial consumption does not follow antimicrobial resistance. Clinics 2023, 78, 100231. [Google Scholar] [CrossRef]
- Tetaj, N.; Capone, A.; Stazi, G.V.; Marini, M.C.; Garotto, G.; Busso, D.; Scarcia, S.; Caravella, I.; Macchione, M.; De Angelis, G.; et al. Epidemiology of ventilator-associated pneumonia in ICU COVID-19 patients: An alarming high rate of multidrug-resistant bacteria. J. Anesth. Analg. Crit. Care 2022, 2, 36. [Google Scholar] [CrossRef]
- Rodriguez Oviedo, A.; Duque, S.; García-Gallo, E.; Bastidas Goyes, A.R.; Reyes, L.F.; Fuentes Barreiro, Y.V.; Ferrer, R. Risk factors for developing ventilator-associated lower respiratory tract infection in patients with severe COVID-19: A multinational, multicentre study, prospective, observational study. Sci. Rep. 2023, 13, 6553. [Google Scholar] [CrossRef]
- Kothari, A.; Kherdekar, R.; Mago, V.; Uniyal, M.; Mamgain, G.; Kalia, R.B.; Kumar, S.; Jain, N.; Pandey, A.; Omar, B.J. Age of Antibiotic Resistance in MDR/XDR Clinical Pathogen of Pseudomonas aeruginosa. Pharmaceuticals 2023, 16, 1230. [Google Scholar] [CrossRef]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Mylonakis, C.E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef]
- Manatrey-Lancaster, J.J.; Bushman, A.M.; Caligiuri, M.E.; Rosa, R. Impact of BioFire FilmArray respiratory panel results on antibiotic days of therapy in different clinical settings. Antimicrob. Steward. Healthc. Epidemiol. 2021, 1, e4. [Google Scholar] [CrossRef]
- Moore, L.S.P.; Villegas, M.V.; Wenzler, E.; Rawson, T.M.; Oladele, R.O.; Doi, Y. Rapid Diagnostic Test Value and Implementation in Antimicrobial Stewardship Across Low-to-Middle and High-Income Countries: A Mixed-Methods Review. Infect. Dis. Ther. 2023, 12, 1445–1463. [Google Scholar] [CrossRef]
- Gatti, M.; Viaggi, B.; Rossolini, G.M.; Pea, F.; Viale, P. An Evidence-Based Multidisciplinary Approach Focused on Creating Algorithms for Targeted Therapy of Infection-Related Ventilator-Associated Complications (IVACs) Caused by Pseudomonas aeruginosa and Acinetobacter baumannii in Critically Ill Adult Patients. Antibiotics 2022, 11, 33. [Google Scholar] [CrossRef]
- Pandolfo, A.M.; Horne, R.; Jani, Y.; Reader, T.W.; Bidad, N.; Brealey, D.; Enne, V.I.; Livermore, D.M.; Gant, V.; Brett, S.J.; et al. Intensivists’ beliefs about rapid multiplex molecular diagnostic testing and its potential role in improving prescribing decisions and antimicrobial stewardship: A qualitative study. Antimicrob. Resist. Infect. Control 2021, 10, 95. [Google Scholar] [CrossRef]
- Iregui, M.; Ward, S.; Sherman, G.; Fraser, V.J.; Kollef, M.H. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator- associated pneumonia. Chest 2002, 122, 262–268. [Google Scholar] [CrossRef]
- Kitano, T.; Nishikawa, H.; Suzuki, R.; Onaka, M.; Nishiyama, A.; Kitagawa, D.; Oka, M.; Masuo, K.; Yoshida, S. The impact analysis of a multiplex PCR respiratory panel for hospitalized pediatric respiratory infections in Japan. J. Infect. Chemother. 2020, 26, 82–85. [Google Scholar] [CrossRef]
- Kuitunen, I.; Renko, M. The effect of rapid point-of-care respiratory pathogen testing on antibiotic prescriptions in acute infections—A systematic review and meta-analysis of randomized controlled trials. Open Forum Infect. Dis. 2023, 10, ofad443. [Google Scholar] [CrossRef]
- Walsh, T.L.; Bremmer, D.N.; Moffa, M.A.; Trienski, T.L.; Buchanan, C.; Stefano, K.; Hand, C.; Taylor, T.; Kasarda, K.; Shively, N.R.; et al. Impact of an Antimicrobial Stewardship Program-bundled initiative utilizing Accelerate Pheno™ system in the management of patients with aerobic Gram-negative bacilli bacteremia. Infection 2021, 49, 511–519. [Google Scholar] [CrossRef]
- World Health Organization (WHO). DDD Indicators. Available online: https://www.who.int/tools/atc-ddd-toolkit/indicators#:~:text=DDD%20per%20100%20bed%20days,stays%20overnight%20in%20a%20hospital (accessed on 1 August 2023).
