Abstract
Neuroblastoma (NB) is the most common extracranial tumor of early childhood and accounts for 15% of all pediatric cancer mortalities. However, the precise pathways and genes underlying its progression are unknown. Therefore, we performed a differential gene expression analysis of neuroblastoma stage 1 and stage 4 + 4S to discover biological processes associated with NB progression. From this preliminary analysis, we found that NB samples (stage 4 + 4S) are characterized by altered expression of some proteins involved in mitochondria function and mitochondria–ER contact sites (MERCS). Although further analyses remain necessary, this review may provide new hints to better understand NB molecular etiopathogenesis, by suggesting that MERCS alterations could be involved in the progression of NB.
1. Introduction
Neuroblastoma (NB) is a heterogeneous malignancy originating from the embryonic cells composing the neural crest [1,2]. NB is the most widespread extracranial solid tumor in children, accounting for nearly 10% of all childhood cancers [3,4]. NB is also the leading cause of cancer death in children younger than 5 years old, accounting for 15% of all pediatric cancer fatalities [5]. About 90% of NB cases are diagnosed before 5 years of age, while a third of those are diagnosed within the first year of life [4,5]. On the other hand, NB is a comparatively rare cancer with an incidence of 10.2 cases per one million children younger than 15 years; it affects 12 infants per 100,000 births [4,5,6].
NB belongs to embryonal neuroendocrine tumors of the peripheral nervous system [2]. It originates from the neural crest progenitor cells, particularly from the sympathoadrenal cell lineage [7]. Therefore, NB can grow anywhere along the sympathetic nervous system. The majority of NB (65%) develops in the abdomen, usually originating in the adrenal gland [8]. However, other sites of NB include the chest (20%), the neck (5%), or pelvis (5%) [8]. Due to a number of primary tumor locations, NB exhibits heterogenous biological, clinical, and morphological characteristics. Clinical symptoms of NB are often vague (e.g., fever, fatigues, loss of appetite) and diverse, depending on the site of malignancy, such as constipation and abdominal distention (NB in the abdomen) or breathing problems (NB in the chest) [9]. Similarly, the clinical outcomes of NB are variable, ranging from spontaneous regression to major treatment-resistant metastatic cancer with poor patient prognosis [10].
NB is classified (by the International Neuroblastoma Staging System) into six stages, in relation to the outcome of surgery used to remove the tumor [11]. In our work, we focused on three stages: (i) stage 1: the tumor is localized in the area of its origin and can be completely removed by surgery; (ii) stage 4: the tumor is widespread to distant lymph nodes, bone marrow, skin, liver, or other organs (except as defined for stage 4S); (iii) stage 4S (applicable only to infants under 1 year of age): the tumor mass locates as defined for stage 1, but with propagation limited to skin, liver, or bone marrow [11].
The pathogenesis of NB and biological processes leading to NB development from normal cells within the neural crest have not been fully described and elucidated. Only little is known about the potential processes and biological pathways that may lead to the initiation, development, and progression of NB. However, it seems there is no direct cause of NB pathogenesis; it rather requires multiplicity and cooperation of several effects, leading altogether to tumorigenesis [2]. The vast majority of neuroblastoma is sporadic and non-familial [12]. Only approximately 1% of cases are familial; germline mutations in several genes, such as anaplastic lymphoma receptor tyrosine kinase (ALK), paired mesoderm homeobox protein 2B (PHOX2B), and kinesin family member 1B (KIF1B) have been identified in patients with familial NB [12,13]. Interestingly, defects in ALK (anaplastic lymphoma kinase) genes have been demonstrated to occur in about 15% of NB cases [13,14]. Additionally, only the activating mutations in ALK and amplification of v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog (MYCN) have been shown to be oncogenic de novo in mice [15,16,17]. The presence of MYCN oncogene amplification highly correlates to advanced NB stages [17]. MYCN, a member of the myc proto-oncogene family, acts as a transcriptional factor for control of cellular differentiation and proliferation and plays an important role in the survival of neuroblastoma cells.
Mitochondria are dynamic organelles responsible for several cellular functions. They establish complex networks in the cells that can rapidly rearrange to react to the needs and metabolic state of the cell, through specific mitochondria fission and fusion processes [18,19,20,21]. Mitochondrial dynamics is regulated by their interactions with other organelles as well as the cellular cytoskeleton [18,22,23]. Evidence of the former are mitochondria–endoplasmic reticulum (ER) contact sites (MERCS), in which their surfaces are separated by a 10–80 nm gap [24,25,26,27,28,29]. Importantly, MERCS regulate crucial cell processes [27,30], such as Ca2+ and lipid homeostasis [31,32,33], mitochondrial fission [34], and apoptosis [35]. Due to their valuable role in mitochondrial biology and dynamics, MERCS have recently gained careful attention from biologists; however, all of their biological functions have not yet been fully described [36]. Similarly, mitochondria can also directly associate with microtubules and actin, the components of the cytoskeleton [22]. While microtubules serve for long-range mitochondria transport, the actin filaments regulate rather short-distance mitochondrial movement [37], their docking [38], and fission [39,40].
Mitochondria play a valuable role in the control of several crucial cellular processes, such as calcium homeostasis [41,42], ATP production [43,44,45], and apoptosis [46,47]. Thus, it is not surprising that mitochondrial dynamics dysfunction has been linked with the development and/or progression of tumors [48] including glioblastoma [49,50], melanoma [51], hepatocellular carcinoma [52], and pancreatic [53], breast [54], ovarian [55], and prostate cancers [56]. Regarding NB, interestingly, Çoku et al. has associated the reduction in the number of MERCS with aggressive NB displaying chemoresistance. This suggests that decreased mitochondria–ER interaction promotes neuroblastoma multidrug resistance [57]. Hence, the above-mentioned pieces of knowledge pose the challenging question of whether MERCS participate in NB development and progression.
To tackle this question, we used previously published datasets to identify genes differentially expressed in non-metastatic (stage 1) and metastatic neuroblastoma (stage 4 + 4S, which are characterized by metastases on skin, liver, and bone marrow) involved in MERCS structure and mitochondrial dynamics. We found that MERCS proteins previously shown to alter several aspects of cancer progression, i.e., cell metabolism, proliferation, and division, are upregulated in NB stage 4 + 4S, indicating the possibility that MERCS could be involved in the progression of this malignancy.
2. Materials and Methods
We used the GEO dataset GSE45547. This dataset includes 649 NB tumor samples whose gene expression was studied by a single-color Agilent-020382 Human Microarray composed of 44 K oligonucleotide probes. Tumor samples were classified according to the International Neuroblastoma Staging System. Data from NB stage 1 (153 samples) have been compared to stage 4 + 4S (292 samples). Raw data were logarithmic-scaled and quantile-normalized using the R Limma 3.26.8 package [58] in the R suite (Supplemental Figure S1); normalized data were used to calculate differentially expressed genes (DEGs) using the R Limma 3.26.8 package, which bases the identification of DEGs on linear models. The Benjamini–Hochberg false discovery rate method was used to correct for multiple tests and only genes with adjusted p-values under 0.05 were considered differentially expressed. Differentially expressed genes were classified according to Gene Ontology (GO) definitions using the WEB-based GEne SeT AnaLysis Toolkit [59], correcting statistical significance for multiple tests with the Benjamini–Hochberg method, considering enrichment significant when FDR < 0.05, and using 5 as the minimum number of IDs in the category and 2000 as the maximum number of IDs in the category to allow the consideration of categories. GO terms are either close in the GO hierarchy (sibling terms) or are related by inheritance (child and parent terms). Therefore, they may consist of a redundant list difficult to interpret. Revigo is a web tool used to reduce GO redundancy [60]. The tree map implemented in the Revigo web tool was used to summarize GO categories. We maintained default parameters (remove absolute GO terms, SimRel as semantic similarity measure) and used Homo sapiens as a species to be used. GO terms and corresponding p-values were considered for analyses with the Revigo web tool.
