Effect of the Enrichment in c-Kit Stem Cell Potential of Foetal Human Amniotic Fluid Cells: Characterization from Single Cell Analysis to the Secretome Content
Abstract
:1. Introduction
2. Results
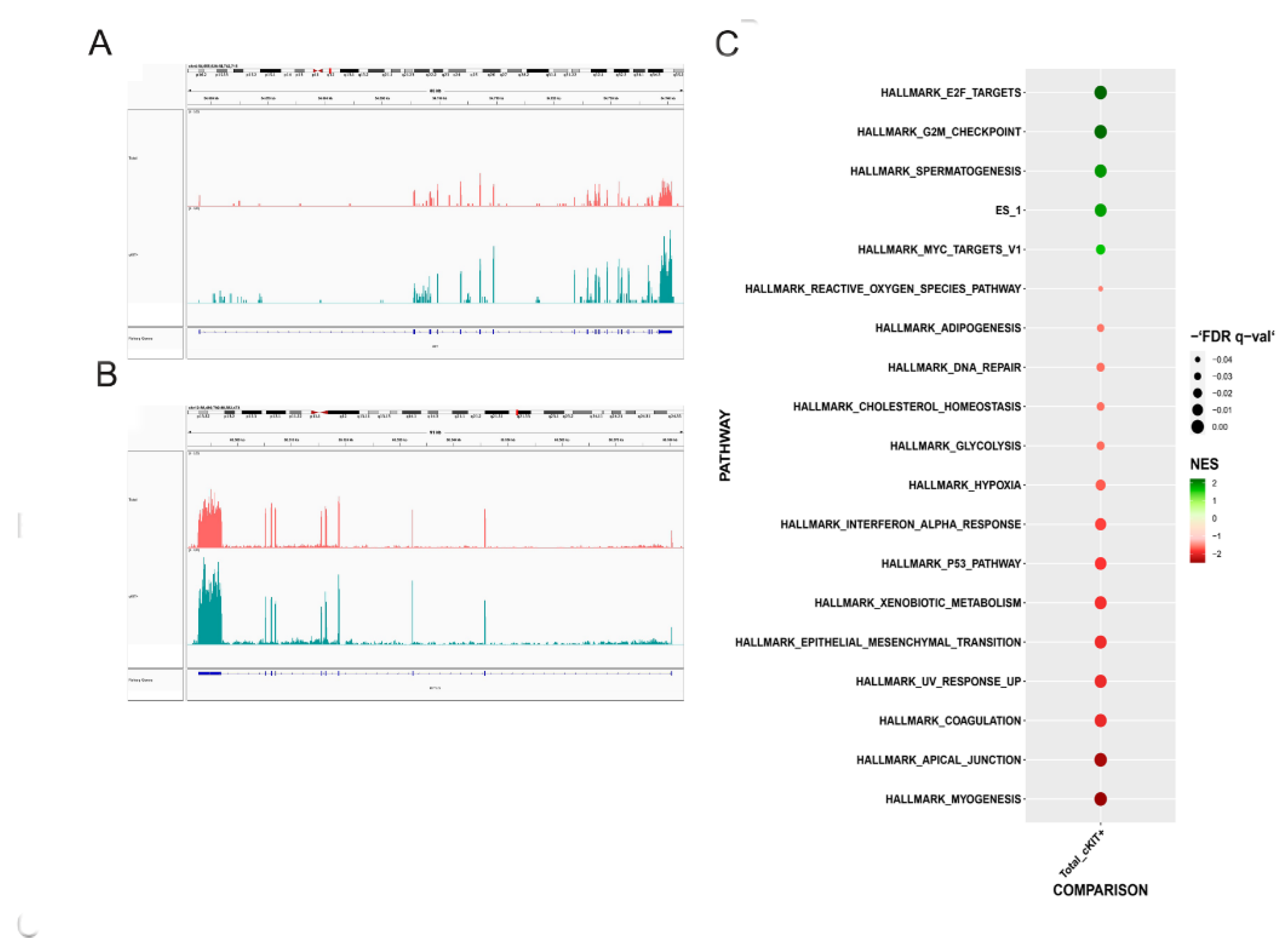
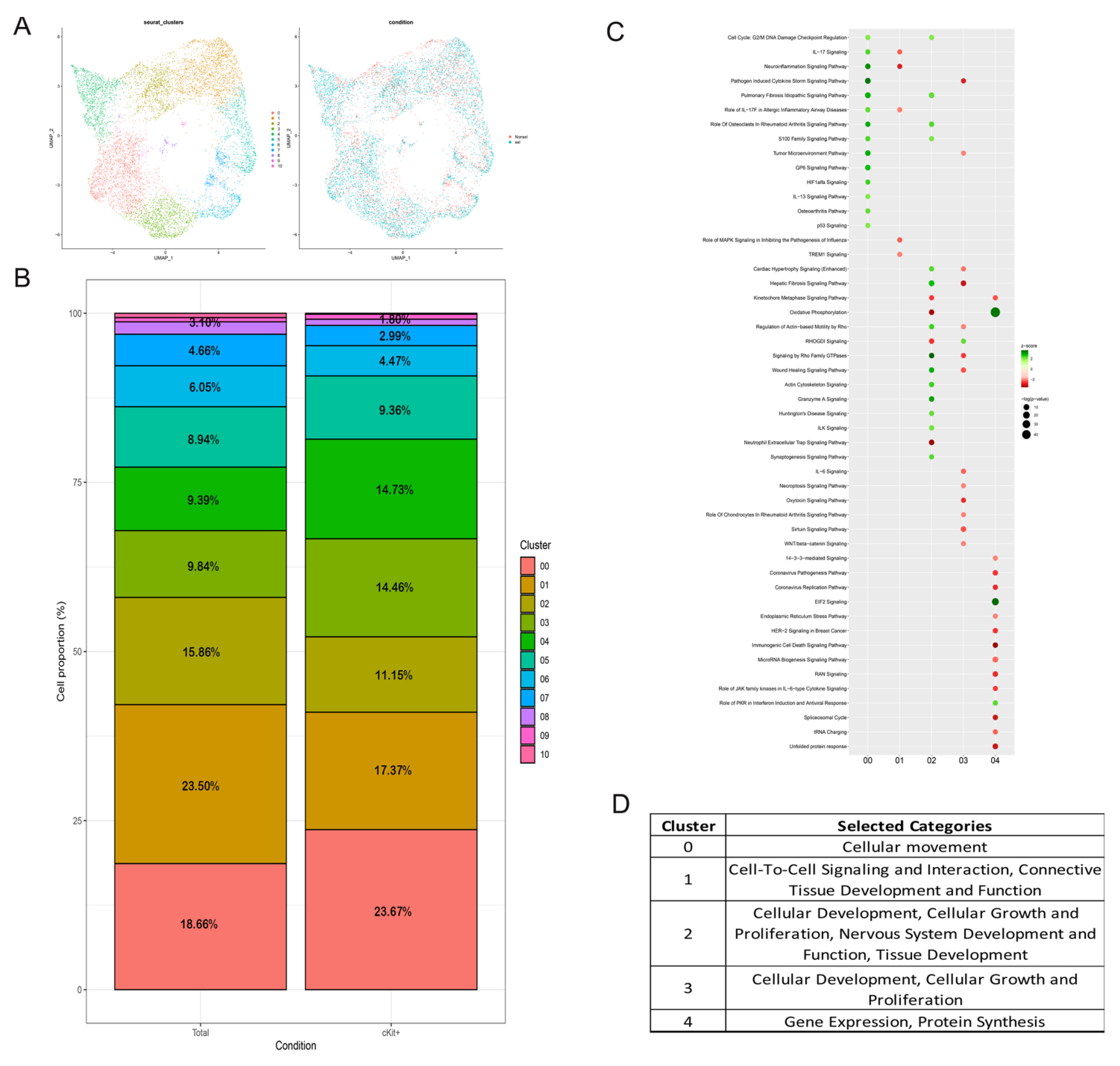
2.1. Transcriptomic Profiles of the Two Different hAFSCs Populations
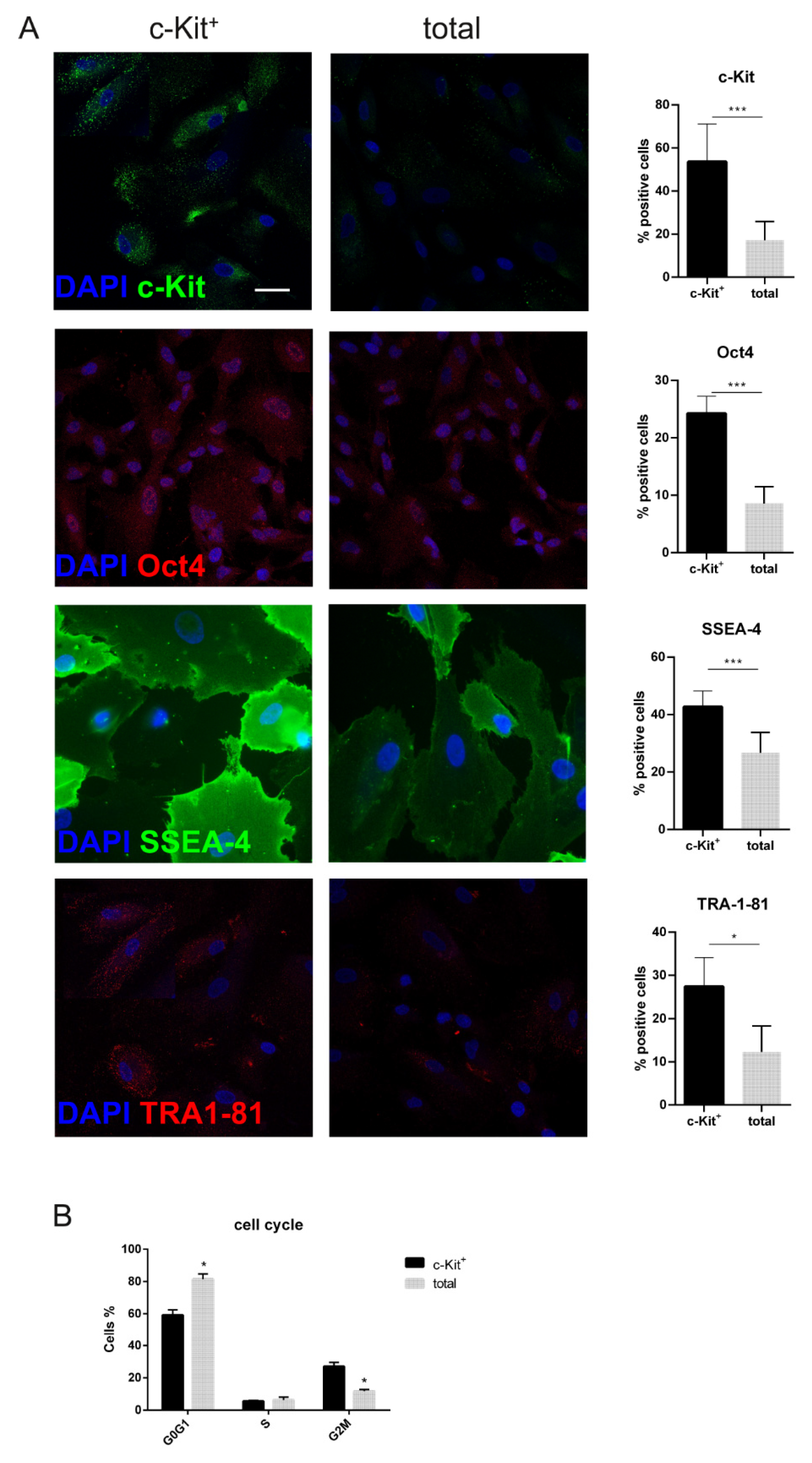
2.2. Phenotypic Characterization of Human Amniotic Fluid Cell Cultures: Effect of c-Kit Selection
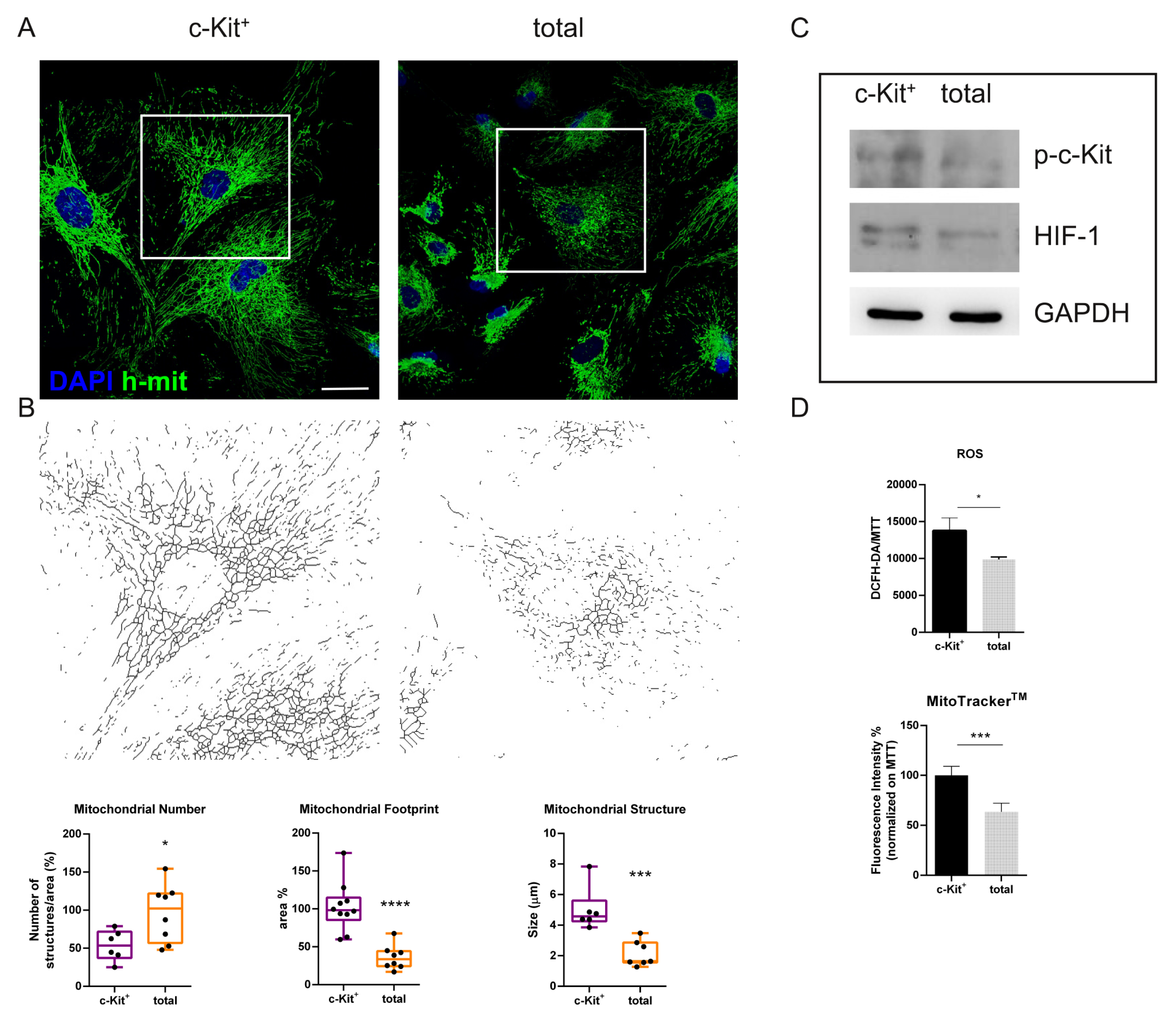
2.3. hAFSC Populations and Their Mitochondrial Phenotype
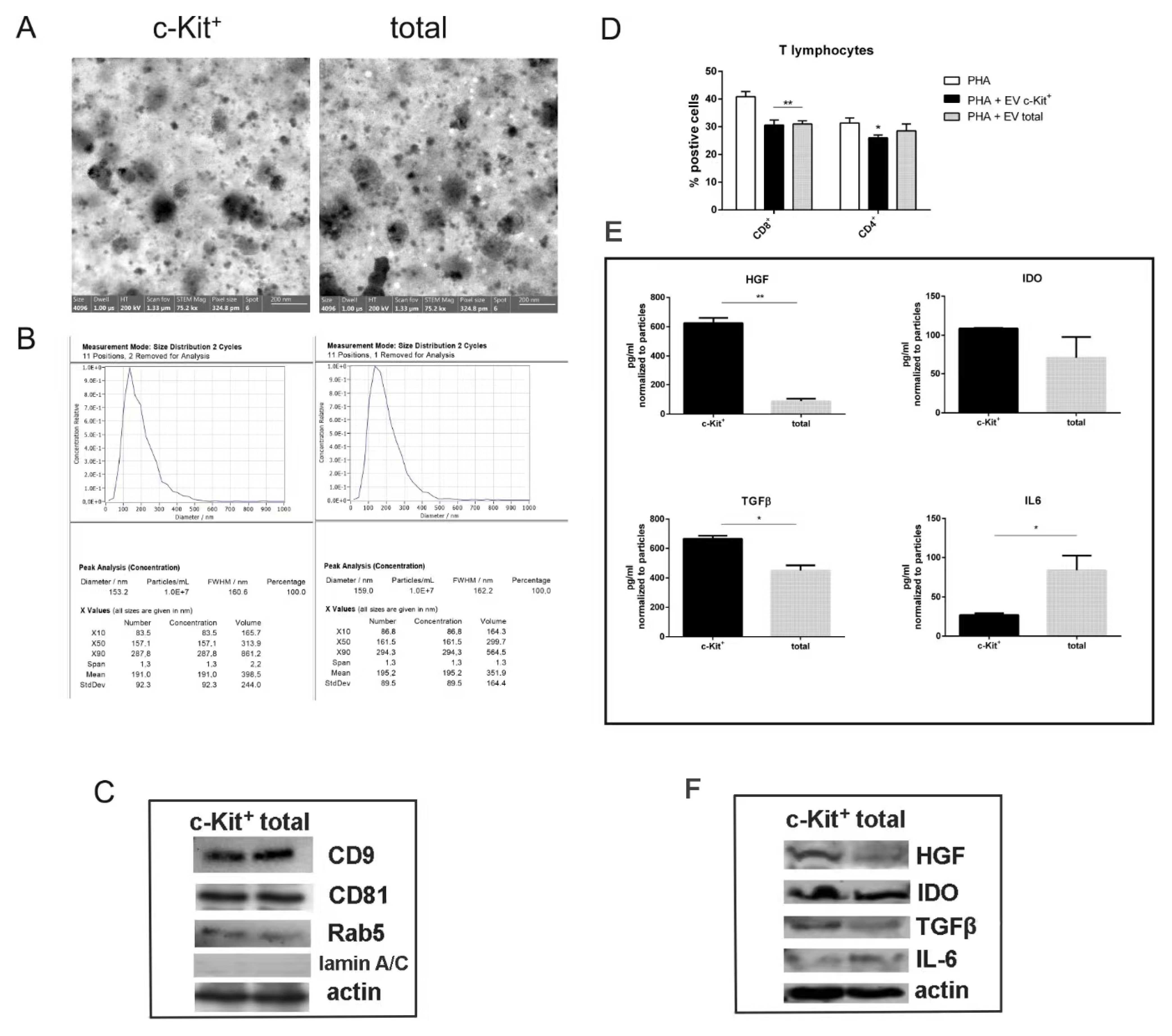
2.4. C-Kit Positive hAFSCs and Total hAFSCs: Comparison of the Secretome
3. Discussion
4. Materials and Methods
4.1. Amniotic Fluid Collection
4.2. Human Amniotic Fluid Cell Culture
4.3. Bulk RNA Transcription Profiling
4.4. Single Cell RNA-seq and Bioinformatic Analysis
4.5. Cell Cycle Analysis
4.6. Immunofluorescence and Confocal Microscopy
4.7. Mitochondria Analysis
4.8. ROS Detection
4.9. Extracellular Vesicle Isolation from Conditioned Medium
4.10. NTA Analysis
4.11. EV Characterization by Transmission Electron Microscopy
4.12. Protein Quantification in Purified EVs
4.13. ELISA Assays
4.14. PBMC Exposure to hAFSC EVs: Flow Cytometer Immune-Assay
4.15. Cellular and EV Extracts Preparation
4.16. SDS PAGE and Western Blot
