The Iron Content of Human Serum Albumin Modulates the Susceptibility of Acinetobacter baumannii to Cefiderocol
Abstract
1. Introduction
2. Results and Discussion
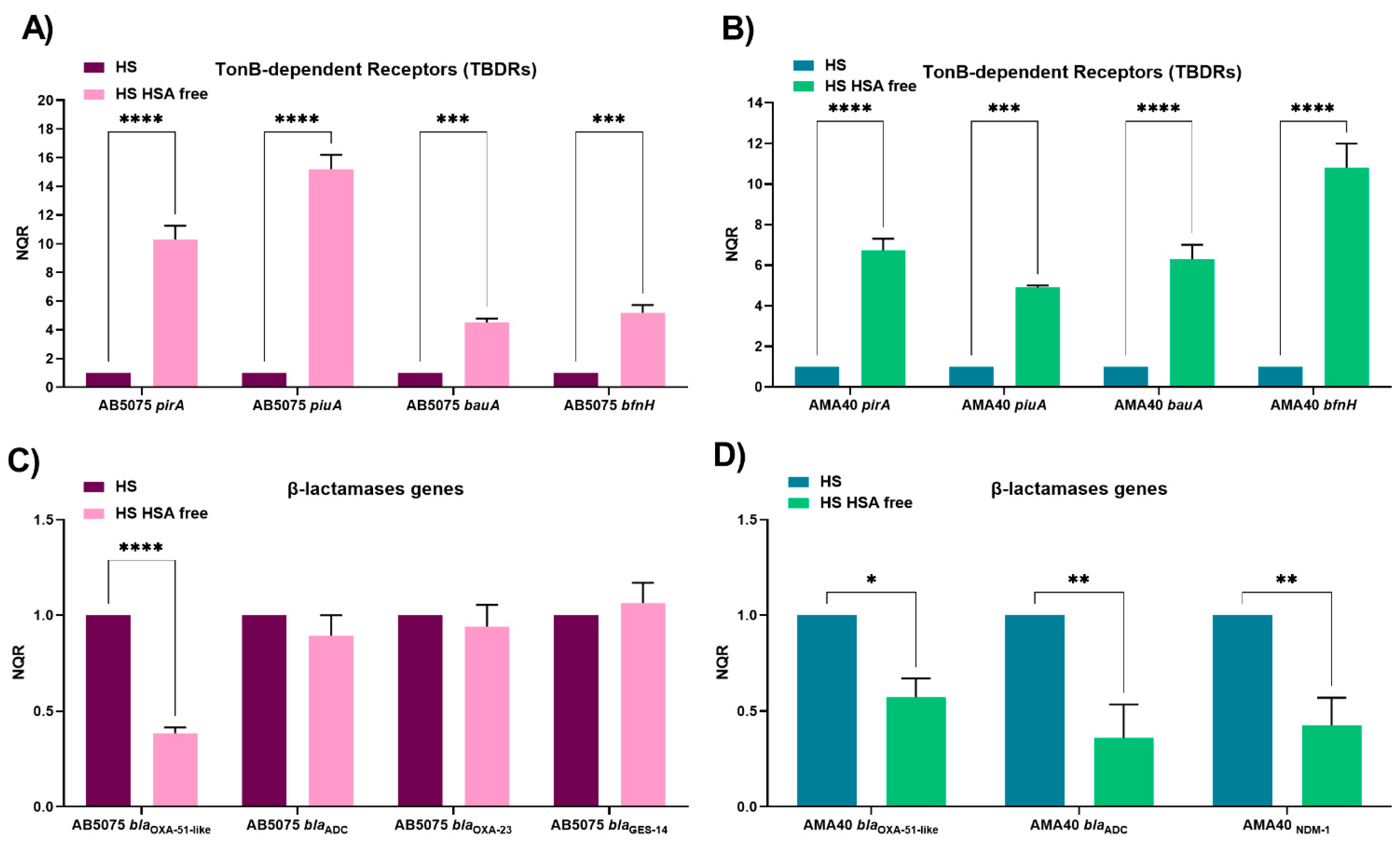
2.1. HSA Present in Human Fluids Alters CFDC MICs via a Global Transcriptional Response
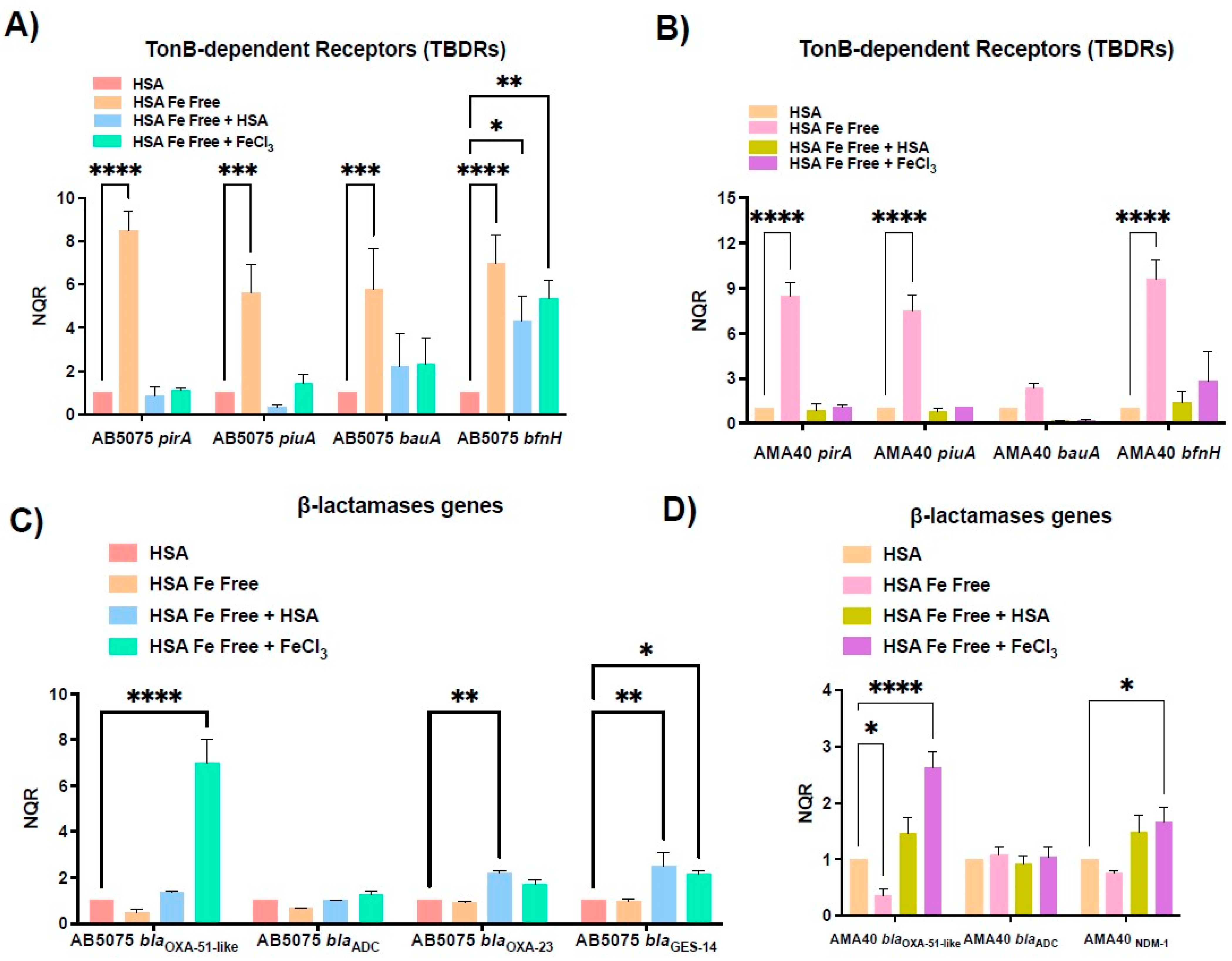
2.2. Role of Ferric HSA on A. baumannii Response Affecting CFDC Susceptibility
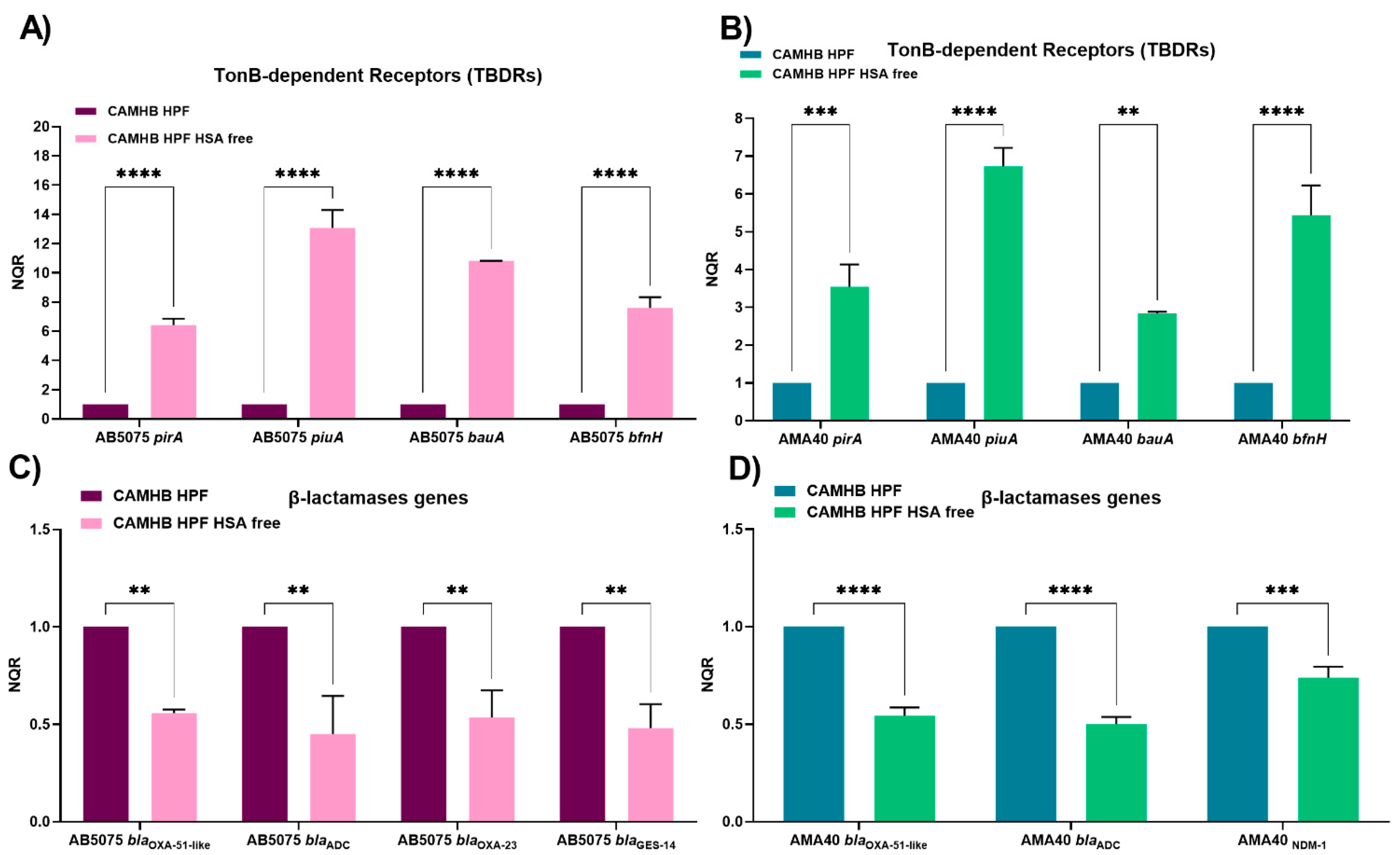
2.3. Changes in the Expression of Genes Coding for Iron Uptake Functions and β-Lactam Resistance in Cerebrospinal Fluid (CSF), a Low HSA Content Fluid
3. Concluding Remarks
4. Materials and Methods
4.1. Bacterial Strains
4.2. RNA Extraction, Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.3. HSA Removal
4.4. Iron Removal
4.5. Antimicrobial Susceptibility Testing
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- CDC. Antibiotic Resistance Threats in the United States; US Department of Health and Human Services, Centres for Disease Control and Prevention, Ed.; Centers for Disease Control: Atlanta, Georgia, 2019. [Google Scholar]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019, 63, e01110-18. [Google Scholar] [CrossRef]
- Karaiskos, I.; Lagou, S.; Pontikis, K.; Rapti, V.; Poulakou, G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health 2019, 7, 151. [Google Scholar] [CrossRef]
- Bassetti, M.; Echols, R.; Matsunaga, Y.; Ariyasu, M.; Doi, Y.; Ferrer, R.; Lodise, T.P.; Naas, T.; Niki, Y.; Paterson, D.L.; et al. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect. Dis. 2021, 21, 226–240. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef]
- Bonomo, R.A. Cefiderocol: A Novel Siderophore Cephalosporin Defeating Carbapenem-resistant Pathogens. Clin. Infect. Dis. 2019, 69 (Suppl. 7), S519–S520. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef]
