Occurrence, Role, and Challenges of MicroRNA in Human Breast Milk: A Scoping Review
Abstract
:1. Introduction
- What microRNA subtypes occur in human breast milk and what are their functions?
- What conditions affect the quantities and types of microRNA in breast milk?
2. Methods
2.1. Protocol
2.2. Data Sources
2.3. Eligibility Criteria
2.4. Source Selection
2.5. Data Extraction and Charting
2.6. Synthesis of Results
3. Results
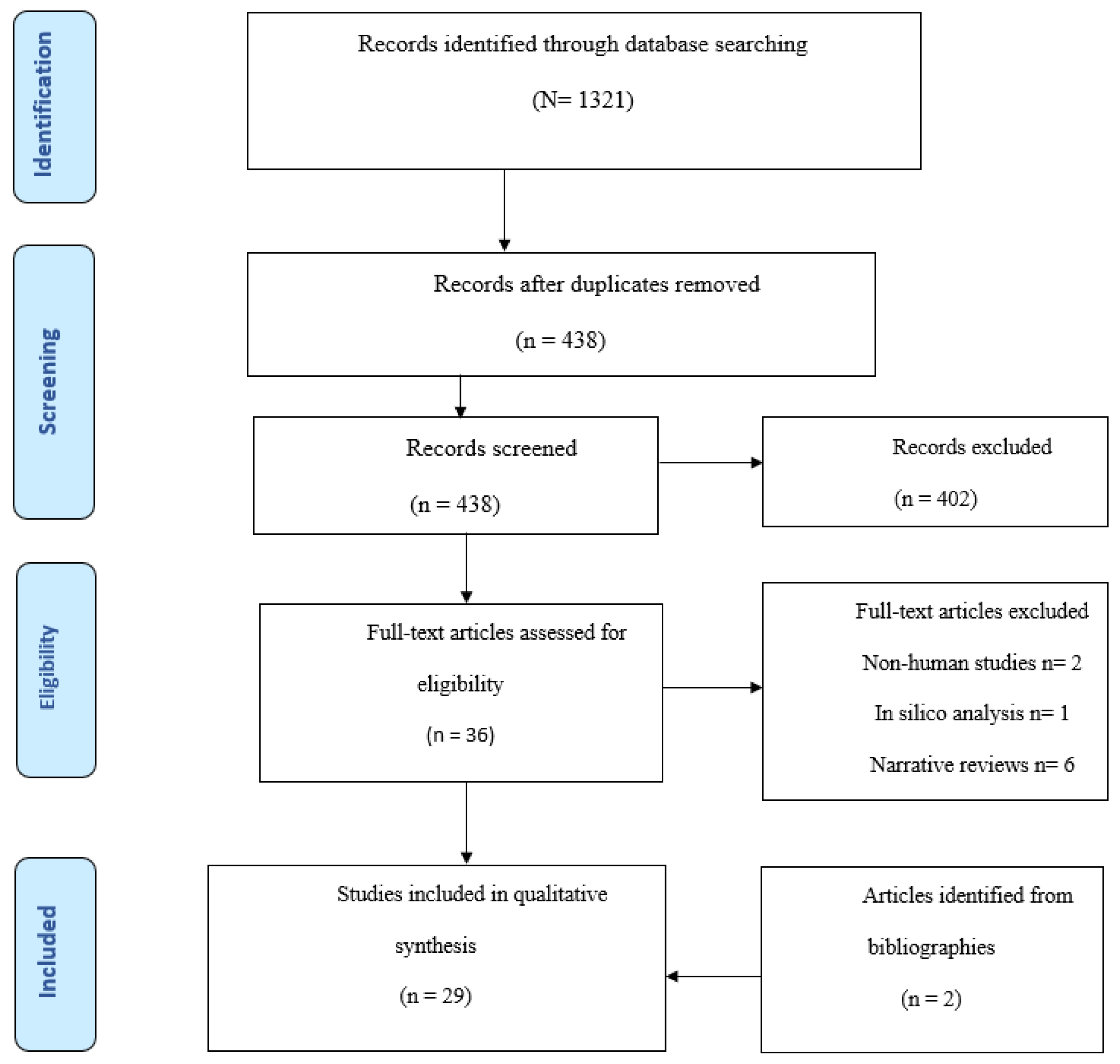
3.1. Selection of Sources
3.2. Study Characteristics
3.3. MicroRNA Isolation and Quantification
3.4. Types and Functions of microRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhi, X.; Xu, R.; Liang, Z.; Wang, F.; Li, X.; Li, Y.; Sun, B. Exploration of microrna profiles in human colostrum. Ann. Transl. Med. 2020, 8, 1170. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. MicroRNA as a new immune-regulatory agent in Breast Milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Vélez-Ixta, J.M.; Benítez-Guerrero, T.; Aguilera-Hernández, A.; Martínez-Corona, H.; Corona-Cervantes, K.; Juárez-Castelán, C.J.; Rangel-Calvillo, M.N.; García-Mena, J. Detection and quantification of immunoregulatory mirnas in human milk and infant milk formula. BioTech 2022, 11, 11. [Google Scholar] [CrossRef]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating micrornas in breast milk and their potential impact on the infant. Nutrients 2020, 12, 3066. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L.; et al. Prisma extension for scoping reviews (PRISMA-SCR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [Green Version]
- Ahlberg, E.; Jenmalm, M.C.; Tingö, L. Evaluation of five column-based isolation kits and their ability to extract MIRNA from human milk. J. Cell. Mol. Med. 2021, 25, 7973–7979. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hepworth, A.R.; Lefèvre, C.; Hartmann, P.E.; Geddes, D.T.; Hassiotou, F. Human milk microrna and total RNA differ depending on milk fractionation. J. Cell. Biochem. 2015, 116, 2397–2407. [Google Scholar] [CrossRef] [Green Version]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel mirnas, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef] [Green Version]
- Bozack, A.K.; Colicino, E.; Rodosthenous, R.; Bloomquist, T.R.; Baccarelli, A.A.; Wright, R.O.; Wright, R.J.; Lee, A.G. Associations between maternal lifetime stressors and negative events in pregnancy and breast milk-derived extracellular vesicle micrornas in the programming of Intergenerational Stress Mechanisms (prism) pregnancy cohort. Epigenetics 2020, 16, 389–404. [Google Scholar] [CrossRef]
- Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Silveyra, P.; Podany, A.; Birch, L.L.; Paul, I.M.; Kelleher, S.; Hicks, S.D. Metabolism-related micrornas in maternal breast milk are influenced by premature delivery. Pediatr. Res. 2017, 82, 226–236. [Google Scholar] [CrossRef]