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cananzi, M.; De Coppi, P. CD117+ amniotic fluid stem cells: State of the art and future perspectives. Organogenesis 2012, 8, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.G.; Lee, H.; Lee, S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. CMLS 2018, 75, 3159–3180. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Montazeri, F.; Bahrami, A.R.; Kalantar, S.M.; Rahmani, S.; Zarein, F.; Matin, M.M. Investigating the expression of pluripotency-related genes in human amniotic fluid cells: A semi-quantitative comparison between different subpopulations, from primary to cultured amniocytes. Reprod. Biol. 2020, 20, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Lesage, F.; Zia, S.; Jiménez, J.; Deprest, J.; Toelen, J. The amniotic fluid as a source of mesenchymal stem cells with lung-specific characteristics. Prenat. Diagn. 2017, 37, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Gaggi, G.; Di Credico, A.; Guarnieri, S.; Mariggiò, M.A.; Di Baldassarre, A.; Ghinassi, B. Human mesenchymal amniotic fluid stem cells reveal an unexpected neuronal potential differentiating into functional spinal motor neurons. Front. Cell Dev. Biol. 2022, 10, 936990. [Google Scholar] [CrossRef]
- Romani, R.; Fallarino, F.; Pirisinu, I.; Calvitti, M.; Caselli, A.; Fiaschi, T.; Gamberi, T.; Matino, D.; Talesa, V.N.; Donti, E.; et al. Comparative proteomic analysis of two distinct stem-cell populations from human amniotic fluid. Mol. bioSyst. 2015, 11, 1622–1632. [Google Scholar] [CrossRef]