- Available online: https://seimc.org/contenidos/documentoscientificos/procedimientosmicrobiologia/seimc-procedimientomicrobiologia25.pdf (accessed on 25 November 2023).
- Available online: https://www.eucast.org/clinical_breakpoints (accessed on 25 November 2023).
- Martin-Loeches, I.; Povoa, P.; Rodríguez, A.; Curcio, D.; Suarez, D.; Mira, J.-P.; Cordero, M.L.; Lepecq, R.; Girault, C.; Candeias, C.; et al. Incidence and prognosis of ventilator-associated tracheobronchitis (TAVeM): A multicentre, prospective, observational study. Lancet Respir. Med. 2015, 3, 859–868. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance. (WHO Fact Sheet). World Health Organization: Geneva, Switzerland, February 2018. Available online: https://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 9 August 2022).
- Fridkin, S.K.; Edwards, J.R.; Courval, J.M.; Hill, H.; Tenover, F.C.; Lawton, R.; Gaynes, R.P.; McGowan, J.E., Jr.; Intensive Care Antimicrobial Resistance Epidemiology (ICARE) Project and the National Nosocomial Infections Surveillance (NNIS) System Hospitals. The effect of vancomycin and thirdgeneration cephalosporins on prevalence of vancomycin-resistant enterococci in 126 U.S. adult intensive care units. Ann. Intern. Med. 2001, 135, 175–183. [Google Scholar] [CrossRef]
- Costelloe, C.; Metcalfe, C.; Lovering, A.; Mant, D.; Hay, A.D. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: Systematic review and meta-analysis. BMJ 2010, 340, c2096. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Sakr, Y.; Singer, M.; Martin-Loeches, I.; Machado, F.R.; Marshall, J.C.; Finfer, S.; Pelosi, P.; Brazzi, L.; Aditianingsih, D.; et al. Prevalence and Outcomes of Infection Among Patients in Intensive Care Units in 2017. JAMA 2020, 323, 1478–1487. [Google Scholar] [CrossRef]
- Ansari, S.; Hays, J.P.; Kemp, A.; Okechukwu, R.; Murugaiyan, J.; Ekwanzala, M.D.; Alvarez, M.J.R.; Paul-Satyaseela, M.; Iwu, C.D.; Balleste-Delpierre, C.; et al. The potential impact of the COVID-19 pandemic on global antimicrobial and biocide resistance: An AMR Insights global perspective. JAC-Antimicrob. Resist. 2021, 3, dlab038. [Google Scholar] [CrossRef]
- Tomczyk, S.; Taylor, A.; Brown, A.; de Kraker, M.E.A.; El-Saed, A.; Alshamrani, M.; Hendriksen, R.S.; Jacob, M.; Löfmark, S.; Perovic, O.; et al. Impact of the COVID-19 pandemic on the surveillance, prevention and control of antimicrobial resistance: A global survey. J. Antimicrob. Chemother. 2021, 76, 3045–3058. [Google Scholar] [CrossRef] [PubMed]
- CDC. COVID-19 & Antibiotic Resistance. 2022. Available online: https://www.cdc.gov/drugresistance/covid19.html (accessed on 27 May 2022).