- Floris, I.; Billard, H.; Boquien, C.Y.; Joram-Gauvard, E.; Simon, L.; Legrand, A.; Boscher, C.; Rozé, J.C.; Bolaños-Jiménez, F.; Kaeffer, B. MIRNA analysis by quantitative PCR in preterm human breast milk reveals daily fluctuations of HSA-Mir-16-5P. PLoS ONE 2015, 10, e0140488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H. Transcriptome profiling of microrna by Next-Gen Deep Sequencing reveals known and novel Mirna species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Bustamante, M.; Hernandez-Ferrer, C.; Fernandez-Orth, D.; Pantano, L.; Sarria, Y.; Piqué-Borras, M.; Vellve, K.; Agramunt, S.; Carreras, R.; et al. Circulating mirnas, isomirs and small RNA clusters in human plasma and breast milk. PLoS ONE 2018, 13, e0193527. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Confair, A.; Warren, K.; Chandran, D. Levels of breast milk micrornas and other non-coding RNAS are impacted by milk maturity and maternal diet. Front. Immunol. 2022, 12, 5754. [Google Scholar] [CrossRef]
- Leiferman, A.; Shu, J.; Upadhyaya, B.; Cui, J.; Zempleni, J. Storage of extracellular vesicles in human milk, and MicroRNA profiles in human milk exosomes and infant formulas. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 235–238. [Google Scholar] [CrossRef]
- Raymond, F.; Lefebvre, G.; Texari, L.; Pruvost, S.; Metairon, S.; Cottenet, G.; Zollinger, A.; Mateescu, B.; Billeaud, C.; Picaud, J.C.; et al. Longitudinal human milk mirna composition over the first 3 mo of lactation in a cohort of healthy mothers delivering term infants. J. Nutr. 2021, 152, 94–106. [Google Scholar] [CrossRef]
- Shiff, Y.E.; Reif, S.; Marom, R.; Shiff, K.; Reifen, R.; Golan-Gerstl, R. Mirna-320A is less expressed and MIRNA-148a more expressed in preterm human milk compared to term human milk. J. Funct. Foods 2019, 57, 68–74. [Google Scholar] [CrossRef]
- Shah, K.B.; Chernausek, S.D.; Garman, L.D.; Pezant, N.P.; Plows, J.F.; Kharoud, H.K.; Demerath, E.W.; Fields, D.A. Human milk exosomal microrna: Associations with maternal overweight/obesity and infant body composition at 1 month of life. Nutrients 2021, 13, 1091. [Google Scholar] [CrossRef]
- Smyczynska, U.; Bartlomiejczyk, M.A.; Stanczak, M.M.; Sztromwasser, P.; Wesolowska, A.; Barbarska, O.; Pawlikowska, E.; Fendler, W. Impact of processing method on donated human breast milk microrna content. PLoS ONE 2020, 15, e0236126. [Google Scholar] [CrossRef]
- Wang, H.; Wu, D.; Sukreet, S.; Delaney, A.; Belfort, M.B.; Zempleni, J. Quantitation of exosomes and their MicroRNA cargos in Frozen Human Milk. JPGN Rep. 2022, 3, e172. [Google Scholar] [CrossRef]
- Yang, L.; Hu, R.; Li, J.; Mo, X.; Xu, L.; Shen, N.; Sheng, W.; Li, Y. Exosomal micrornas in human breast milk: Potential effect on neonatal breast milk jaundice. Res. Sq. 2020, PPR223039. [Google Scholar]
- Pisano, C.; Galley, J.; Elbahrawy, M.; Wang, Y.; Farrell, A.; Brigstock, D.; Besner, G.E. Human breast milk-derived extracellular vesicles in the protection against experimental necrotizing enterocolitis. J. Pediatr. Surg. 2020, 55, 54–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.Q.; Gong, G.; Xu, Z.L.; Wang, L.Y.; Fang, M.L.; Zhou, H.; Xing, H.; Wang, K.R.; Sun, L. MIRNA profiling reveals a potential role of milk stasis in breast carcinogenesis. Int. J. Mol. Med. 2014, 33, 1243–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human breast milk mirna, maternal probiotic supplementation and atopic dermatitis in offspring. PLoS ONE 2015, 10, e0143496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, B.; Kim, Y.; Park, D.J.; Oh, S. Comparative analysis of dietary exosome-derived micrornas from human, bovine and Caprine Colostrum and mature milk. J. Anim. Sci. Technol. 2021, 63, 593–602. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related micrornas are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef]