3. Results
The role of mitochondrial dynamics in NB progression is still poorly explored, despite a reduction in the number of MERCS being associated with aggressive NB phenotypes characterized by chemoresistance. To study their involvement in NB progression, we took advantage of published transcriptomic datasets and analyzed the expression changes occurring during NB progression. Using NB stage 1 as control, we found out that 1971 genes were upregulated in stage 1 and 1529 were upregulated in stage 4 + 4S (Supplemental Table S1 and Figure 1). Interestingly, we found that genes involved in the movement of subcellular vesicles, in neurogenesis, and in synaptic signaling were downregulated in stage 4 + 4S compared to controls (Figure 2 and Supplemental Table S2). At variance, upregulated genes were involved in the control of the cell cycle (Figure 3 and Supplemental Table S2). This is in agreement with the evidence that suggests dysregulation of the cell cycle results in NB formation [61]. Of note, various mechanisms involved in the control of cell cycle progression dictate whether NB cells undergo neural differentiation or enter into cell cycle arrest and adopt senescence-like state [62].
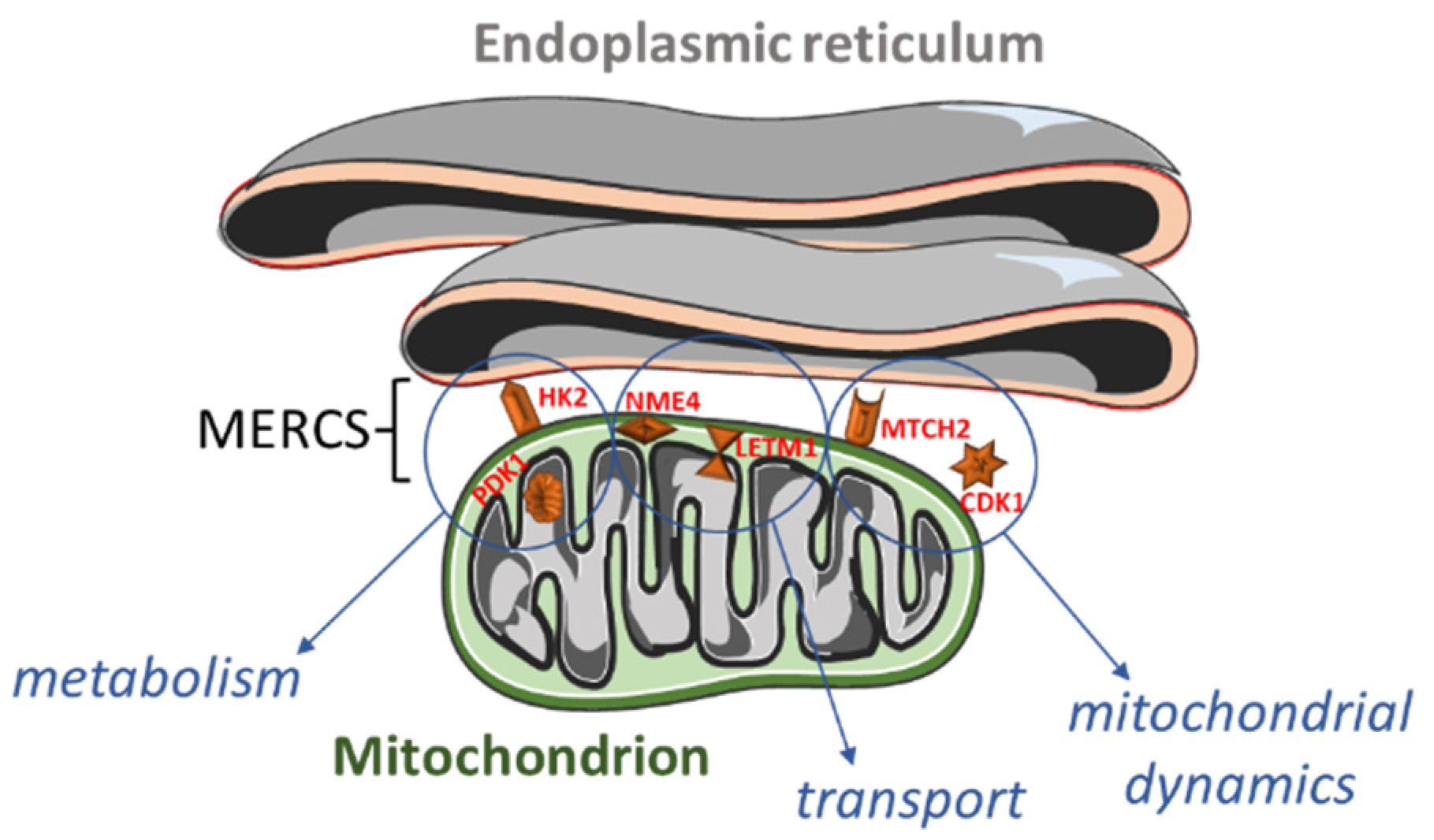
Figure 1.
Overview of the proteins localized within or close to mitochondria–endoplasmic reticulum (ER) contact sites (MERCS) and their functions. Hexokinase 2 (HK2; outer mitochondrial membrane) and pyruvate dehydrogenase kinase 1 (PDK1; mitochondrial matrix) are involved in the (carbohydrate) metabolism. NME/NM23 nucleoside diphosphate kinase 4 (NME4; mitochondrial intermembrane space) and leucine zipper and EF-hand-containing transmembrane protein 1 (LETM1; inner mitochondrial membrane) participate in the cardiolipin and calcium transport. Finally, mitochondrial carrier 2 (MTCH2; outer mitochondrial membrane) and cyclin B1-dependent kinase 1 (CDK1; cytoplasm, mitochondrion, nucleus) regulate mitochondrial dynamics.