- Weiner-Lastinger, L.M.; Pattabiraman, V.; Konnor, R.Y.; Patel, P.R.; Wong, E.; Xu, S.Y.; Smith, B.; Edwards, J.R.; Dudeck, M.A. The impact of coronavirus disease 2019 (COVID-19) on healthcare-associated infections in 2020: A summary of data reported to the National Healthcare Safety Network. Infect. Control Hosp. Epidemiol. 2021, 43, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; Soucy, J.P.; Leung, V.; So, M.; Kwan, A.T.; Portnoff, J.S.; Bertagnolio, S.; Raybardhan, S.; MacFadden, D.R.; Daneman, N. Antibiotic resistance associated with the COVID-19 pandemic: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2023, 29, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Darie, A.M.; Khanna, N.; Jahn, K.; Osthoff, M.; Bassetti, S.; Osthoff, M.; Schumann, D.M.; Albrich, W.C.; Hirsch, H.; Brutsche, M.; et al. Fast multiplex bacterial PCR of bronchoalveolar lavage for antibiotic stewardship in hospitalised patients with pneumonia at risk of Gram-negative bacterial infection (Flagship II): A multicentre, randomised controlled trial. Lancet Respir. Med. 2022, 10, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Bergese, S.; Fox, B.; García-Allende, N.; Elisiri, M.E.; Schneider, A.E.; Ruiz, J.; Gonzalez-Fraga, S.; Rodriguez, V.; Fernandez-Canigia, L. Impact of the multiplex molecular FilmArray Respiratory Panel on antibiotic prescription and clinical management of immunocompromised adults with suspected acute respiratory tract infections: A retrospective before-after study. Rev. Argent. De Microbiol. 2023, in press. [Google Scholar] [CrossRef]
- Qian, Y.; Ai, J.; Wu, J.; Yu, S.; Cui, P.; Gao, Y.; Jin, J.; Weng, X.; Zhang, W. Rapid detection of respiratory organisms with FilmArray respiratory panel and its impact on clinical decisions in Shanghai, China, 2016–2018. Influenza Other Respi Viruses 2020, 14, 142–149. [Google Scholar] [CrossRef]