- Kahn, S.; Liao, Y.; Du, X.; Xu, W.; Li, J.; Lönnerdal, B. Exosomal MicroRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2018, 62, 1701050. [Google Scholar] [CrossRef]
- Na, R.S.; GX, E.; Sun, W.; Sun, X.W.; Qiu, X.Y.; Chen, L.P.; Huang, Y.F. Expressional analysis of immune-related mirnas in breast milk. Genet. Mol. Res. 2015, 14, 11371–11376. [Google Scholar] [CrossRef]
- Karlsson, O.; Rodosthenous, R.S.; Jara, C.; Brennan, K.J.; Wright, R.O.; Baccarelli, A.A.; Wright, R.J. Detection of long non-coding RNAS in human breastmilk extracellular vesicles: Implications for early child development. Epigenetics 2016, 11, 721–729. [Google Scholar] [CrossRef] [Green Version]
- Kupsco, A.; Prada, D.; Valvi, D.; Hu, L.; Petersen, M.S.; Coull, B.; Grandjean, P.; Weihe, P.; Baccarelli, A.A. Human milk extracellular vesicle MIRNA expression and associations with maternal characteristics in a population-based cohort from the Faroe Islands. Sci. Rep. 2021, 11, 5840. [Google Scholar] [CrossRef]
- Lukasik, A.; Brzozowska, I.; Zielenkiewicz, U.; Zielenkiewicz, P. Detection of plant mirnas abundance in human breast milk. Int. J. Mol. Sci. 2017, 19, 37. [Google Scholar] [CrossRef] [PubMed]
| First Author, Year (Country) | Study Aims | Methods | Sample Size | Findings |
|---|---|---|---|---|
| 1. microRNA EXTRACTION TECHNIQUES | ||||
| Ahlberg, 2021 (Sweden) [6] | To determine the efficiency of five RNA extraction kits in retrieving microRNA from human breast milk. | Five internationally available RNA extraction kits were used to retrieve the microRNA hsa-miR-39-3p and an exogenous control microRNA from four samples of skim milk. Total and specific RNA extracts were quantified using a Qubit microRNA assay kit, and performance was compared using statistical analysis in GraphPad Prism. | Five international kits were compared to determine the extraction efficiency: Promega, Zymo, Norgen RNA, Norgen Cell, and Sigma-Aldrich. | Two of the extraction kits, Promega and Zymo, had the highest yields of mRNA extracts. Variations in kit performance highlight the need for a standardized protocol for research on mRNA in milk. |
| Alsaweed, 2015 (Australia) [7] | To standardize microRNA isolation from three fractions of centrifuged human milk using different methods. | Human milk samples were fractionated, and microRNA content was extracted from each layer (lipid, skim milk and cells) using 8 commercially available kits. | 49 milk samples from 29 lactating women. | All three layers of fractionated milk had microRNA content, with the cellular and lipid layers having the highest concentrations. |
| 2. TYPES AND QUANTITIES OF ISOLATED microRNAS | ||||
| Alsaweed, 2016 (Australia) [8] | To determine the types of microRNAs present in human milk after different lactation periods. | Milk samples were collected at 2, 4, and 6 months of lactation. Bioinformatics analysis of microRNAs extracted from the lipid and cell layers was conducted to identify known and predicted microRNAs. | Breast milk was obtained from 10 lactating women | MicroRNA content in cell fractions was higher than in lipid fractions. The most common microRNA species from both columns was let-7f-5p. Across the three lactation stages, the profile (amount and species) of microRNA in breast milk changed significantly, suggesting adaptation to infant’s needs. |