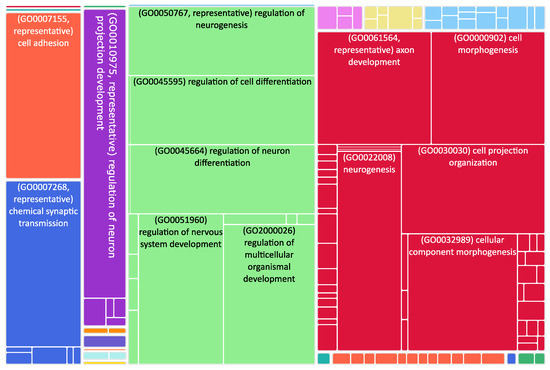
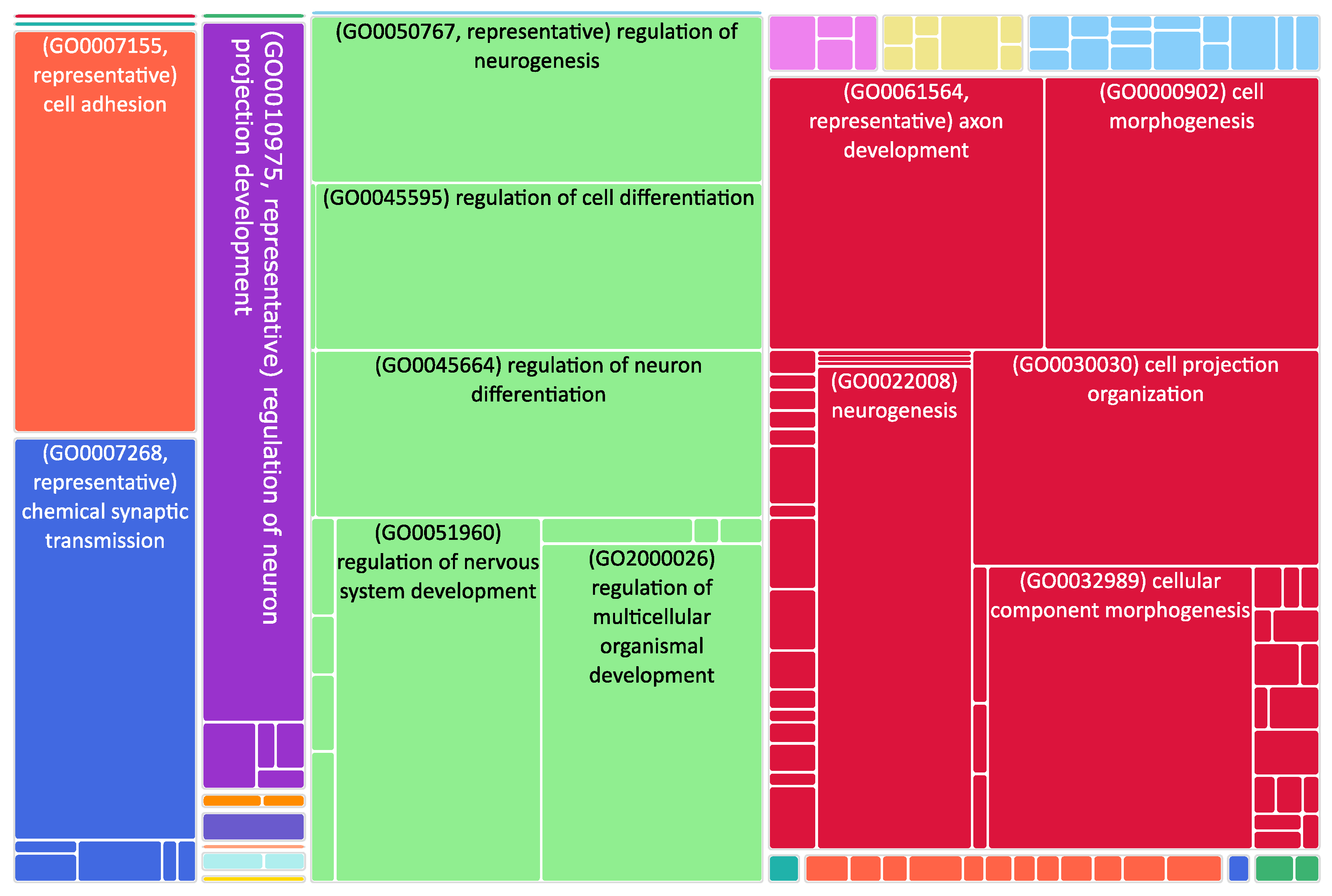
Figure 2.
Tree map of downregulated genes in the stage 4 + 4S. Each rectangle is a single cluster representative. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Sizes of the rectangles are adjusted to reflect the p-value of the GO term (“biological process”). Most representative terms are indicated. Several terms associated with neuronal function are enriched.
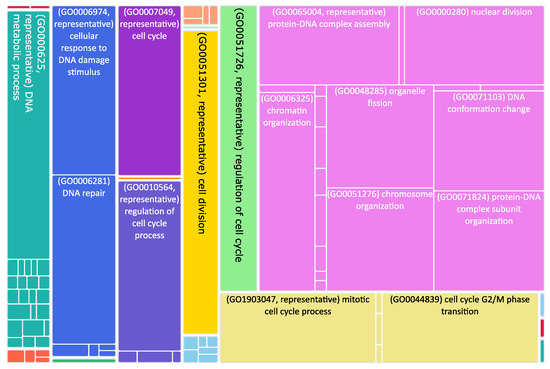
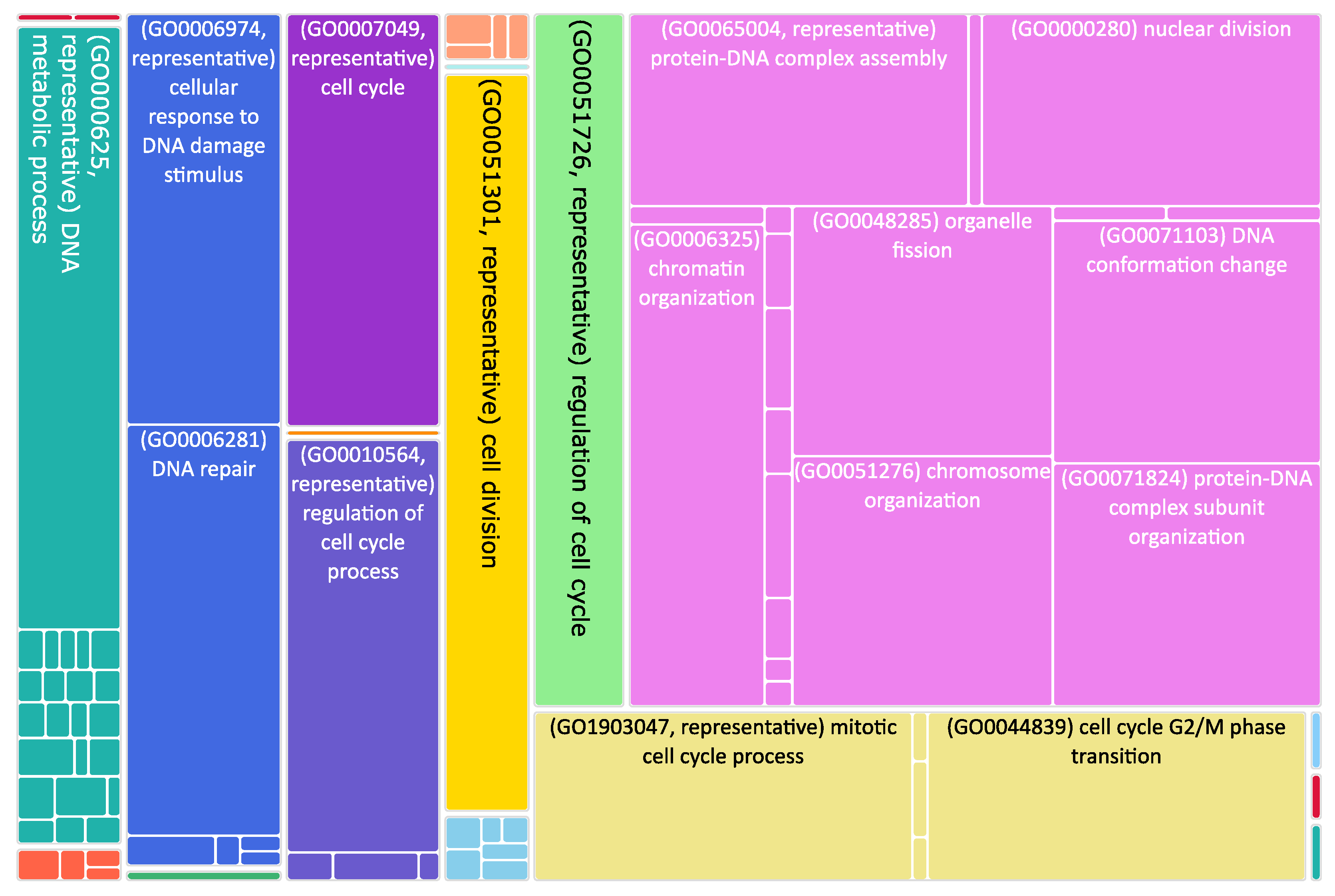
Figure 3.