- Rizk, N.A.; Zahreddine, N.; Haddad, N.; Ahmadieh, R.; Hannun, A. The Impact of Antimicrobial Stewardship and Infection Control Interventions on Acinetobacter baumannii Resistance Rates in the ICU of a Tertiary Care Center in Lebanon. Antibiotics 2022, 11, 911. [Google Scholar] [CrossRef]
- High, J.; Enne, V.I.; Barber, J.A.; Brealey, D.; Turner, D.A.; Horne, R.; Peters, M.; Dhesi, Z.; Wagner, A.P.; Pandolfo, A.M.; et al. INHALE: The impact of using FilmArray Pneumonia Panel molecular diagnostics for hospital-acquired and ventilator-associated pneumonia on antimicrobial stewardship and patient outcomes in UK Critical Care—Study protocol for a multicentre randomised controlled trial. Trials 2021, 22, 680. [Google Scholar] [CrossRef]
- Carbonell, R.; Urgelés, S.; Rodríguez, A.; Bodí, M.; Martín-Loeches, I.; Solé-Violán, J.; Díaz, E.; Gómez, J.; Trefler, S.; Vallverdú, M.; et al. Mortality comparison between the first and second/third waves among 3795 critical COVID-19 patients with pneumonia admitted to the ICU: A multicentre retrospective cohort study. Lancet Reg. Health–Eur. 2021, 11, 100243. [Google Scholar] [CrossRef]
- Kariyawasam, R.M.; Julien, D.A.; Jelinski, D.C.; Larose, S.L.; Rennert-May, E. Antimicrobial resistance (AMR) in COVID-19 patients: A systematic review and meta-analysis (November 2019–June 2021). Antimicrob. Resist. Infect. Control. 2022, 11, 45. [Google Scholar] [CrossRef]
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, J.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’Grady, N.P.; Bartlett, J.G.; Carratalà, J.; et al. Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef]
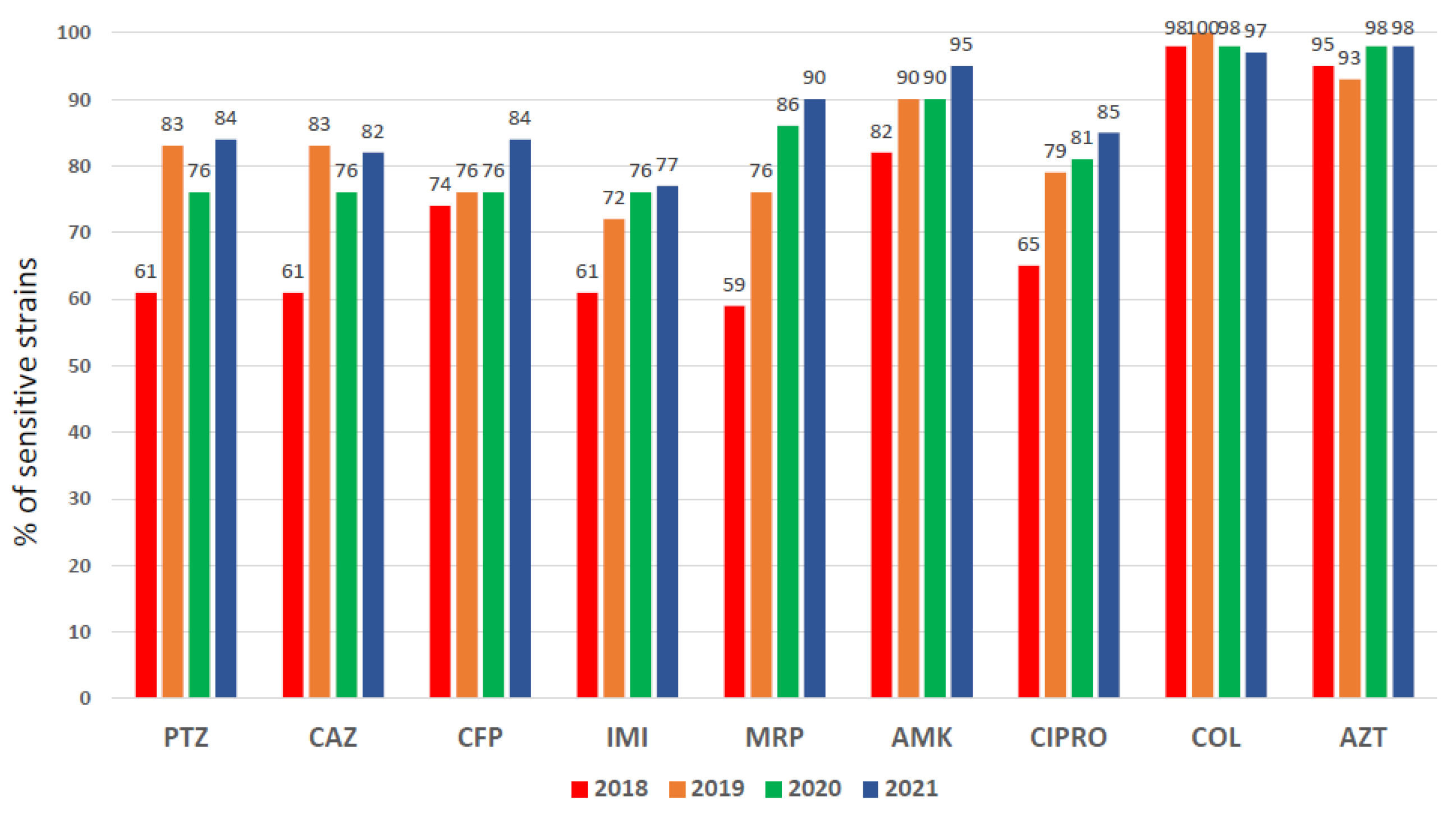

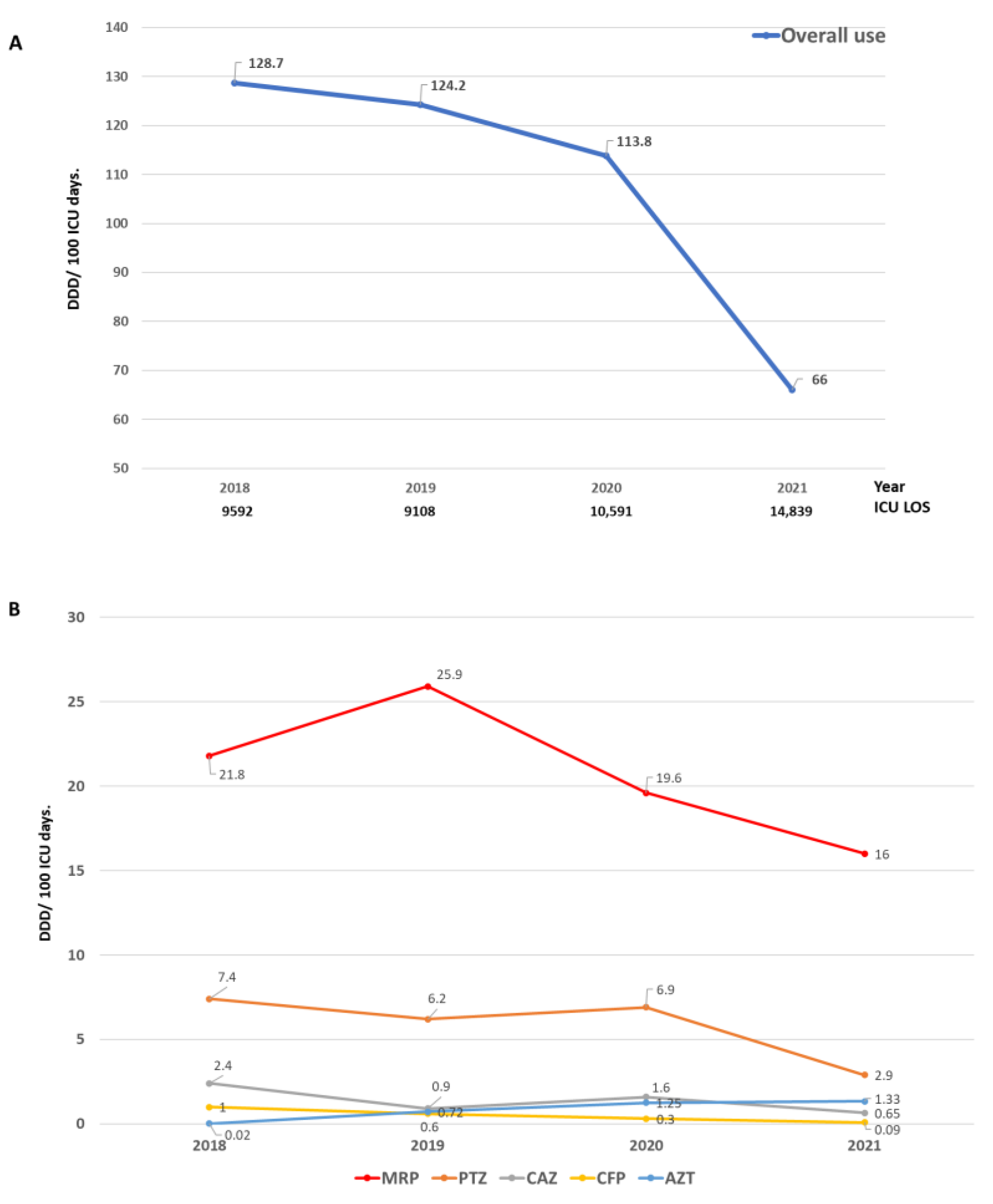
| Study Period | Overall | Pre-Intervention | Intervention | p-Value | ||
|---|---|---|---|---|---|---|
| Variable | n = 3635 | 2018 (n = 987) | 2019 (n = 979) | 2020 (n = 804) | 2021 (n = 865) | |
| Demographics and Severity | ||||||
