Network Analysis for the Discovery of Common Oncogenic Biomarkers in Liver Cancer Experimental Models
Abstract
:1. Introduction
2. Materials and Methods
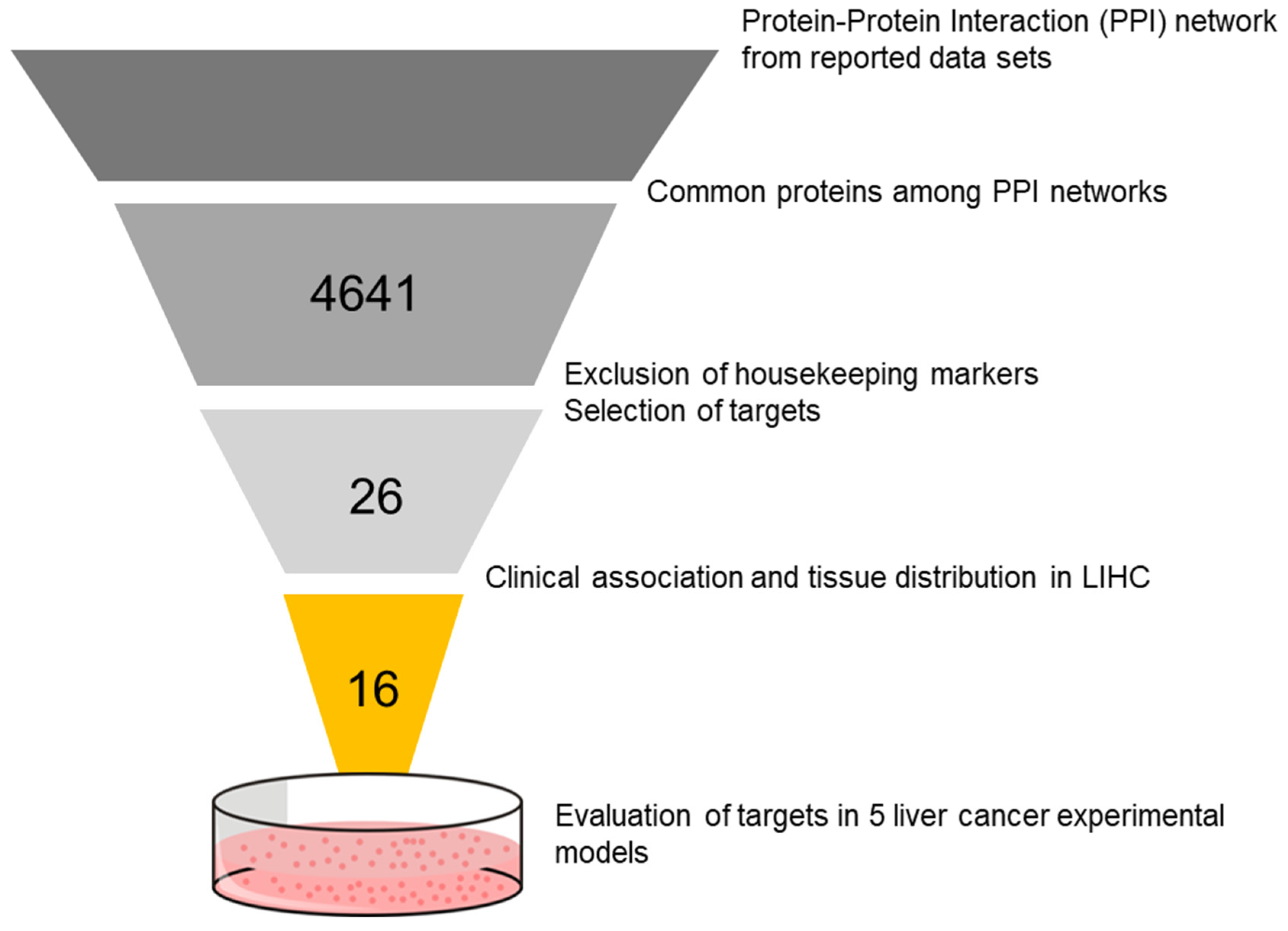
2.1. Selection of Targets
2.2. Classification and Characterization of In Vitro Models
2.3. Evaluation of Targeted Therapies in In Vitro Models
2.4. Collection of Total RNA and Protein from Treated Cells
2.5. Quantitative Real-Time PCR (RT-qPCR)
2.6. Western Blot Analysis
2.7. Statistical Analysis
3. Results
3.1. Identification of Candidate Targets
3.2. Expression of Targets in the Cell Populations
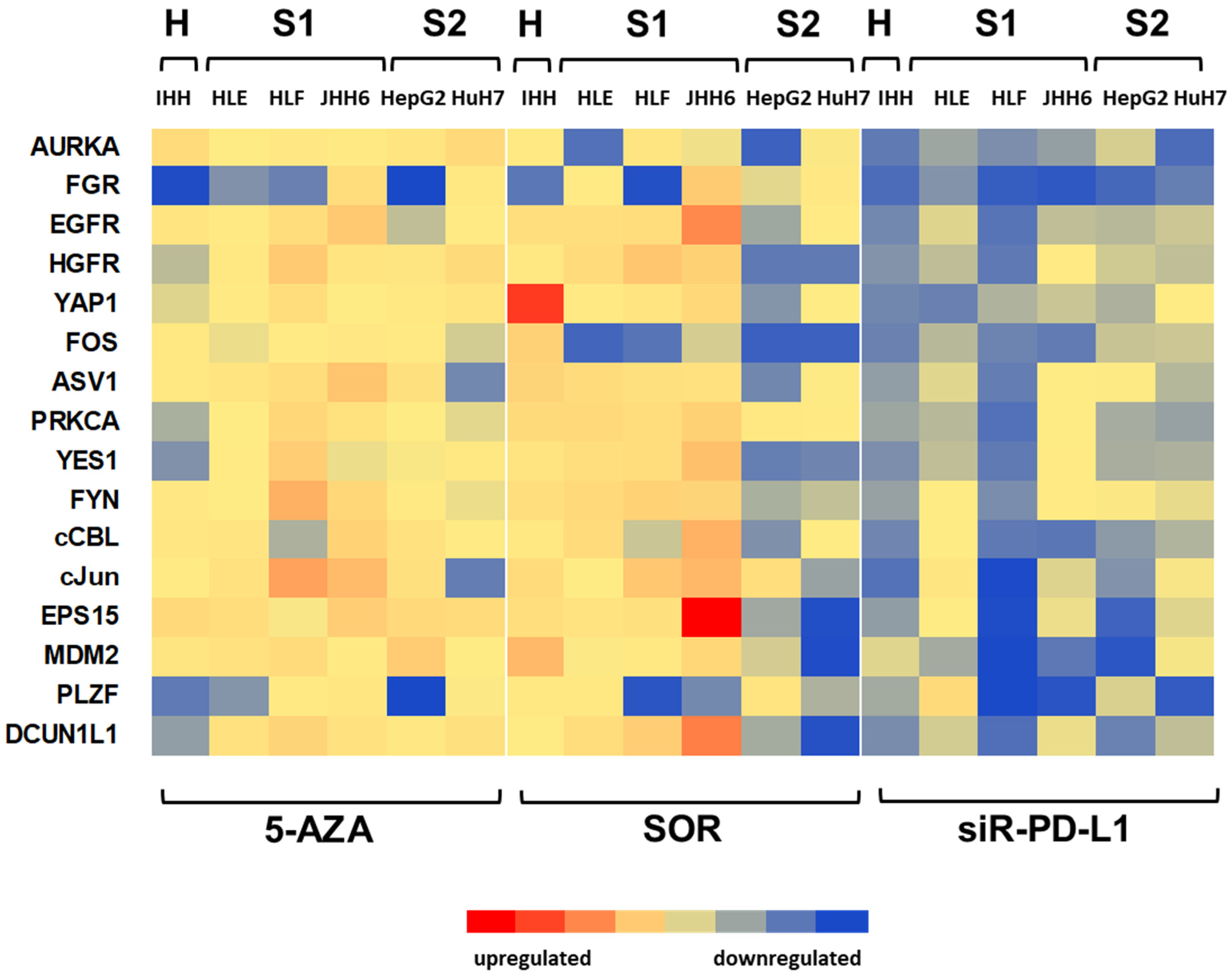
3.3. Effect of Targeted Treatments on Different Cell Populations
3.4. Effect of Treatments on the Dysregulations of Targets
3.5. Dysregulation Effects of Various Treatments on FGR Protein Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular Carcinoma. Nat. Rev. Dis. Prim. 2016, 2, 16018. [Google Scholar] [CrossRef]
- Caruso, S.; Calatayud, A.-L.; Pilet, J.; La Bella, T.; Rekik, S.; Imbeaud, S.; Letouzé, E.; Meunier, L.; Bayard, Q.; Rohr-Udilova, N.; et al. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology 2019, 157, 760–776. [Google Scholar] [CrossRef]
- Cabral, L.K.D.; Tiribelli, C.; Sukowati, C.H.C. Sorafenib Resistance in Hepatocellular Carcinoma: The Relevance of Genetic Heterogeneity. Cancers 2020, 12, 1576. [Google Scholar] [CrossRef]
- Belghiti, J.; Fuks, D. Liver Resection and Transplantation in Hepatocellular Carcinoma. Liver Cancer 2012, 1, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in Advanced Hepatocellular Carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus Sorafenib in First-Line Treatment of Patients with Unresectable Hepatocellular Carcinoma: A Randomised Phase 3 Non-Inferiority Trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.-Y.; Choo, S.-P.; Trojan, J.; Welling, T.H.; et al. Nivolumab in Patients with Advanced Hepatocellular Carcinoma (CheckMate 040): An Open-Label, Non-Comparative, Phase 1/2 Dose Escalation and Expansion Trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.-Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Zhang, T.; Merle, P.; Wang, H.; Zhao, H.; Kudo, M. Combination Therapy for Advanced Hepatocellular Carcinoma: Do We See the Light at the End of the Tunnel? Hepatobiliary Surg. Nutr. 2021, 10, 180–192. [Google Scholar] [CrossRef]