Tree map of upregulated genes in the stage 4 + 4S. Each rectangle is a single cluster representative. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Sizes of the rectangles are adjusted to reflect the p-value of the GO term (“biological process”). Most representative terms are indicated. Upregulated genes in the stage 4 + 4S are prevalently associated with cell cycle regulation.
A significant number of altered genes encoded for cytoskeletal proteins, or localized in mitochondria or in the endoplasmic reticulum (Figure 4 and Supplementary Table S3). As speculated, we also found some differentially expressed genes (DEGs) encoding for MERCS proteins (Figure 1).
Among the identified MERCS DEGs, HK2 (hexokinase 2) was 1.4 times upregulated in stage 4 + 4S (Supplemental Table S1). It is interesting to note that PDK1 (pyruvate dehydrogenase kinase 1) was also upregulated. PDK1 and HK2 are involved in cell metabolism, which is known to be affected in neuroblastoma. Indeed, in N-Myc (MYCN)-amplified NB cells, altered energy metabolism due to direct or indirect activation of genes involved in glycolysis, glutamine and fatty acid metabolism, and mitochondrial dysfunction has been reported [63]. It has been shown that PDK1 is specifically required for the metabolic response to hypoxia and nutrient deprivation in some cancer types; PDK1 phosphorylates pyruvate dehydrogenase (PDH) to inhibit its activity, thereby reducing the level of pyruvic acid in the tricarboxylic acid cycle, which affects the rates of oxidative phosphorylation [64]. The outcome of upregulated PDK1 activity on metabolic reprogramming might be amplified by the concomitant increase in HK2 expression observed in stage 4 + 4S (Supplemental Table S1). HK2 catalyzes the first step of glycolysis and it has been found upregulated in several types of cancer; its activity has been associated with the Warburg effect, e.g., high lactate production in the presence of oxygen [65]. Notably, upregulation of PDK1 and HK2 has been associated with an increased proliferation and poor prognosis in MYCN-amplified NB [66].
As mentioned before, many upregulated genes in grade 4 + 4S were involved in the regulation of the cell cycle (Figure 3). Interestingly, several stages of the cell cycle are accompanied and/or controlled by changes in mitochondria dynamics; for example, the cell cycle regulator CDK5 (cyclin-dependent kinase 5) is necessary for mitochondrial movement [67]. Interestingly, increased CDK5 activity was shown to be involved in several cancers [68,69,70]. Here, we found that CCNB1 (cyclin B1) and CDK1 (cyclin-dependent kinase 1) were twice upregulated and NME4 (NME/NM23 nucleoside diphosphate kinase 4) was 1.6 times upregulated (Supplementary Table S2). CDK1 is essential for cell division in mammals; CDK1 combines with cyclin B1 to form the cyclin B1–CDK1 complex, which is required for early mitotic events such as spindle assembly, nuclear envelope breakdown, and chromosome condensation. Genomic aberrations of cyclin B1 and CDK1 genes are associated with a dysregulated G1 entry checkpoint and have been described in NB [71]. NME4 belongs to a multifunctional NDPK/NME protein family. Its members are predominantly found in the mitochondrial intermembrane space, tethered to the inner membrane via anionic phospholipids, such as cardiolipin (CL) [72]. Here, NDPK-D plays two roles crucial for proper mitochondrial physiology. Firstly, it transfers phosphate from ATP (generated by oxidative phosphorylation pathway) to other NDPs (mostly to GDP). This phosphotransfer reaction, thus, generates GTP necessary for powering mitochondrial GTPases, such as optic atrophy 1 (OPA1) [73], a dynamin protein mediating mitochondrial fusion and maintaining the cristae. Secondly, NDPK-D transports CL from the inner mitochondrial membrane to the outer one; there, CL functions as a pro-apoptotic or pro-mitophagic signal.
NME4 inhibition was recently shown to reduce NB cell migration and to be involved in NB cell differentiation [74]. However, further research needs to be undertaken both to validate the functional NME4 role in NB pathogenesis and to identify the kinase targets and signaling pathways regulated by NME1. If elucidated and confirmed, NME4 and its activity may represent a novel target for NB therapy by inducing NB cell differentiation [75,76,77].
Other mitochondria genes differentially expressed in high-stage NB are LETM1 (leucine zipper and EF-hand-containing transmembrane protein 1; 1.4 times upregulated in stage 4 + 4S) and MTCH2 (mitochondrial carrier 2; 1.5 times upregulated in stage 4 + 4S; Supplemental Table S1).
LETM1 is a mitochondrial proton/calcium antiporter that has been described to mediate proton-dependent calcium efflux from mitochondria [78]. Mitochondria calcium handling is fundamental not only for their activity, but for the overall cell physiology, fostering either ATP production in case of physiological calcium uptake, or promoting permeability transition in case of calcium overload [32,35].
LETM1 is also crucial for the maintenance of mitochondrial tubular networks and for the assembly of respiratory chain super-complexes [79]. LETM1 knockdown caused mitochondria to become dot-like structures, losing their tubular networks to an extent significantly greater than that observed in OPA1-knockdown cells. Images of mitochondria lacking LETM1 were reminiscent of observations following overexpression of pro-fission proteins such as Fis1 or knockdown of pro-fusion proteins such as OPA1 [80,81]. Although the functions and mechanisms of LETM1 with respect to cell viability and tumorigenesis remain controversial, accumulating data suggest that LETM1 is a crucial candidate. Deepening its role will clarify how mitochondria regulate the normal life of the cell and tumor-associated metabolic reprogramming.
MTCH2 is an outer mitochondrial membrane protein that functions in the process of intrinsic cell death as well as in the regulation of fatty acid metabolism. MTCH2 interacts with the truncated BH3-interacting domain death agonist (tBID) to regulate cell apoptosis [82]. Previous studies demonstrate that loss of MTCH2 impairs mitochondrial architecture and functions, including enlarged size [83], reduced motility [84], and elevated oxidative stress [85]. MTCH2 expression was associated with several types of tumors.
Altogether, our data suggest that several MERCS-resident proteins, regulating (carbohydrate) metabolism, mitochondrial dynamics, and molecular transport, may be involved in the etiopathogenesis of NB.