| Bozack, 2020 (USA) [9] | To investigate the association between maternal psychological stress and the microRNA amount and quantity expressed in breast milk. | Psychological stress was assessed using Life Stressor Checklist-Revised, while microRNA profiling was performed using the TaqMan Open Array Human miRNA panel. Statistical analysis based on regression was used to determine association. | Breast milk was obtained from 80 lactating women | 744 microRNAs were detected in total. The quantity of microRNA in breast milk was negatively correlated with psychological stress scores. The types of microRNA extracted also differed depending on psychological stress, with six species demonstrating the highest association with high stress scores. |
| Carney, 2017 (USA) [10] | To determine the correlation between gestational age at delivery and maternal milk microRNA profiles. | MicroRNA content from breast milk from mothers of infants born prematurely was compared with breast milk collected from mothers of term babies. | Colostrum and hind milk samples were obtained from the total of 44 mothers. | The quantities of nine microRNAs differed across the two groups. The affected microRNAs had target genes related to metabolism and lipid formation. |
| Floris, 2015 (France) [11] | To investigate changes in breast milk microRNA content in a 24-h period. | MicroRNA content from samples collected at different times were analyzed to determine expression of 4 reference species. | 84 milk samples were collected at different times from 22 healthy mothers. | Two microRNA species, hsa-miR-16-5p, hsa-miR-21-5p, were stably expressed throughout the day, supporting their potential for use as endogenous reference species in future studies. |
| Munch, 2013 (USA) [12] | To determine the amount and types of microRNAs in human breast milk. | Deep sequence analysis of human breast milk to identify known and novel microRNAs. | 5 lactating women | Generated sequences matched 308 known mature microRNAs targeting 9074 genes; and 21 novel microRNAs. A number of the novel microRNAs had origins from enriched foods. |
| Rubio, 2018 (Spain) [13] | To characterize the profiles of small RNA species in milk and plasma. | Isolation and quantitation of clusters of small RNAs from plasma and breast milk samples. | 15 healthy postpartum mothers | Both milk and plasma expressed microRNAs, piwi-interfering RNAs (piRNAs), tRNAs, and small nucleolar RNAs. The concentrations of different small RNAs varied between the two biofluids. |
| 3. EFFECTS OF PROCESSING, TIMING, ENVIRONMENTAL, AND MATERNAL CONDITIONS | ||||
| Hicks, 2022 (USA) [14] | To identify the most abundant microRNAs in milk and determine their variation with time. | Deep sequence analysis to identify microRNAs in samples collected at different times. | 503 samples obtained from 192 mothers | The quantity of most microRNA species increases with breast milk maturity, with nearly half being influenced by maternal diet. |
| Leiferman, 2019 (USA) [15] | To assess exosome and microRNA expression in human breast milk and commercial infant formulas. | RNA sequencing of fresh and stored breast milk and infant formulas | Breast milk was obtained from 5 lactating mothers. | Fresh human breast milk had 21 microRNAs stored in exosomes, while infant formulas had none. Storage at 4 °C resulted in a 49% decrease in the microRNA content in breast milk. |