- Casciaro, F.; Zia, S.; Forcato, M.; Zavatti, M.; Beretti, F.; Bertucci, E.; Zattoni, A.; Reschiglian, P.; Alviano, F.; Bonsi, L.; et al. Unravelling Heterogeneity of Amplified Human Amniotic Fluid Stem Cells Sub-Populations. Cells 2021, 10, 158. [Google Scholar] [CrossRef]
- Martin, M.M.; Chan, M.; Antoine, C.; Bar-El, L.; Bornstein, E.; Young, B.K. Clinical potential of human amniotic fluid stem cells. J. Perinat. Med. 2022, 51, 117–124. [Google Scholar] [CrossRef]
- Moschidou, D.; Mukherjee, S.; Blundell, M.P.; Drews, K.; Jones, G.N.; Abdulrazzak, H.; Nowakowska, B.; Phoolchund, A.; Lay, K.; Ramasamy, T.S.; et al. Valproic Acid Confers Functional Pluripotency to Human Amniotic Fluid Stem Cells in a Transgene-free Approach. Mol. Ther. 2012, 20, 1953–1967. [Google Scholar] [CrossRef]
- Pogozhykh, O.; Pogozhykh, D.; Neehus, A.L.; Hoffmann, A.; Blasczyk, R.; Müller, T. Molecular and cellular characteristics of human and non-human primate multipotent stromal cells from the amnion and bone marrow during long term culture. Stem Cell Res. Ther. 2015, 6, 150. [Google Scholar] [CrossRef] [Green Version]
- Cucco, C.; Zhang, Z.; Botero, T.M.; Chiego, D.J.; Castilho, R.M.; Nör, J.E. SCF/C-Kit Signaling Induces Self-Renewal of Dental Pulp Stem Cells. J. Endod. 2020, 46, S56–S62. [Google Scholar] [CrossRef]