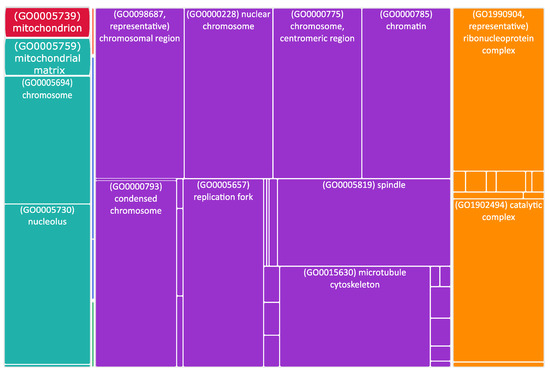
Figure 4.
Tree map of upregulated genes in the stage 4 + 4S. Each rectangle is a single cluster representative. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Sizes of the rectangles are adjusted to reflect the p-value of the GO term (“cellular component”). Most representative terms are indicated. Cytoskeleton and mitochondria are enriched terms.
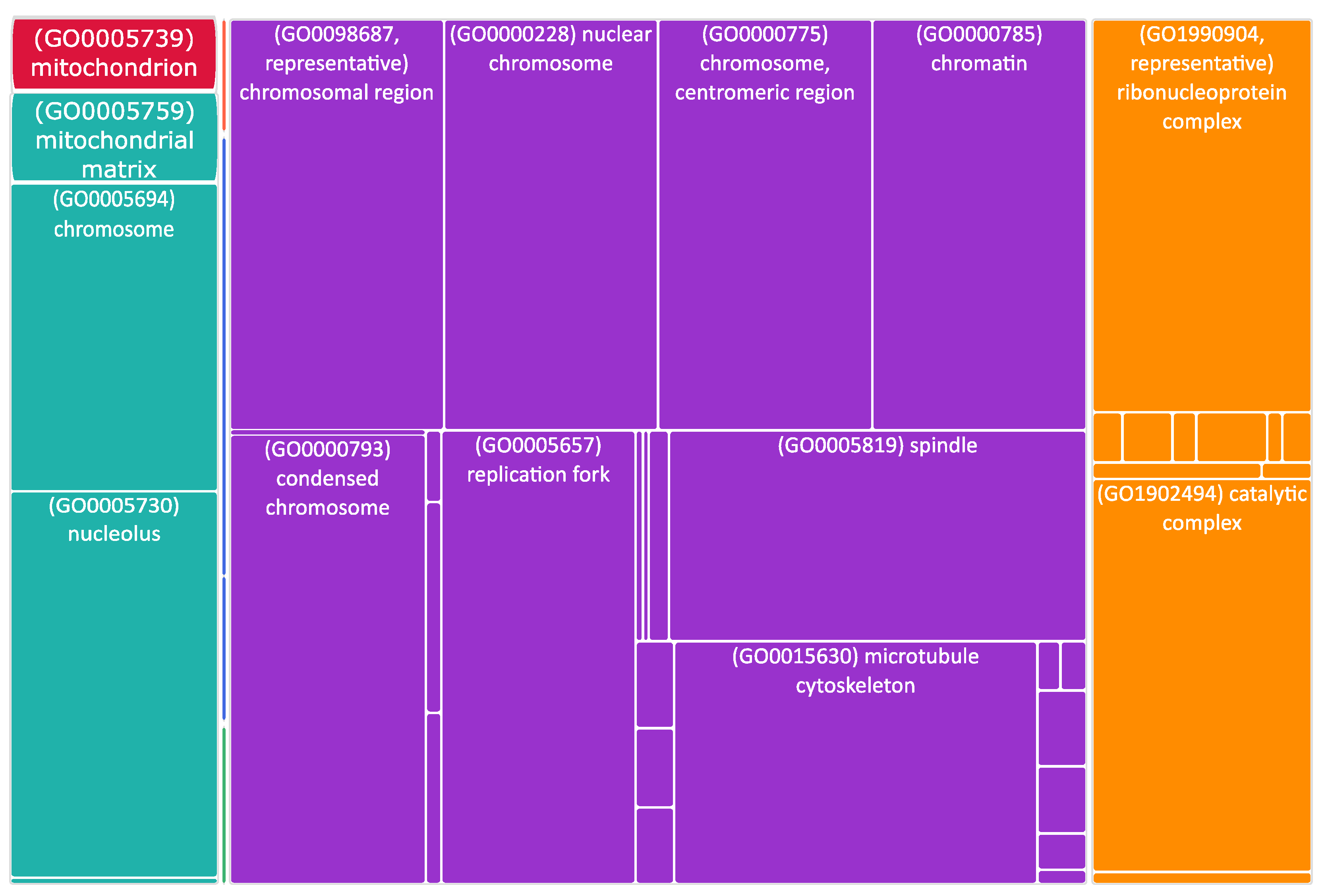
Figure 4.
Tree map of upregulated genes in the stage 4 + 4S. Each rectangle is a single cluster representative. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Sizes of the rectangles are adjusted to reflect the p-value of the GO term (“cellular component”). Most representative terms are indicated. Cytoskeleton and mitochondria are enriched terms.
4. Discussion
Neuroblastoma is the most common extracranial tumor of early childhood and accounts for 15% of all pediatric cancer mortalities. This tumor is characterized by high clinical and biological heterogeneity. Indicating several genetic aspects, besides external factors, might cooperate to define the phenotypic outcomes. A tremendous effort has been made to elucidate the molecular mechanisms implicated both in the etiology and pathogenesis of NB, which eventually resulted in identification of novel therapeutic targets. Whole-genome-based methods, such as high-throughput genome analysis, genome-wide association studies, and genome sequencing, have revealed genetic alterations and disrupted pathways that participate in NB growth and development. However, the precise pathways and genes involved during NB progression are still largely unknown. Metabolic reprogramming accompanies development and progression of many cancer types; thus, mitochondria, which are central for cells’ energy production, fundamentally contribute to tumorigenesis. A great deal of evidence in the last years has shown that the shape of these organelles, along with their interplay with other subcellular compartments, control their function and energy production ability.
In the context of NB, the role of mitochondria in shaping the phenotypic outcomes and progression of the disease has not been fully elucidated. Here, we performed a differential gene expression analysis of NB stage 1 and stage 4 + 4S to discover NB-related biological processes driven by mitochondrial dynamics. Among many differentially expressed genes, we found some interesting genes coding for proteins either residing at mitochondria or modulating their function: HK2, PDK1, NME4, LETM1, MTCH2, and CDK1. Based on the current literature, we propose and explain that these proteins could cooperate to define the metabolic adaptations needed to sustain cancer progression based on enhanced glycolysis and to promote cell cycle progression.
Although further analyses remain necessary, this brief report provides new hints in NB molecular etiopathogenesis, suggesting that alterations of mitochondria dynamics could participate in the development and worsening of the disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines11020596/s1, Table S1: Differentially expressed genes; Table S2: Gene Ontology analysis based on Biological Processes; Table S3: Gene Ontology analysis based on Cellular Components. Figure S1: Normalized data used to identify DEGs. In green stage 1 tumor samples and in violet stage 4 + 4S. Total number of samples (box plots) is 445. This makes difficult the visualization of the sample names but it is possible to see that the median of the fluorescence intensity is the same for all the samples (lane in the middle of each box).
Author Contributions
Conceptualization, S.C., T.K., A.D.M. and M.G.; methodology, S.C.; writing—original draft preparation, review and editing: S.C., T.K., C.V., A.P.M.R., A.D.M. and M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Padua STARS@Unipd Consolidator grant FIRMESs to M.G. T.K. is grateful to the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement for financial support (No. 896745).