- Kelley, R.K.; W Oliver, J.; Hazra, S.; Benzaghou, F.; Yau, T.; Cheng, A.-L.; Rimassa, L. Cabozantinib in Combination with Atezolizumab versus Sorafenib in Treatment-Naive Advanced Hepatocellular Carcinoma: COSMIC-312 Phase III Study Design. Future Oncol. 2020, 16, 1525–1536. [Google Scholar] [CrossRef]
- Kelley, R.K.; Yau, T.; Cheng, A.-L.; Kaseb, A.; Qin, S.; Zhu, A.X.; Chan, S.; Sukeepaisarnjaroen, W.; Breder, V.; Verset, G.; et al. VP10-2021: Cabozantinib (C) plus Atezolizumab (A) versus Sorafenib (S) as First-Line Systemic Treatment for Advanced Hepatocellular Carcinoma (AHCC): Results from the Randomized Phase III COSMIC-312 Trial. Ann. Oncol. 2022, 33, 114–116. [Google Scholar] [CrossRef]
- Lee, M.; Ryoo, B.-Y.; Hsu, C.-H.; Numata, K.; Stein, S.; Verret, W.; Hack, S.; Spahn, J.; Liu, B.; Abdullah, H.; et al. LBA39—Randomised Efficacy and Safety Results for Atezolizumab (Atezo) + Bevacizumab (Bev) in Patients (Pts) with Previously Untreated, Unresectable Hepatocellular Carcinoma (HCC). Ann. Oncol. 2019, 30, v875. [Google Scholar] [CrossRef]
- Roxburgh, P.; Evans, T.R.J. Systemic Therapy of Hepatocellular Carcinoma: Are We Making Progress? Adv Ther. 2008, 25, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.B.; Kobayashi, M.; Chan, J.A.; Brunet, J.-P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative Transcriptome Analysis Reveals Common Molecular Subclasses of Human Hepatocellular Carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef]
- Boyault, S.; Rickman, D.S.; Reyniès, A.d.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome Classification of HCC Is Related to Gene Alterations and to New Therapeutic Targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Portela, A.; Sayols, S.; Battiston, C.; Hoshida, Y.; Méndez-González, J.; Imbeaud, S.; Letouzé, E.; Hernandez-Gea, V.; Cornella, H.; et al. DNA Methylation-Based Prognosis and Epidrivers in Hepatocellular Carcinoma. Hepatology 2015, 61, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- GTEx Consortium The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Cabral, K.; Reyes, P.; Croce, S.; Tiribelli, C.; Sukowati, C. The Relevance of SOCS1 Methylation and Epigenetic Therapy in Diverse Cell Populations of Hepatocellular Carcinoma. Diagnostics 2021, 11, 1825. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Cottini, F.; Hideshima, T.; Xu, C.; Sattler, M.; Dori, M.; Agnelli, L.; ten Hacken, E.; Bertilaccio, M.T.; Antonini, E.; Neri, A.; et al. Rescue of Hippo Coactivator YAP1 Triggers DNA Damage–Induced Apoptosis in Hematological Cancers. Nat. Med. 2014, 20, 599–606. [Google Scholar] [CrossRef]
- Joosten, M.; Ginzel, S.; Blex, C.; Schmidt, D.; Gombert, M.; Chen, C.; Linka, R.M.; Gräbner, O.; Hain, A.; Hirsch, B.; et al. A Novel Approach to Detect Resistance Mechanisms Reveals FGR as a Factor Mediating HDAC Inhibitor SAHA Resistance in B-Cell Lymphoma. Mol. Oncol. 2016, 10, 1232–1244. [Google Scholar] [CrossRef]
- Mamo, M.; Ye, I.C.; DiGiacomo, J.W.; Park, J.Y.; Downs, B.; Gilkes, D.M. Hypoxia Alters the Response to Anti-EGFR Therapy by Regulating EGFR Expression and Downstream Signaling in a DNA Methylation-Specific and HIF-Dependent Manner. Cancer Res. 2020, 80, 4998–5010. [Google Scholar] [CrossRef]
- Laidlaw, A.M.; Copeland, B.; Ross, C.M.; Hardingham, J.E. Extent of Over-Expression of Hepatocyte Growth Factor Receptor in Colorectal Tumours Is Dependent on the Choice of Normaliser. Biochem. Biophys. Res. Commun. 2006, 341, 1017–1021. [Google Scholar] [CrossRef]
- Wan, X.; Yeung, C.; Heske, C.; Mendoza, A.; Helman, L.J. IGF-1R Inhibition Activates a YES/SFK Bypass Resistance Pathway: Rational Basis for Co-Targeting IGF-1R and Yes/SFK Kinase in Rhabdomyosarcoma. Neoplasia 2015, 17, 358–366. [Google Scholar] [CrossRef]
- Hui, A.W.-H.; Lau, H.-W.; Cao, C.Y.; Zhou, J.-W.; Lai, P.B.-S.; Tsui, S.K.-W. Downregulation of PLZF in Human Hepatocellular Carcinoma and Its Clinical Significance. Oncol. Rep. 2015, 33, 397–402. [Google Scholar] [CrossRef]
- Huang, G.; Stock, C.; Bommeljé, C.C.; Weeda, V.B.; Shah, K.; Bains, S.; Buss, E.; Shaha, M.; Rechler, W.; Ramanathan, S.Y.; et al. SCCRO3 (DCUN1D3) Antagonizes the Neddylation and Oncogenic Activity of SCCRO (DCUN1D1)*. J. Biol. Chem. 2014, 289, 34728–34742. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Fan, C.-W.; Maa, M.-C.; Leu, T.