- Aoki, T.; Yoshizawa, H.; Yamawaki, K.; Yokoo, K.; Sato, J.; Hisakawa, S.; Hasegawa, Y.; Kusano, H.; Sano, M.; Sugimoto, H.; et al. Cefiderocol (S-649266), A new siderophore cephalosporin exhibiting potent activities against Pseudomonas aeruginosa and other gram-negative pathogens including multi-drug resistant bacteria: Structure activity relationship. Eur. J. Med. Chem. 2018, 155, 847–868. [Google Scholar] [CrossRef]
- Ito, A.; Nishikawa, T.; Matsumoto, S.; Yoshizawa, H.; Sato, T.; Nakamura, R.; Tsuji, M.; Yamano, Y. Siderophore Cephalosporin Cefiderocol Utilizes Ferric Iron Transporter Systems for Antibacterial Activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 7396–7401. [Google Scholar] [CrossRef]
- McCreary, E.K.; Heil, E.L.; Tamma, P.D. New Perspectives on Antimicrobial Agents: Cefiderocol. Antimicrob. Agents Chemother. 2021, 65, e0217120. [Google Scholar] [CrossRef]
- Parsels, K.A.; Mastro, K.A.; Steele, J.M.; Thomas, S.J.; Kufel, W.D. Cefiderocol: A novel siderophore cephalosporin for multidrug-resistant Gram-negative bacterial infections. J. Antimicrob. Chemother. 2021, 76, 1379–1391. [Google Scholar] [CrossRef]
- Wunderink, R.G.; Matsunaga, Y.; Ariyasu, M.; Clevenbergh, P.; Echols, R.; Kaye, K.S.; Kollef, M.; Menon, A.; Pogue, J.M.; Shorr, A.F.; et al. Cefiderocol versus high-dose, extended-infusion meropenem for the treatment of Gram-negative nosocomial pneumonia (APEKS-NP): A randomised, double-blind, phase 3, non-inferiority trial. Lancet Infect. Dis. 2021, 21, 213–225. [Google Scholar] [CrossRef]
- Jacobs, M.R.; Abdelhamed, A.M.; Good, C.E.; Rhoads, D.D.; Hujer, K.M.; Hujer, A.M.; Domitrovic, T.N.; Rudin, S.D.; Richter, S.S.; van Duin, D.; et al. ARGONAUT-I: Activity of Cefiderocol (S-649266), a Siderophore Cephalosporin, against Gram-Negative Bacteria, Including Carbapenem-Resistant Nonfermenters and Enterobacteriaceae with Defined Extended-Spectrum beta-Lactamases and Carbapenemases. Antimicrob. Agents Chemother. 2019, 63, e01801-18. [Google Scholar] [CrossRef]