| Age, mean (Q1–Q3) | 60 (50–72) | 64 (52–73) | 64 (50–73) | 63 (50–72) | 61 (49–72) *** | 0.009 |
| Male, n (%) | 2350 (64.6) | 634 (64.2) | 620 (63.3) | 536 (66.7) | 560 (64.7) | 0.52 |
| APACHE, mean (Q1–Q3) | 19 (14–25) | 20 (15–25) | 20 (15–25) | 18.5 (14–24) *** | 19 (14–25) | 0.003 |
| SOFA, mean (Q1–Q3) | 3 (1–6) | 3.0 (2.0–5.1) | 2.2 (1.0–5.0) *** | 3.0 (2.0–6.0) | 3 (2.0–6.0) | <0.001 |
| Patients Type | ||||||
| Surgical, n (%) | 955 (26.3) | 275 (27.9) | 300 (30.7) *** | 199 (24.7) | 181 (21.0) ** | <0.001 |
| Medical, n (%) | 2680 (73.7) | 712 (72.1) | 679 (69.3) | 605 (75.3) | 684 (79.0) ** | <0.001 |
| COVID-19, n (%) within medical patients | 398 (14.8) | 0 (0.0) | 0 (0.0) | 173 (28.5) | 225 (32.9) | NA |
| Comorbidities | ||||||
| Obesity, n (%) | 569 (15.6) | 123 (12.4) | 146 (14.9) ** | 135 (16.8) *** | 165 (19.0) *** | <0.001 |
| Diabetes, n (%) | 825 (22.7) | 239 (24.2) | 229 (23.4) | 174 (21.6) | 183 (21.1) | 0.35 |
| Chronic heart disease, n (%) | 161 (4.4) | 61 (6.2) | 35 (3.6) ** | 30 (3.7) * | 35 (4.0) | 0.01 |
| COPD, n (%) | 400 (11.0) | 126 (12.7) | 120 (12.2) | 89 (11.0) | 65 (7.5) *** | 0.001 |
| Chronic Rennal failure, n (%) | 339 (9.3) | 93 (9.4) | 88 (9.0) | 78 (9.7) | 80 (9.2) | 0.96 |
| Immunosupression, n (%) | 159 (4.4) | 43 (4.3) | 33 (3.4) | 47 (5.8) | 36 (4.1) | 0.29 |
| Laboratory | ||||||
| Hemoglobin g/dL, median (Q1–Q3) | 10.0 (8.5–12.0) | 10.3 (8.6–12.2) | 10.1(8.6–12.0) | 9.7 (8.4–11.5) *** | 10.1 (8.6–12.1) | <0.001 |
| WBC count 103/uL, median (Q1–Q3) | 10.4 (8.0–13.6) | 10.8 (8.1–13.9) | 10.3 (7.9–13.5) | 10.4 (8.3–13.5) | 10.7 (8.2–13.8) | 0.21 |
| Serum creatinine mg/dL, median (Q1–Q3) | 0.7 (0.5–1.1) | 0.7 (0.6–1.1) | 0.7 (0.5–1.1) | 0.7 (0.5–1.1) | 0.7 (0.5–1.1) | 0.19 |
| PCT ng/mL, median (Q1–Q3) | 2.25(0.57–7.27) | 3.17 (1.27–8.65) | 2.65(0.94–8.34) | 1.60(0.34–6.0) | 1.11(0.26–5.23) | <0.001 |