| Raymond, 2021 (France) [16] | To characterize the microRNA profile of breast milk from week 2 to the second month post-partum | Next generation sequencing of breast milk samples collected at different times. | 44 mothers | 685 microRNAs were isolated, 35 of which were present in stable quantities throughout the lactation period. |
| Shiff, 2019 (Israel) [17] | To investigate the differences in the microRNA profiles of breast milk collected from mothers of premature and term infants. | Quantitative real-time PCR detection and comparison of four common microRNA species | 38 healthy mothers | MicroRNA 320 occurred more commonly in colostrum of term infant mothers, while microRNA 148a occurred more commonly in premature human breast milk. |
| Shah, 2021 (USA) [18] | To examine the association between maternal obesity/overweight and expression of specific microRNAs in breast milk. | Real-time quantitative PCR for 6 specific microRNAs, followed by regression analysis to determine association with different maternal characteristics. | 60 lactating mothers | Two of the six microRNAs, miR-148a and miR-30b, were lower in overweight mothers than in normal weight mothers. |
| Smyczynska, 2020 (Poland) [19] | To determine the effects of pasteurization on the microRNA content in human breast milk. | Milk samples were pasteurized using either Holder pasteurization or high-pressure processing, or left unpasteurized. Subsequently, they were analyzed for microRNA content using quantitative sequencing techniques. | Milk samples were collected from 3 volunteers | Pasteurization resulted in significant reduction in microRNA content in the breast milk samples. |
| Wang, 2022 (USA) [20] | To assess the feasibility of microRNA analysis in frozen human breast milk. | Breast milk samples were frozen at -80o C, then analyzed for microRNAs and exosomes using PCR and immunoblot techniques. | Milk samples were obtained from three mothers of preterm infants. | While freezing resulted in significant reductions in microRNA and exosome levels, analysis was still feasible. |
| Wu, 2020 (China) [1] | To investigate the expression of microRNA in human colostrum. | Microarray analysis of colostrum samples and prediction of microRNA targets. | 18 lactating volunteers | The microRNA composition of human milk changes through the lactation period. 49 microRNAs were consistently found in colostrum in higher concentrations than in milk. |
| 4. ROLE OF microRNAS IN DISEASES | ||||
| Yang, 2020 (China) [21] | To determine whether microRNAs in breast milk contribute to breast milk jaundice. | Flow cytometry, Western blotting, and nanoparticle tracking were used to monitor exosomes in breast milk, and association analysis was used to find association with breast milk jaundice. | 32 mother-infant dyads | Based on the expression of a few microRNA species, the evidence supports a role for microRNAs in breast milk jaundice. |