- Martinez, J.; Fernandez, J.S.; Liu, C.; Hoard, A.; Mendoza, A.; Nakanouchi, J.; Rodman, N.; Courville, R.; Tuttobene, M.R.; Lopez, C.; et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019, 9, 17251. [Google Scholar] [CrossRef]
- Martinez, J.; Razo-Gutierrez, C.; Le, C.; Courville, R.; Pimentel, C.; Liu, C.; Fung, S.E.; Tuttobene, M.R.; Phan, K.; Vila, A.J.; et al. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci. Rep. 2021, 11, 4737. [Google Scholar] [CrossRef] [PubMed]
- Quinn, B.; Rodman, N.; Jara, E.; Fernandez, J.S.; Martinez, J.; Traglia, G.M.; Montana, S.; Cantera, V.; Place, K.; Bonomo, R.A.; et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018, 8, 14741. [Google Scholar] [CrossRef] [PubMed]
- Rodman, N.; Martinez, J.; Fung, S.; Nakanouchi, J.; Myers, A.L.; Harris, C.M.; Dang, E.; Fernandez, J.S.; Liu, C.; Mendoza, A.M.; et al. Human Pleural Fluid Elicits Pyruvate and Phenylalanine Metabolism in Acinetobacter baumannii to Enhance Cytotoxicity and Immune Evasion. Front. Microbiol. 2019, 10, 1581. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Bartoletti, M.; Cojutti, P.G.; Gaibani, P.; Conti, M.; Giannella, M.; Viale, P.; Pea, F. A descriptive case series of pharmacokinetic/pharmacodynamic target attainment and microbiological outcome in critically ill patients with documented severe extensively drug-resistant Acinetobacter baumannii bloodstream infection and/or ventilator-associated pneumonia treated with cefiderocol. J. Glob. Antimicrob. Resist. 2021, 27, 294–298. [Google Scholar]