| RCP mg/dL, median (Q1–Q3) | 9.7 (4.2–18.9) | 9.9 (5.3–18) | 9.5 (4.9–16.4) | 9.4 (4.8–17.0) | 8.5 (4.2–16.0) *** | 0.01 |
| Microbiologically Confirmed Infections During ICU Stay | ||||||
| Total number of infections, n (%) | 463 (100) | 83 (17.9) | 55 (11.9) | 119 (25.7) | 206 (44.5) | <0.001 |
| Ventilator-associated pneumonia (VAP), n (%) | 163 (35.2) | 21 (25.3) | 12 (21.8) | 38 (32.0) | 92 (44.6) *** | 0.01 |
| Bacteraemia secondary to other septic foci (BS), n (%) | 57 (12.3) | 17 (20.5) | 9 (16.6) | 14 (11.8) | 17 (8.3) ** | 0.02 |
| Bacteraemia of unknown origin (BUNK), n (%) | 69 (14.9) | 9 (10.8) | 12 (21.8) | 19 (16.0) | 29 (14.0) | 0.33 |
| Catheter-associated urinary tract infection (CAUTI), n (%) | 50 (10.8) | 8 (9.6) | 3 (5.4) | 10 (8.4) | 29 (14.0) | 0.19 |
| Ventilator-associated tracheobronchitis (VAT), n (%) | 37 (8.0) | 5 (6.0) | 3 (5.4) | 16 (13.4) | 13 (6.4) | 0.08 |
| Catheter-related bacteraemia (CRB), n (%) | 51 (11.0) | 4 (4.9) | 3 (5.4) | 22 (18.4) ** | 22 (10.7) | 0.008 |
| Intra-abdominal infections (IAI), n (%) | 10 (2.1) | 4 (4.9) | 5 (9.1) | 0 (0%) | 1 (0.5) * | <0.001 |
| Skin and soft tissue infection (SSTI), n (%) | 9 (2.0) | 4 (4.8) | 3 (5.4) | 0 (0%) | 2 (1.0) * | 0.01 |
| Others, n (%) | 17 (3.7) | 11 (13.2) | 5 (9.1) | 0 (0%) | 1 (0.5) *** | <0.001 |
| Main Micro-Organisms Isolated During ICU Stay | ||||||
| Total number of microorganisms isolated, n (%) | 602 (100) | 102 (17.0) | 75 (12.4) | 159 (26.4) | 266 (44.2) | <0.01 |
| Staphylococcus aureus | 86 (14.4) | 16 (15.7) | 9 (12.0) | 23 (14.5) | 38 (14.2) | 0.85 |
| Escherichia coli | 62 (10.4) | 13 (12.7) | 7 (9.3) | 12 (7.5) | 30 (11.3) | 0.50 |
| Klebsiella pneumoniae | 68 (11.3) | 10 (9.8) | 10 (13.3) | 18 (11.3) | 30 (11.3) | 0.91 |
| Pseudomonas aeruginosa | 87 (14.4) | 9 (8.8) | 9 (12.0) | 20 (12.6) | 49 (18.4) * | 0.07 |
| Enterobacter aerogenes | 19 (3.1) | 7 (6.8) | 0 (0%) | 5 (3.1) | 7 (2.6) | 0.06 |