-H. Lipopolysaccharide-Promoted Proliferation of Caco-2 Cells Is Mediated by c-Src Induction and ERK Activation. Biomedicine 2015, 5, 5. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, J.; Liu, S.; Qu, D.; Jin, H.; Zhu, L.; Yang, J.; Zhang, J.; Li, Q.; Zhang, Y.; et al. MiR-216a Promotes Breast Cancer Cell Apoptosis by Targeting PKCα. Fundam. Clin. Pharmacol. 2019, 33, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Schlott, T.; Reimer, S.; Jahns, A.; Ohlenbusch, A.; Ruschenburg, I.; Nagel, H.; Droese, M. Point Mutations and Nucleotide Insertions in the Mdm2 Zinc Finger Structure of Human Tumours. J. Pathol. 1997, 182, 54–61. [Google Scholar] [CrossRef]
- Fan, Q.; He, M.; Deng, X.; Wu, W.K.K.; Zhao, L.; Tang, J.; Wen, G.; Sun, X.; Liu, Y. Derepression of C-Fos Caused by MicroRNA-139 down-Regulation Contributes to the Metastasis of Human Hepatocellular Carcinoma. Cell Biochem. Funct. 2013, 31, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Hewagama, A.; Gorelik, G.; Patel, D.; Liyanarachchi, P.; Joseph McCune, W.; Somers, E.; Gonzalez-Rivera, T.; The Michigan Lupus Cohort; Strickland, F.; Richardson, B. Overexpression of X-Linked Genes in T Cells from Women with Lupus. J. Autoimmun. 2013, 41, 60–71. [Google Scholar] [CrossRef]
- MicroRNA-125a-3p Reduces Cell Proliferation and Migration by Targeting Fyn|Journal of Cell Science|The Company of Biologists. Available online: https://journals.biologists.com/jcs/article/126/13/2867/53648/microRNA-125a-3p-reduces-cell-proliferation-and (accessed on 22 November 2022).
- Knirsh, R.; Ben-Dror, I.; Spangler, B.; Matthews, G.D.; Kuphal, S.; Bosserhoff, A.K.; Vardimon, L. Loss of E-Cadherin–Mediated Cell–Cell Contacts Activates a Novel Mechanism for Up-Regulation of the Proto-Oncogene c-Jun. MBoC 2009, 20, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Roxrud, I.; Raiborg, C.; Pedersen, N.M.; Stang, E.; Stenmark, H. An Endosomally Localized Isoform of Eps15 Interacts with Hrs to Mediate Degradation of Epidermal Growth Factor Receptor. J. Cell Biol. 2008, 180, 1205–1218. [Google Scholar] [CrossRef]
- Uhercik, M.; Sanders, A.J.; Owen, S.; Davies, E.L.; Sharma, A.K.; Jiang, W.G.; Mokbel, K. Clinical Significance of PD1 and PDL1 in Human Breast Cancer. Anticancer Res. 2017, 37, 4249–4254. [Google Scholar] [CrossRef]