- Falcone, M.; Tiseo, G.; Leonildi, A.; Della Sala, L.; Vecchione, A.; Barnini, S.; Farcomeni, A.; Menichetti, F. Cefiderocol- Compared to Colistin-Based Regimens for the Treatment of Severe Infections Caused by Carbapenem-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2022, 66, e0214221. [Google Scholar] [CrossRef]
- Le, C.; Pimentel, C.; Pasteran, F.; Tuttobene, M.R.; Subils, T.; Escalante, J.; Nishimura, B.; Arriaga, S.; Carranza, A.; Mezcord, V.; et al. Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport. Biomedicines 2022, 10, 600. [Google Scholar] [CrossRef]
- Nishimura, B.; Escalante, J.; Tuttobene, M.R.; Subils, T.; Mezcord, V.; Pimentel, C.; Georgeos, N.; Pasteran, F.; Rodriguez, C.; Sieira, R.; et al. Acinetobacter baumannii response to cefiderocol challenge in human urine. Sci. Rep. 2022, 12, 8763. [Google Scholar] [CrossRef]
- Pinsky, M.; Roy, U.; Moshe, S.; Weissman, Z.; Kornitzer, D. Human Serum Albumin Facilitates Heme-Iron Utilization by Fungi. mBio 2020, 11, e00607-20. [Google Scholar] [CrossRef]
- Egesten, A.; Frick, I.M.; Morgelin, M.; Olin, A.I.; Bjorck, L. Binding of albumin promotes bacterial survival at the epithelial surface. J. Biol. Chem. 2011, 286, 2469–2476. [Google Scholar] [CrossRef] [PubMed]
- Gonyar, L.A.; Gray, M.C.; Christianson, G.J.; Mehrad, B.; Hewlett, E.L. Albumin, in the Presence of Calcium, Elicits a Massive Increase in Extracellular Bordetella Adenylate Cyclase Toxin. Infect. Immun. 2017, 85, e00198-17. [Google Scholar] [CrossRef] [PubMed]
- Ledger, E.K.; Mesnage, S.; Edwards, A.M. Human serum triggers antibiotic tolerance in Staphylococcus aureus. Nat. Commun. 2022, 13, 2041. [Google Scholar] [CrossRef] [PubMed]
- Pekmezovic, M.; Kaune, A.K.; Austermeier, S.; Hitzler, S.U.J.; Mogavero, S.; Hovhannisyan, H.; Gabaldón, T.; Gresnigt, M.S.; Hube, B. Human albumin enhances the pathogenic potential of Candida glabrata on vaginal epithelial cells. PLoS Pathog 2021, 17, e1010037. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y.; et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. MBio 2014, 5, e01076-14. [Google Scholar] [CrossRef]