| Serratia marcescens | 26 (4.3) | 6 (5.8) | 1 (1.3) | 5 (3.1) | 14 (5.3) | 0.34 |
| Haemophilus influenzae | 28 (4.6) | 5 (4.9) | 4 (5.3) | 8 (5.0) | 11 (4.1) | 0.94 |
| Enterococcus faecium | 13 (2.1) | 4 (3.9) | 5 (6.7) | 1 (0.6) | 3 (1.1) | 0.79 |
| Klebsiella oxytoca | 15 (2.5) | 4 (3.9) | 1 (1.3) | 4 (2.5) | 6 (2.6) | 0.72 |
| Proteus mirabilis | 11 (1.8) | 3 (2.9) | 0 (0%) | 5 (3.1) | 3 (1.1) | 0.22 |
| Citrobacter spp. | 16 (2.6) | 3 (2.9) | 0 (0%) | 4 (2.5) | 9 (3.4) | 0.45 |
| Enterobacter cloacae | 32 (5.3) | 3 (2.9) | 7 (9.3) | 10 (6.3) | 12 (4.5) | 0.24 |
| Enterococcus faecalis | 33 (5.5) | 3 (2.9) | 2 (2.7) | 18 (11.3) * | 10 (3.7) | 0.02 |
| Others | 106 (17.7) | 16 (15.6) | 20 (26.6) | 26 (16.3) | 44 (16.4) | 0.18 |
| Complications and Outcome | ||||||
| Invasive Mechanical ventilation, n (%) | 1802 (49.6) | 425 (43.1) | 421 (43.0) | 476 (59.2) *** | 480 (55.5) *** | <0.001 |
| LOS ICU, mean (Q1–Q3) | 4.1(2.0–10.2) | 4.0 (2.0–8.0) | 3.6 (1.8–7.7) ** | 4.8 (2.2–14.0) *** | 5.4(2.2–14.1) *** | <0.001 |
| Crude ICU Mortality, n (%) | 625 (17.2) | 165 (16.7) | 148 (15.1) | 158 (19.7) | 154 (17.8) | 0.08 |
| (A) | |||||||
|---|---|---|---|---|---|---|---|
| Study Period | Pre-Intervention | Intervention | RR | 95%ICRR | |||
| Variable | 2018 (n = 987) (1) | 2019–21 (n = 2648) (2) | 2 vs. 1 | ||||
| Incidence density of reported ICU-associated infections | |||||||
| VAP episodes/1000 mechanical ventilation days | 5.5 | 7.33 | 1.33 | 0.4–4.1 | |||
| CAUTI episodes/1000 urinary catheter days | 1.30 | 1.55 | 1.19 | 0.1–11.2 | |||
| CRB and BUNK episodes/1000 catheter days | 1.7 | 2.8 | 1.64 | 0.2–11.0 | |||
| BS episodes/1000 ICU days | 2.3 | 1.3 | 0.56 | 0.1–4.8 | |||
| (B) | |||||||
| Pre-Intervention | Intervention | RR (95% CI) | |||||
| Variable | 2018 (1) (n= 987) | 2019 (2) (n = 979) | 2020 (3) (n = 804) | 2021 (4) (n = 865) | RR 2 vs. 1 (95% CI) | RR 3 vs. 1 (95% CI) | RR 4 vs. 1 (95% CI) |