| Pisano, 2020 (USA) [22] | To determine whether extracellular vesicles (EVs) from human breast milk can protect against experimentally induced necrotizing (NEC) enterocolitis in mice. | Lactating mice were randomized to either breastfeeding without NEC, NEC with no breastfeeding, NEC with intraperitoneal EVs, and NEC with oral EVs. Histological analysis was done to determine the extent of NEC. | Not indicated | Breastfed rats had 0% NEC, while 62% of those with experimentally induced disease experienced it. Intraperitoneal EV reduced NEC to 29%, while oral administration reduced it to 11.9%. |
| Gu, 2014 (China) [23] | To determine the association between milk stasis and breast carcinogenesis and the potential role of microRNAs as biomarkers. | Milk samples from patients with milk stasis and patients with both stasis and breast cancer were analyzed for microRNA profiles. | A total of 20 patients | MicroRNA profiles differed between samples from patients with and without a neoplasm. In addition, microRNAs with roles in tumor suppression and oncogenesis had pro-neoplastic expression profiles. |
| Simpson, 2015 (Norway) [24] | To investigate the potential mediating role of breast milk microRNAs in the anti-dermatitis effects of probiotic supplements. | Small RNA sequencing for microRNAs in breast milk samples, followed by statistical analysis to determine correlations between expression profiles and probiotic effects. | 54 lactating mothers | The biological significance of microRNAs in atopic dermatitis was not apparent. |
| 5. PHYSIOLOGICAL FUNCTIONS OF microRNAS | ||||
| Yun, 2021 (South Korea) [25] | To explore microRNA profiles of human, bovine, and caprine milk. | RNA sequencing of fractionated milk samples. | Milk samples were obtained from 2 human volunteers, 6 cows, and 3 goats. | Several dietary micro-RNAs are expressed in human, cow, and caprine milk. These species are mostly involved in immune regulation. |
| Zhou, 2012 (China) [26] | To investigate microRNAs in human breast milk. | Deep sequence analysis of samples for microRNA identification. | Breast milk was collected from 4 healthy mothers. | 602 unique microRNAs were isolated from the milk samples. Resistance to harsh conditions suggests transfer to infants via the digestive tract. |
| Kahn, 2018 (USA) [27] | To determine the effect of in vitro digestion on microRNA from breast milk of mothers of preterm infants. | Exosomes from milk samples were isolated and lysed using commercial RNAses, exposed to in vitro digestive enzymes, and subsequently sequenced to determine surviving contents. | Milk samples were collected from 10 mothers. | MicroRNAs that are specific to preterm infant milk were not affected by in vitro digestion, suggesting increased susceptibility of preterm infants to maternal microRNA-driven epigenetic effects. |
| Na, 2015 (China) [28] | To screen for presence of 5 immune-related microRNAs in human, goat, and cow milk. | Real-time PCR of breast milk samples to identify MiR-146, miR-155, miR-181a, miR-223, and miR-150 species. | Milk samples were collected from 3 human volunteers, 2 lactating goats, and 3 dairy cows. | All five microRNA species were found in human and goat milk, and four were present in cow milk. |
| Karlsson, 2016 (USA) [29] | Screening for developmentally important lncRNAs in breast milk. | Quantitative analysis was used to determine the presence of specific non-coding RNAs in breast milk samples. | Breast milk was collected from 30 participants. | 55 of the 87 non-coding RNAs screened for were found, demonstrating a possible role in childhood development. |