- Czarna, A.; Sanada, F.; Matsuda, A.; Kim, J.; Signore, S.; Pereira, J.D.; Sorrentino, A.; Kannappan, R.; Cannatà, A.; Hosoda, T.; et al. Single-cell analysis of the fate of c-kit-positive bone marrow cells. NPJ Regen. Med. 2017, 2, 27. [Google Scholar] [CrossRef]
- Resca, E.; Zavatti, M.; Bertoni, L.; Maraldi, T.; De Biasi, S.; Pisciotta, A.; Nicoli, A.; La Sala, G.B.; Guillot, P.V.; David, A.L.; et al. Enrichment in c-Kit+ enhances mesodermal and neural differentiation of human chorionic placental cells. Placenta 2013, 34, 526–535. [Google Scholar] [CrossRef]
- Resca, E.; Zavatti, M.; Maraldi, T.; Bertoni, L.; Beretti, F.; Guida, M.; La Sala, G.B.; Guillot, P.V.; David, A.L.; Sebire, N.J.; et al. Enrichment in c-Kit improved differentiation potential of amniotic membrane progenitor/stem cells. Placenta 2015, 36, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wang, Y.; Liu, L.; Chen, J.; Yang, W.; Gao, L.; Wang, Y. Human amniotic fluid-derived c-kit+ and c-kit− stem cells: Growth characteristics and some differentiation potential capacities comparison. Cytotechnology 2012, 64, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Ježek, J.; Cooper, K.F.; Strich, R. Reactive Oxygen Species and Mitochondrial Dynamics: The Yin and Yang of Mitochondrial Dysfunction and Cancer Progression. Antioxidants 2018, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Beretti, F.; Zavatti, M.; Casciaro, F.; Comitini, G.; Franchi, F.; Barbieri, V.; La Sala, G.B.; Maraldi, T. Amniotic fluid stem cell exosomes: Therapeutic perspective. BioFactors 2018, 44, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, I.; Pantalone, A.; Tete, S.; Salini, V.; Borlongan, C.V.; Hess, D.; Stuppia, L. Amniotic fluid stem cells: A promising therapeutic resource for cell-based regenerative therapy. Curr. Pharm. Des. 2012, 18, 1846–1863. [Google Scholar] [CrossRef]
- Cananzi, M.; Atala, A.; De Coppi, P. Stem cells derived from amniotic fluid: New potentials in regenerative medicine. Reprod. Biomed. Online 2009, 18 (Suppl. 1), 17–27. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, Y.H.; Choi, G.E.; Kim, J.S.; Chae, C.W.; Han, H.J. Role of HIF1 α Regulatory Factors in Stem Cells. Int. J. Stem Cells 2019, 12, 8–20. [Google Scholar] [CrossRef] [Green Version]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, Q.; Hu, H.; Zhang, D.; Li, J.; Ma, G.; Bhat, K.; Wu, E. Stem cell factor/c-kit signaling enhances invasion of pancreatic cancer cells via HIF-1α under normoxic condition. Cancer Lett. 2011, 303, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Kiani, A.A.; Abdi, J.; Halabian, R.; Roudkenar, M.H.; Amirizadeh, N.; Soltanpour, M.S.; Kazemi, A. Over expression of HIF-1α in human mesenchymal stem cells increases their supportive functions for hematopoietic stem cells in an experimental co-culture model. Hematology 2014, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Sadek, H.A. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid. Redox Signal. 