Institutional Review Board Statement
Ethical review and approval were not necessary for this study because biological samples were not taken from patients directly, but data deposited in a public gene expression database were used.
Informed Consent Statement
No necessary because used public gene expression data.
Data Availability Statement
Gene expression data are available at GEO database (GSE45547).
Conflicts of Interest
C.V., A.D.M., and M.G. are Guest Editors of the Special Issue. All other authors declare no conflict of interest.
References
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, J.I.; Dyberg, C.; Wickström, M. Neuroblastoma-A Neural Crest Derived Embryonal Malignancy. Front. Mol. Neurosci. 2019, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Eggert, A.; Caron, H. Neuroblastoma: Biology, Prognosis, and Treatment. Pediatr. Clin. N. Am. 2008, 55, 97–120. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M. Recent Advances in Neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Eggert, A.; Caron, H. Neuroblastoma: Biology, Prognosis, and Treatment. Hematol. Oncol. Clin. N. Am. 2010, 24, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M. Neuroblastoma: Biological Insights into a Clinical Enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Tsubota, S.; Kadomatsu, K. Origin and Initiation Mechanisms of Neuroblastoma. Cell Tissue Res. 2018, 372, 211–221. [Google Scholar] [CrossRef]
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and Adolescent Cancer Statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef]
- Haase, G.M.; LaQuaglia, M.P. Neuroblastoma. In Operative Pediatric Surgery; Ziegler, M., Azizkhan, R.G., von Allmen, D., Weber, T.R., Eds.; McGraw Hill: New York, NY, USA, 2003; pp. 1181–1192. ISBN 978-0-07-121239-7. [Google Scholar]
- Colon, N.C.; Chung, D.H. Neuroblastoma. Adv. Pediatr. 2011, 58, 297–311. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F. Revisions of the International Criteria for Neuroblastoma Diagnosis, Staging, and Response to Treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef]
- Mathew, P.; Valentine, M.B.; Bowman, L.C.; Rowe, S.T.; Nash, M.B.; Valentine, V.A.; Cohn, S.L.; Castleberry, R.P.; Brodeur, G.M.; Look, A.T. Detection of MYCN Gene Amplification in Neuroblastoma by Fluorescence in Situ Hybridization: A Pediatric Oncology Group Study. Neoplasia 2001, 3, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Mossé, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a Major Familial Neuroblastoma Predisposition Gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Carén, H.; Abel, F.; Kogner, P.; Martinsson, T. High Incidence of DNA Mutations and Gene Amplifications of the ALK Gene in Advanced Sporadic Neuroblastoma Tumours. Biochem. J. 2008, 416, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J.M. Targeted Expression of MYCN Causes Neuroblastoma in Transgenic Mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef]
- Heukamp, L.C.; Thor, T.; Schramm, A.; De Preter, K.; Kumps, C.; De Wilde, B.; Odersky, A.; Peifer, M.; Lindner, S.; Spruessel, A.; et al. Targeted Expression of Mutated ALK Induces Neuroblastoma in Transgenic Mice. Sci. Transl. Med. 2012, 4, 141ra91. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-Myc in Untreated Human Neuroblastomas Correlates with Advanced Disease Stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Detmer, S.A.; Chan, D.C. Functions and Dysfunctions of Mitochondrial Dynamics. Nat. Rev. Mol. Cell Biol. 2007, 8, 870–879. [Google Scholar] [CrossRef]
- Liesa, M.; Palacín, M.; Zorzano, A. Mitochondrial Dynamics in Mammalian Health and Disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef]
- De Mario, A.; Peggion, C.; Massimino, M.L.; Norante, R.P.; Zulian, A.; Bertoli, A.; Sorgato, M.C. The Link of the Prion Protein with Ca2+ Metabolism and ROS Production, and the Possible Implication in Aβ Toxicity. Int. J. Mol. Sci. 2019, 20, E4640. [Google Scholar] [CrossRef]
- De Mario, A.; Peggion, C.; Massimino, M.L.; Viviani, F.; Castellani, A.; Giacomello, M.; Lim, D.; Bertoli, A.; Sorgato, M.C. The Prion Protein Regulates Glutamate-Mediated Ca2+ Entry and Mitochondrial Ca2+ Accumulation in Neurons. J. Cell Sci. 2017, 130, 2736–2746. [Google Scholar] [CrossRef]
- Moore, A.S.; Holzbaur, E.L.F. Mitochondrial-Cytoskeletal Interactions: Dynamic Associations That Facilitate Network Function and Remodeling. Curr. Opin. Physiol. 2018, 3, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Murley, A.; Nunnari, J. The Emerging Network of Mitochondria-Organelle Contacts. Mol. Cell 2016, 61, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Copeland, D.E.; Dalton, A.J. An Association between Mitochondria and the Endoplasmic Reticulum in Cells of the Pseudobranch Gland of a Teleost. J. Biophys. Biochem. Cytol. 1959, 5, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Renken, C.; Várnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Mannella, C.A.; Hajnóczky, G. Structural and Functional Features and Significance of the Physical Linkage between ER and Mitochondria. J. Cell Biol. 2006, 174, 915–921. [Google Scholar] [CrossRef]
- Sood, A.; Jeyaraju, D.V.; Prudent, J.; Caron, A.; Lemieux, P.; McBride, H.M.; Laplante, M.; Tóth, K.; Pellegrini, L. A Mitofusin-2-Dependent Inactivating Cleavage of Opa1 Links Changes in Mitochondria Cristae and ER Contacts in the Postprandial Liver. Proc. Natl. Acad. Sci. USA 2014, 111, 16017–16022. [Google Scholar] [CrossRef] [PubMed]
- Giacomello, M.; Pellegrini, L. The Coming of Age of the Mitochondria-ER Contact: A Matter of Thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnóczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming Together to Define Membrane Contact Sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef]
- De Mario, A.; Quintana-Cabrera, R.; Martinvalet, D.; Giacomello, M. (Neuro)Degenerated Mitochondria-ER Contacts. Biochem. Biophys. Res. Commun. 2017, 483, 1096–1109. [Google Scholar] [CrossRef]
- Rowland, A.A.; Voeltz, G.K. Endoplasmic Reticulum-Mitochondria Contacts: Function of the Junction. Nat. Rev. Mol. Cell Biol. 2012, 13, 607–625. [Google Scholar] [CrossRef]