- Ling, S.; Hu, Z.; Yang, Z.; Yang, F.; Li, Y.; Lin, P.; Chen, K.; Dong, L.; Cao, L.; Tao, Y.; et al. Extremely High Genetic Diversity in a Single Tumor Points to Prevalence of Non-Darwinian Cell Evolution. Proc. Natl. Acad. Sci. USA 2015, 112, E6496–E6505. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, X.; Wang, Z.; Wang, L.; Xia, Q.; Zhang, W. The Role of MDM2–P53 Axis Dysfunction in the Hepatocellular Carcinoma Transformation. Cell Death Discov. 2020, 6, 53. [Google Scholar] [CrossRef]
- Wang, B.; Hsu, C.-J.; Chou, C.-H.; Lee, H.-L.; Chiang, W.-L.; Su, C.-M.; Tsai, H.-C.; Yang, S.-F.; Tang, C.-H. Variations in the AURKA Gene: Biomarkers for the Development and Progression of Hepatocellular Carcinoma. Int. J. Med. Sci. 2018, 15, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Guo, T.-F.; Jing, Z.-T.; Yang, Z.; Liu, L.; Yang, Y.-P.; Lin, X.; Tong, Q.-Y. Hepatitis B Virus Core Protein Promotes Hepatocarcinogenesis by Enhancing Src Expression and Activating the Src/PI3K/Akt Pathway. FASEB J. 2018, 32, 3033–3046. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Chen, Y.; Liu, L.; Yin, X.; Yang, Y.; Tang, L. PLZF and PLZF-MAPK10 Can Predict the Prognosis of Postoperative Patients with Hepatocellular Carcinoma. Int. J. Clin. Exp. Pathol. 2020, 13, 3158–3166. [Google Scholar]
- Vincent, A.; Omura, N.; Hong, S.-M.; Jaffe, A.; Eshleman, J.; Goggins, M. Genome-Wide Analysis of Promoter Methylation Associated with Gene Expression Profile in Pancreatic Adenocarcinoma. Clin. Cancer Res. 2011, 17, 4341–4354. [Google Scholar] [CrossRef]
- Kim, B.H.; Park, J.-W.; Kim, J.S.; Lee, S.-K.; Hong, E.K. Stem Cell Markers Predict the Response to Sorafenib in Patients with Hepatocellular Carcinoma. Gut Liver 2019, 13, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, X.; Zhang, W.; Liu, Y.; Chen, B.; Xiang, Y.; Zhang, R.; Zhang, M.; Feng, J.; Liu, S.; et al. Correlation of PD-L1 and SOCS3 Co-Expression with the Prognosis of Hepatocellular Carcinoma Patients. J. Cancer 2020, 11, 5440–5448. [Google Scholar] [CrossRef]