- Rodgers, D.; Pasteran, F.; Calderon, M.; Jaber, S.; Traglia, G.M.; Albornoz, E.; Corso, A.; Vila, A.J.; Bonomo, R.A.; Adams, M.D.; et al. Characterisation of ST25 NDM-1-producing Acinetobacter spp. strains leading the increase in NDM-1 emergence in Argentina. J. Glob. Antimicrob. Resist. 2020, 23, 108–110. [Google Scholar] [CrossRef]
- Le, C.; Pimentel, C.; Tuttobene, M.R.; Subils, T.; Nishimura, B.; Traglia, G.M.; Perez, F.; Papp-Wallace, K.M.; Bonomo, R.A.; Tolmasky, M.E.; et al. Interplay between meropenem and human serum albumin on expression of carbapenem resistance genes and natural competence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021, 65, AAC0101921. [Google Scholar] [CrossRef]
- Martinez, J.; Liu, C.; Rodman, N.; Fernandez, J.S.; Barberis, C.; Sieira, R.; Perez, F.; Bonomo, R.A.; Ramirez, M.S. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn Microbiol. Infect. Dis. 2018, 93, 183–187. [Google Scholar] [CrossRef]
- Pimentel, C.; Le, C.; Tuttobene, M.R.; Subils, T.; Martinez, J.; Sieira, R.; Papp-Wallace, K.M.; Keppetipola, N.; Bonomo, R.A.; Actis, L.A.; et al. Human Pleural Fluid and Human Serum Albumin Modulate the Behavior of a Hypervirulent and Multidrug-Resistant (MDR) Acinetobacter baumannii Representative Strain. Pathogens 2021, 10, 471. [Google Scholar] [CrossRef]
- Bal, W.; Sokolowska, M.; Kurowska, E.; Faller, P. Binding of transition metal ions to albumin: Sites, affinities and rates. Biochim. Biophys Acta 2013, 1830, 5444–5455. [Google Scholar] [CrossRef]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the mysteries of serum albumin-more than just a serum protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef]