2014, 20, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, C.; D’Aprile, A.; Scrima, R.; Ripoli, M.; Boffoli, D.; Tabilio, A.; Capitanio, N. Role of reactive oxygen species as signal molecules in the pre-commitment phase of adult stem cells. Proc. Ital. J. Biochem. 2007, 56, 295–301. [Google Scholar]
- Bonello, S.; Zähringer, C.; BelAiba, R.S.; Djordjevic, T.; Hess, J.; Michiels, C.; Kietzmann, T.; Görlach, A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 755–761. [Google Scholar] [CrossRef]
- Beckervordersandforth, R.; Ebert, B.; Schäffner, I.; Moss, J.; Fiebig, C.; Shin, J.; Moore, D.L.; Ghosh, L.; Trinchero, M.F.; Stockburger, C.; et al. Role of Mitochondrial Metabolism in the Control of Early Lineage Progression and Aging Phenotypes in Adult Hippocampal Neurogenesis. Neuron 2017, 93, 560–573.e6. [Google Scholar] [CrossRef]
- Santel, A.; Fuller, M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001, 114, 867–874. [Google Scholar] [CrossRef]
- Bahat, A.; Goldman, A.; Zaltsman, Y.; Khan, D.H.; Halperin, C.; Amzallag, E.; Krupalnik, V.; Mullokandov, M.; Silberman, A.; Erez, A.; et al. MTCH2-mediated mitochondrial fusion drives exit from naïve pluripotency in embryonic stem cells. Nat. Commun. 2018, 9, 5132. [Google Scholar] [CrossRef]