- Jung, H.I.; Jeong, D.; Ji, S.; Ahn, T.S.; Bae, S.H.; Chin, S.; Chung, J.C.; Kim, H.C.; Lee, M.S.; Baek, M.-J. Overexpression of PD-L1 and PD-L2 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res. Treat 2017, 49, 246–254. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, S.S.; Wang, H.J.; Kim, B.W.; Cho, H.; Jung, J.; Cho, S.S.; Kim, J.K.; Lee, J.H.; Kim, Y.B.; et al. Detection of Novel Genomic Markers for Predicting Prognosis in Hepatocellular Carcinoma Patients by Integrative Analysis of Copy Number Aberrations and Gene Expression Profiles: Results from a Long-Term Follow-Up. DNA Cell Biol. 2016, 35, 71–80. [Google Scholar] [CrossRef]



| Target | Sequence F (5′ → 3′) | Sequence R (5′ → 3′) | Ref. |
|---|---|---|---|
| YAP1 | CAATAGCTCAGATCCTTTCCT | TAGTATCACCTGTATCCATCTC | [24] |
| AURKA | GAGAATTGTGCTACTTATACTG | GGTACTAGGAAGGTTATTGC | ts |
| FGR | GGCCCGGCCTGCAT | TTGATGGCCTGAGAGGAGAAG | [25] |
| EGFR | AGGCACGAGTAACAAGCTCAC | ATGAGGGACATAACCAGCCACC | [26] |
| MET, HGFR | GGGCACCGAAAGATAAACCTCT | GACATTCTGGATGGGTGTTTCC | [27] |
| YES1 | ACAGCAAGACAAGGTGCAAA | GTAAACCGACCATACAGTGCAG | [28] |
| PLZF, ZBTB16 | TCACATACAGGCGACCACC | CTTGAGGCTGAACTTCTTGC | [29] |
| DCUN1D1 | CTGGAGGACACCAACATG | TTCACTAGATTGTGTGAAGATC | [30] |
| ASV, SRC1 | CGCTGGCCGGTGGAGTG | CCAGCTTGCGGATCTTGTAGT | [31] |
| PRKCA | GTGGCAAAGGAGCAGAGAAC | TGTAAGATGGGGTGCACAAA | [32] |
| MDM2 | TTATTAAAGTCTGTTGGTGCA | TGAAGGTTTCTCTTCCTGAAG | [33] |
| FOS | CCGGGGATAGCCTCTCTTAC | GTGGGAATGAAGTTGGCACT | [34] |
| CBL | TGCCAAAACTGCCACCTGGGG | GGGCTGCGGCCAAATTCCCT | [35] |
| FYN | GGACATGGCAGCACAGGTG | TTTGCTGATCGCAGATCTCTATG | [36] |
| JUN | AAGTAAGAGTGCGGGAGGCA3 | GGGCATCGTCATAGAAGGTCG | [37] |
| EPS15 | CCTGTTGCAGATTTCTCTG | TCATCTTGAAGATCCTGAAC | [38] |
| ACTB | CGCCGCCAGCTCACCATG | CACGATGGAGGGGAAGACGG | ts |
| PD-L1 | AAAGTCAATGCCCCATACAA | ACATGTCAGTTCATGTTCAGAG | [39] |
| UNIPROT ID | Protein Name | Gene | Gene Name | * Gene Expression |
|---|---|---|---|---|
| P46937 | Transcriptional coactivator YAP1 | YAP1 | yes-associated protein 1 | no difference |
| O14965 | Aurora kinase A | AURKA | aurora kinase A | upregulated |
| P09769 | Tyrosine-protein kinase Fgr | FGR | FGR proto-oncogene | down-regulated |
| P00533 | Epidermal growth factor receptor | EGFR | epidermal growth factor receptor | no difference |