- Rusiñol, A.E.; Cui, Z.; Chen, M.H.; Vance, J.E. A Unique Mitochondria-Associated Membrane Fraction from Rat Liver Has a High Capacity for Lipid Synthesis and Contains Pre-Golgi Secretory Proteins Including Nascent Lipoproteins. J. Biol. Chem. 1994, 269, 27494–27502. [Google Scholar] [CrossRef]
- Csordás, G.; Várnai, P.; Golenár, T.; Roy, S.; Purkins, G.; Schneider, T.G.; Balla, T.; Hajnóczky, G. Imaging Interorganelle Contacts and Local Calcium Dynamics at the ER-Mitochondrial Interface. Mol. Cell 2010, 39, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Giacomello, M.; Drago, I.; Bortolozzi, M.; Scorzeto, M.; Gianelle, A.; Pizzo, P.; Pozzan, T. Ca2+ Hot Spots on the Mitochondrial Surface Are Generated by Ca2+ Mobilization from Stores, but Not by Activation of Store-Operated Ca2+ Channels. Mol. Cell 2010, 38, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Iwasawa, R.; Mahul-Mellier, A.-L.; Datler, C.; Pazarentzos, E.; Grimm, S. Fis1 and Bap31 Bridge the Mitochondria-ER Interface to Establish a Platform for Apoptosis Induction. EMBO J. 2011, 30, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Martinvalet, D. The Role of the Mitochondria and the Endoplasmic Reticulum Contact Sites in the Development of the Immune Responses. Cell Death Dis. 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Quintero, O.A.; DiVito, M.M.; Adikes, R.C.; Kortan, M.B.; Case, L.B.; Lier, A.J.; Panaretos, N.S.; Slater, S.Q.; Rengarajan, M.; Feliu, M.; et al. Human Myo19 Is a Novel Myosin That Associates with Mitochondria. Curr. Biol. 2009, 19, 2008–2013. [Google Scholar] [CrossRef] [PubMed]
- Pathak, D.; Sepp, K.J.; Hollenbeck, P.J. Evidence That Myosin Activity Opposes Microtubule-Based Axonal Transport of Mitochondria. J. Neurosci. 2010, 30, 8984–8992. [Google Scholar] [CrossRef] [PubMed]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef]
- Moore, A.S.; Wong, Y.C.; Simpson, C.L.; Holzbaur, E.L.F. Dynamic Actin Cycling through Mitochondrial Subpopulations Locally Regulates the Fission-Fusion Balance within Mitochondrial Networks. Nat. Commun. 2016, 7, 12886. [Google Scholar] [CrossRef]
- De Mario, A.; Scarlatti, C.; Costiniti, V.; Primerano, S.; Lopreiato, R.; Calì, T.; Brini, M.; Giacomello, M.; Carafoli, E. Calcium Handling by Endoplasmic Reticulum and Mitochondria in a Cell Model of Huntington’s Disease. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
- De Mario, A.; Tosatto, A.; Hill, J.M.; Kriston-Vizi, J.; Ketteler, R.; Vecellio Reane, D.; Cortopassi, G.; Szabadkai, G.; Rizzuto, R.; Mammucari, C. Identification and Functional Validation of FDA-Approved Positive and Negative Modulators of the Mitochondrial Calcium Uniporter. Cell Rep. 2021, 35, 109275. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G.; Pratt, C.W. Fundamentals of Biochemistry: Life at the Molecular Level, 2nd ed.; Wiley: Hoboken, NJ, USA, 2006; ISBN 978-0-471-21495-3. [Google Scholar]
- Alexandre, A.; Reynafarje, B.; Lehninger, A.L. Stoichiometry of Vectorial H+ Movements Coupled to Electron Transport and to ATP Synthesis in Mitochondria. Proc. Natl. Acad. Sci. USA 1978, 75, 5296–5300. [Google Scholar] [CrossRef] [PubMed]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and Molecular Mechanisms of Mitochondrial Function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Budihardjo, I.; Oliver, H.; Lutter, M.; Luo, X.; Wang, X. Biochemical Pathways of Caspase Activation during Apoptosis. Annu. Rev. Cell Dev. Biol. 1999, 15, 269–290. [Google Scholar] [CrossRef] [PubMed]
- van Loo, G.; Saelens, X.; van Gurp, M.; MacFarlane, M.; Martin, S.J.; Vandenabeele, P. The Role of Mitochondrial Factors in Apoptosis: A Russian Roulette with More than One Bullet. Cell Death Differ. 2002, 9, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, L.; Jia, R. The Role of Mitochondrial Dynamics in Human Cancers. Am. J. Cancer Res. 2020, 10, 1278–1293. [Google Scholar]
- Xie, Q.; Wu, Q.; Horbinski, C.M.; Flavahan, W.A.; Yang, K.; Zhou, W.; Dombrowski, S.M.; Huang, Z.; Fang, X.; Shi, Y.; et al. Mitochondrial Control by DRP1 in Brain Tumor Initiating Cells. Nat. Neurosci. 2015, 18, 501–510. [Google Scholar] [CrossRef]
- Grespi, F.; Vianello, C.; Cagnin, S.; Giacomello, M.; De Mario, A. The Interplay of Microtubules with Mitochondria-ER Contact Sites (MERCs) in Glioblastoma. Biomolecules 2022, 12, 567. [Google Scholar] [CrossRef]
- Morita, M.; Prudent, J.; Basu, K.; Goyon, V.; Katsumura, S.; Hulea, L.; Pearl, D.; Siddiqui, N.; Strack, S.; McGuirk, S.; et al. MTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 2017, 67, 922–935.e5. [Google Scholar] [CrossRef]
- Li, M.; Wang, L.; Wang, Y.; Zhang, S.; Zhou, G.; Lieshout, R.; Ma, B.; Liu, J.; Qu, C.; Verstegen, M.M.A.; et al. Mitochondrial Fusion Via OPA1 and MFN1 Supports Liver Tumor Cell Metabolism and Growth. Cells 2020, 9, E121. [Google Scholar] [CrossRef]
- Kashatus, J.A.; Nascimento, A.; Myers, L.J.; Sher, A.; Byrne, F.L.; Hoehn, K.L.; Counter, C.M.; Kashatus, D.F. Erk2 Phosphorylation of Drp1 Promotes Mitochondrial Fission and MAPK-Driven Tumor Growth. Mol. Cell 2015, 57, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Lyu, Z.; Chen, Y.; Ji, X.; Cao, H.; Jin, M.; Zhu, J.; Yang, J.; Ling, R.; et al. Positive Feedback Loop between Mitochondrial Fission and Notch Signaling Promotes Survivin-Mediated Survival of TNBC Cells. Cell Death Dis. 2018, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Zhang, X.; Zhao, J.; Zhou, F.; Wang, Y.; Zhao, Z.; Xing, J.; Chen, B.; Li, J.; Liu, S. SIK2 Promotes Reprogramming of Glucose Metabolism through PI3K/AKT/HIF-1α Pathway and Drp1-Mediated Mitochondrial Fission in Ovarian Cancer. Cancer Lett. 2020, 469, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Civenni, G.; Bosotti, R.; Timpanaro, A.; Vàzquez, R.; Merulla, J.; Pandit, S.; Rossi, S.; Albino, D.; Allegrini, S.; Mitra, A.; et al. Epigenetic Control of Mitochondrial Fission Enables Self-Renewal of Stem-like Tumor Cells in Human Prostate Cancer. Cell Metab. 2019, 30, 303–318.e6. [Google Scholar] [CrossRef] [PubMed]