- Agarwal, S.; Yadav, A.; Tiwari, S.K.; Seth, B.; Chauhan, L.K.S.; Khare, P.; Ray, R.S.; Chaturvedi, R.K. Dynamin-related Protein 1 Inhibition Mitigates Bisphenol A-mediated Alterations in Mitochondrial Dynamics and Neural Stem Cell Proliferation and Differentiation. J. Biol. Chem. 2016, 291, 15923–15939. [Google Scholar] [CrossRef]
- Wade, S.; Khacho, M. MITOCHONDRIA: Mitochondrial dynamics in the regulation of stem cells. Int. J. Biochem. Cell Biol. 2022, 144, 106158. [Google Scholar] [CrossRef]
- Han, Y.; Yang, J.; Fang, J.; Zhou, Y.; Candi, E.; Wang, J.; Hua, D.; Shao, C.; Shi, Y. The secretion profile of mesenchymal stem cells and potential applications in treating human diseases. Signal Transduct. Target. Ther. 2022, 7, 92. [Google Scholar] [CrossRef]
- Harrell, C.R.; Fellabaum, C.; Arsenijevic, A.; Markovic, B.S.; Djonov, V.; Volarevic, V. Therapeutic Potential of Mesenchymal Stem Cells and Their Secretome in the Treatment of Glaucoma. Stem Cells Int. 2019, 2019, 7869130. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef]
- Marrazzo, P.; Angeloni, C.; Freschi, M.; Lorenzini, A.; Prata, C.; Maraldi, T.; Hrelia, S. Combination of epigallocatechin gallate and sulforaphane counteracts in vitro oxidative stress and delays stemness loss of amniotic fluid stem cells. Oxidative Med. Cell. Longev. 2018, 2018, 5263985. [Google Scholar] [CrossRef]
- Casciaro, F.; Beretti, F.; Zavatti, M.; McCubrey, J.A.; Ratti, S.; Marmiroli, S.; Follo, M.Y.; Maraldi, T. Nuclear Nox4 interaction with prelamin A is associated with nuclear redox control of stem cell aging. Aging 2018, 10, 2911–2934. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casciaro, F.; Beretti, F.; Gatti, M.; Persico, G.; Bertucci, E.; Giorgio, M.; Maraldi, T. Effect of the Enrichment in c-Kit Stem Cell Potential of Foetal Human Amniotic Fluid Cells: Characterization from Single Cell Analysis to the Secretome Content. Biomedicines 2023, 11, 430. https://doi.org/10.3390/biomedicines11020430
Casciaro F, Beretti F, Gatti M, Persico G, Bertucci E, Giorgio M, Maraldi T. Effect of the Enrichment in c-Kit Stem Cell Potential of Foetal Human Amniotic Fluid Cells: Characterization from Single Cell Analysis to the Secretome Content. Biomedicines. 2023; 11(2):430. https://doi.org/10.3390/biomedicines11020430
Chicago/Turabian StyleCasciaro, Francesca, Francesca Beretti, Martina Gatti, Giuseppe Persico, Emma Bertucci, Marco Giorgio, and Tullia Maraldi. 2023. "Effect of the Enrichment in c-Kit Stem Cell Potential of Foetal Human Amniotic Fluid Cells: Characterization from Single Cell Analysis to the Secretome Content" Biomedicines 11, no. 2: 430. https://doi.org/10.3390/biomedicines11020430
APA StyleCasciaro, F., Beretti, F., Gatti, M., Persico, G., Bertucci, E., Giorgio, M., & Maraldi, T. (2023). Effect of the Enrichment in c-Kit Stem Cell Potential of Foetal Human Amniotic Fluid Cells: Characterization from Single Cell Analysis to the Secretome Content. Biomedicines, 11(2), 430. https://doi.org/10.3390/biomedicines11020430