| P08581 | Hepatocyte growth factor receptor | HGFR, MET | MET proto-oncogene, receptor tyrosine kinase | upregulated |
| P07947 | Tyrosine-protein kinase Yes | YES1 | YES proto-oncogene 1, Src family tyrosine kinase | upregulated |
| Q05516 | Zinc finger and BTB domain containing 16 | PLZF, ZBTB16 | zinc finger and BTB domain containing 16 | down-regulated |
| Q96GG9 | DCN1-like protein 1 | DCUN1D1 | defective in cullin neddylation 1 domain containing 1 | upregulated |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | ASV, SRC1 | SRC proto-oncogene, non-receptor tyrosine kinase | upregulated |
| P17252 | Protein kinase C alpha type | PRKCA | protein kinase C alpha | upregulated |
| Q00987 | E3 ubiquitin-protein ligase Mdm2 | MDM2 | MDM2 proto-oncogene | upregulated |
| P01100 | Protein c-Fos | FOS | Fos proto-oncogene, AP-1 transcription factor subunit | down-regulated |
| P22681 | E3 ubiquitin-protein ligase CBL | CBL | Cbl proto-oncogene | upregulated |
| P06241 | Tyrosine-protein kinase Fyn | FYN | FYN proto-oncogene, Src family tyrosine kinase | down-regulated |
| P05412 | Transcription factor Jun | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | upregulated |
| P42566 | Epidermal growth factor receptor substrate 15 | EPS15 | epidermal growth factor receptor pathway substrate 15 | upregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabral, L.K.D.; Giraudi, P.J.; Giannelli, G.; Dituri, F.; Negro, R.; Tiribelli, C.; Sukowati, C.H.C. Network Analysis for the Discovery of Common Oncogenic Biomarkers in Liver Cancer Experimental Models. Biomedicines 2023, 11, 342. https://doi.org/10.3390/biomedicines11020342
Cabral LKD, Giraudi PJ, Giannelli G, Dituri F, Negro R, Tiribelli C, Sukowati CHC. Network Analysis for the Discovery of Common Oncogenic Biomarkers in Liver Cancer Experimental Models. Biomedicines. 2023; 11(2):342. https://doi.org/10.3390/biomedicines11020342
Chicago/Turabian StyleCabral, Loraine Kay D., Pablo J. Giraudi, Gianluigi Giannelli, Francesco Dituri, Roberto Negro, Claudio Tiribelli, and Caecilia H. C. Sukowati. 2023. "Network Analysis for the Discovery of Common Oncogenic Biomarkers in Liver Cancer Experimental Models" Biomedicines 11, no. 2: 342. https://doi.org/10.3390/biomedicines11020342