- Çoku, J.; Booth, D.M.; Skoda, J.; Pedrotty, M.C.; Vogel, J.; Liu, K.; Vu, A.; Carpenter, E.L.; Ye, J.C.; Chen, M.A.; et al. Reduced ER-Mitochondria Connectivity Promotes Neuroblastoma Multidrug Resistance. EMBO J. 2022, 41, e108272. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A More Comprehensive, Powerful, Flexible and Interactive Gene Set Enrichment Analysis Toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell Cycle Control in Cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Wainwright, L.J.; Lasorella, A.; Iavarone, A. Distinct Mechanisms of Cell Cycle Arrest Control the Decision between Differentiation and Senescence in Human Neuroblastoma Cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9396–9400. [Google Scholar] [CrossRef]
- Qing, G.; Skuli, N.; Mayes, P.A.; Pawel, B.; Martinez, D.; Maris, J.M.; Simon, M.C. Combinatorial Regulation of Neuroblastoma Tumor Progression by N-Myc and Hypoxia Inducible Factor HIF-1alpha. Cancer Res. 2010, 70, 10351–10361. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, F.; Tabariès, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.J.; Miyamoto, S. Hexokinase II Integrates Energy Metabolism and Cellular Protection: Akting on Mitochondria and TORCing to Autophagy. Cell Death Differ. 2015, 22, 364. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Yue, M.; Xiao, D.; Xiu, R.; Gan, L.; Liu, H.; Qing, G. ATF4 and N-Myc Coordinate Glutamine Metabolism in MYCN-Amplified Neuroblastoma Cells through ASCT2 Activation. J. Pathol. 2015, 235, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Ratner, N.; Bloom, G.S.; Brady, S.T. A Role for Cyclin-Dependent Kinase(s) in the Modulation of Fast Anterograde Axonal Transport: Effects Defined by Olomoucine and the APC Tumor Suppressor Protein. J. Neurosci. 1998, 18, 7717–7726. [Google Scholar] [CrossRef]
- Feldmann, G.; Mishra, A.; Hong, S.-M.; Bisht, S.; Strock, C.J.; Ball, D.W.; Goggins, M.; Maitra, A.; Nelkin, B.D. Inhibiting the Cyclin-Dependent Kinase CDK5 Blocks Pancreatic Cancer Formation and Progression through the Suppression of Ras-Ral Signaling. Cancer Res. 2010, 70, 4460–4469. [Google Scholar] [CrossRef]
- Hsu, F.-N.; Chen, M.-C.; Chiang, M.-C.; Lin, E.; Lee, Y.-T.; Huang, P.-H.; Lee, G.-S.; Lin, H. Regulation of Androgen Receptor and Prostate Cancer Growth by Cyclin-Dependent Kinase 5. J. Biol. Chem. 2011, 286, 33141–33149. [Google Scholar] [CrossRef]
- Liu, R.; Tian, B.; Gearing, M.; Hunter, S.; Ye, K.; Mao, Z. Cdk5-Mediated Regulation of the PIKE-A-Akt Pathway and Glioblastoma Cell Invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 7570–7575. [Google Scholar] [CrossRef]
- Liu, Z.; Rader, J.; He, S.; Phung, T.; Thiele, C.J. CASZ1 Inhibits Cell Cycle Progression in Neuroblastoma by Restoring PRb Activity. Cell Cycle 2013, 12, 2210–2218. [Google Scholar] [CrossRef]
- Tokarska-Schlattner, M.; Boissan, M.; Munier, A.; Borot, C.; Mailleau, C.; Speer, O.; Schlattner, U.; Lacombe, M.-L. The Nucleoside Diphosphate Kinase D (NM23-H4) Binds the Inner Mitochondrial Membrane with High Affinity to Cardiolipin and Couples Nucleotide Transfer with Respiration. J. Biol. Chem. 2008, 283, 26198–26207. [Google Scholar] [CrossRef]
- Boissan, M.; Montagnac, G.; Shen, Q.; Griparic, L.; Guitton, J.; Romao, M.; Sauvonnet, N.; Lagache, T.; Lascu, I.; Raposo, G.; et al. Membrane Trafficking. Nucleoside Diphosphate Kinases Fuel Dynamin Superfamily Proteins with GTP for Membrane Remodeling. Science 2014, 344, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Adam, K.; Lesperance, J.; Hunter, T.; Zage, P.E. The Potential Functional Roles of NME1 Histidine Kinase Activity in Neuroblastoma Pathogenesis. IJMS 2020, 21, 3319. [Google Scholar] [CrossRef]
- Reiss, M.; Gamba-Vitalo, C.; Sartorelli, A.C. Induction of Tumor Cell Differentiation as a Therapeutic Approach: Preclinical Models for Hematopoietic and Solid Neoplasms. Cancer Treat. Rep. 1986, 70, 201–218. [Google Scholar] [PubMed]
- Zeineldin, M.; Patel, A.G.; Dyer, M.A. Neuroblastoma: When Differentiation Goes Awry. Neuron 2022, 110, 2916–2928. [Google Scholar] [CrossRef] [PubMed]
- Edsjö, A.; Holmquist, L.; Påhlman, S. Neuroblastoma as an Experimental Model for Neuronal Differentiation and Hypoxia-Induced Tumor Cell Dedifferentiation. Semin. Cancer Biol. 2007, 17, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhao, L.; Clapham, D.E. Genome-Wide RNAi Screen Identifies Letm1 as a Mitochondrial Ca 2+ /H + Antiporter. Science 2009, 326, 144–147. [Google Scholar] [CrossRef]
- Tamai, S.; Iida, H.; Yokota, S.; Sayano, T.; Kiguchiya, S.; Ishihara, N.; Hayashi, J.-I.; Mihara, K.; Oka, T. Characterization of the Mitochondrial Protein LETM1, Which Maintains the Mitochondrial Tubular Shapes and Interacts with the AAA-ATPase BCS1L. J. Cell Sci. 2008, 121, 2588–2600. [Google Scholar] [CrossRef]
- Cipolat, S.; de Brito, O.M.; Dal Zilio, B.; Scorrano, L. OPA1 Requires Mitofusin 1 to Promote Mitochondrial Fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef]
- Dimmer, K.S.; Navoni, F.; Casarin, A.; Trevisson, E.; Endele, S.; Winterpacht, A.; Salviati, L.; Scorrano, L. LETM1, Deleted in Wolf Hirschhorn Syndrome Is Required for Normal Mitochondrial Morphology and Cellular Viability. Hum. Mol. Genet. 2007, 17, 201–214. [Google Scholar] [CrossRef]
- Cogliati, S.; Scorrano, L. A BID on Mitochondria with MTCH2. Cell Res. 2010, 20, 863–865. [Google Scholar] [CrossRef]
- Bahat, A.; Goldman, A.; Zaltsman, Y.; Khan, D.H.; Halperin, C.; Amzallag, E.; Krupalnik, V.; Mullokandov, M.; Silberman, A.; Erez, A.; et al. MTCH2-Mediated Mitochondrial Fusion Drives Exit from Naïve Pluripotency in Embryonic Stem Cells. Nat. Commun. 2018, 9, 5132. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Aloni, E.; Korkotian, E.; Zaltsman, Y.; Oni-Biton, E.; Kuperman, Y.; Tsoory, M.; Shachnai, L.; Levin-Zaidman, S.; Brenner, O.; et al. Loss of Forebrain MTCH2 Decreases Mitochondria Motility and Calcium Handling and Impairs Hippocampal-Dependent Cognitive Functions. Sci. Rep. 2017, 7, 44401. [Google Scholar] [CrossRef] [PubMed]
- Buzaglo-Azriel, L.; Kuperman, Y.; Tsoory, M.; Zaltsman, Y.; Shachnai, L.; Zaidman, S.L.; Bassat, E.; Michailovici, I.; Sarver, A.; Tzahor, E.; et al. Loss of Muscle MTCH2 Increases Whole-Body Energy Utilization and Protects from Diet-Induced Obesity. Cell Rep. 2017, 18, 1335–1336. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).