| Kosaka, 2010 (Japan) [2] | To determine the importance of breast milk microRNAs in infant immune regulation. | MicroRNA content of breast milk was screened for species known to play a role in immune function. | 6 breastfeeding women provided milk samples | Breast milk has high levels of immune-regulating microRNAs in the first 6 months of lactation. These microRNAs are stable in acidic conditions, allowing for dietary intake. |
| Kupsco, 2021 (USA) [30] | To characterize breast milk microRNA in population cohort and evaluate association with maternal characteristics. | Small RNA PCR sequencing of human milk samples to identify the most common microRNA species, followed by association analysis with several maternal characteristics. | Breast milk was obtained from 364 mothers | A total of 1523 microRNAs were detected in at least two samples. Three of the most common clusters demonstrated common ontogeny in pathways enriched by such systems as endocrine development and neurodevelopment. Association of microRNA expression with maternal BMI, time of milk collection, and smoking was demonstrated. |
| Lukasik, 2017 (Poland) [31] | To screen human breast milk for presence of specific plant-derived microRNAs. | Real-time quantitative reverse transcription PCR of human breast milk samples to detect 5 specific plant-derived microRNAs. | 6 lactating mothers | Two of the 5 screened microRNAs were isolated in breast milk samples. These microRNAs are believed to be acquired from ingested plant-based foods, and to have a role in infant development. |
| Velez-Ixta, 2022 (Mexico) [3] | To assess the expression of immunoregulatory microRNAs in human breast milk. | Expression of 5 specific microRNAs was assessed using quantitative PCR in milk samples and infant formula. | 60 lactating mothers | All five screened microRNAs (miR-146b-5p, miR148a-3p, miR155-5p, mir181a-5p, and mir200a-3p) were detected in human breast milk, but only very small concentrations of the species were found in infant formulas. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondracka, A.; Gil-Kulik, P.; Kondracki, B.; Frąszczak, K.; Oniszczuk, A.; Rybak-Krzyszkowska, M.; Staniczek, J.; Kwaśniewska, A.; Kocki, J. Occurrence, Role, and Challenges of MicroRNA in Human Breast Milk: A Scoping Review. Biomedicines 2023, 11, 248. https://doi.org/10.3390/biomedicines11020248
Kondracka A, Gil-Kulik P, Kondracki B, Frąszczak K, Oniszczuk A, Rybak-Krzyszkowska M, Staniczek J, Kwaśniewska A, Kocki J. Occurrence, Role, and Challenges of MicroRNA in Human Breast Milk: A Scoping Review. Biomedicines. 2023; 11(2):248. https://doi.org/10.3390/biomedicines11020248
Chicago/Turabian StyleKondracka, Adrianna, Paulina Gil-Kulik, Bartosz Kondracki, Karolina Frąszczak, Anna Oniszczuk, Magda Rybak-Krzyszkowska, Jakub Staniczek, Anna Kwaśniewska, and Janusz Kocki. 2023. "Occurrence, Role, and Challenges of MicroRNA in Human Breast Milk: A Scoping Review" Biomedicines 11, no. 2: 248. https://doi.org/10.3390/biomedicines11020248
APA StyleKondracka, A., Gil-Kulik, P., Kondracki, B., Frąszczak, K., Oniszczuk, A., Rybak-Krzyszkowska, M., Staniczek, J., Kwaśniewska, A., & Kocki, J. (2023). Occurrence, Role, and Challenges of MicroRNA in Human Breast Milk: A Scoping Review. Biomedicines, 11(2), 248. https://doi.org/10.3390/biomedicines11020248