| VAP episodes/1000 mechanical ventilation days | 5.5 | 2.82 | 6.28 | 12.9 | 0.5 (0.3–1.4) | 1.14 (0.7–1.9) | 2.3 ** (1.4–3.7) |
| CAUTI episodes/1000 urinary catheter days | 1.30 | 0.46 | 1.18 | 3.01 | 0.35 (0.1–1.3) | 0.90 (0.3–2.2) | 2.3 ** (1.1–6.1) |
| CRB and BUNK episodes/1000 catheter days | 1.7 | 1.8 | 3.0 | 3.5 | 1.05 (0.4–2.5) | 1.8 (0.5–2.0) | 2.0 (1.0–3.5) |
| BS episodes/1000 ICU days | 2.3 | 1.1 | 1.4 | 1.4 | 0.5 (0.2–1.2) | 0.6 (0.2–1.7) | 0.6 (0.3–1.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, A.; Gómez, F.; Sarvisé, C.; Gutiérrez, C.; Giralt, M.G.; Guerrero-Torres, M.D.; Pardo-Granell, S.; Picó-Plana, E.; Benavent-Bofill, C.; Trefler, S.; et al. Clinical and Microbiological Impact of Implementing a Decision Support Algorithm through Microbiologic Rapid Diagnosis in Critically Ill Patients: An Epidemiological Retrospective Pre-/Post-Intervention Study. Biomedicines 2023, 11, 3330. https://doi.org/10.3390/biomedicines11123330
Rodríguez A, Gómez F, Sarvisé C, Gutiérrez C, Giralt MG, Guerrero-Torres MD, Pardo-Granell S, Picó-Plana E, Benavent-Bofill C, Trefler S, et al. Clinical and Microbiological Impact of Implementing a Decision Support Algorithm through Microbiologic Rapid Diagnosis in Critically Ill Patients: An Epidemiological Retrospective Pre-/Post-Intervention Study. Biomedicines. 2023; 11(12):3330. https://doi.org/10.3390/biomedicines11123330
Chicago/Turabian StyleRodríguez, Alejandro, Frederic Gómez, Carolina Sarvisé, Cristina Gutiérrez, Montserrat Galofre Giralt, María Dolores Guerrero-Torres, Sergio Pardo-Granell, Ester Picó-Plana, Clara Benavent-Bofill, Sandra Trefler, and et al. 2023. "Clinical and Microbiological Impact of Implementing a Decision Support Algorithm through Microbiologic Rapid Diagnosis in Critically Ill Patients: An Epidemiological Retrospective Pre-/Post-Intervention Study" Biomedicines 11, no. 12: 3330. https://doi.org/10.3390/biomedicines11123330