- Kim, B.N.; Peleg, A.Y.; Lodise, T.P.; Lipman, J.; Li, J.; Nation, R.; Paterson, D.L. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect. Dis. 2009, 9, 245–255. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, C.; Ye, S. Acinetobacter baumannii meningitis in children: A case series and literature review. Infection 2019, 47, 643–649. [Google Scholar] [CrossRef]
- Ramström, M.; Zuberovic, A.; Grönwall, C.; Hanrieder, J.; Bergquist, J.; Hober, S. Development of affinity columns for the removal of high-abundance proteins in cerebrospinal fluid. Biotechnol. Appl. Biochem. 2009, 52 Pt 2, 159–166. [Google Scholar] [CrossRef]
- Adams, M.D.; Pasteran, F.; Traglia, G.M.; Martinez, J.; Huang, F.; Liu, C.; Fernandez, J.S.; Lopez, C.; Gonzalez, L.J.; Albornoz, E.; et al. Distinct mechanisms of dissemination of NDM-1 metallo- beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob. Agents Chemother. 2020, 64, e00324-20. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- CLSI Document M100-S30:2020; Performance Standards for Antimicrobial Susceptibility Testing: Thirty Edition Informational Supplement. Clinical Lab Standards Institute: Wayne, PA, USA, 2020.
- Huband, M.D.; Ito, A.; Tsuji, M.; Sader, H.S.; Fedler, K.A.; Flamm, R.K. Cefiderocol MIC quality control ranges in iron-depleted cation-adjusted Mueller-Hinton broth using a CLSI M23-A4 multi-laboratory study design. Diagn Microbiol. Infect. Dis. 2017, 88, 198–200. [Google Scholar] [CrossRef]

| Condition | CFDC MIC (mg/L) | |
|---|---|---|
| AB5075 | AMA40 | |
| CAMHB | 0.5 (S) | 0.5 (S) |
| 4% HPF | 1 (S) | 16 * (R) |
| 4% HPF HSA free ** | 0.5 (S) | 0.25 (S) |
| 100% HS | 1 (S) | 4 * (S) |
| 100% HS HSA free ** | 0.5 (S) | 0.25 (S) |
| Strains | CFDC | |||
|---|---|---|---|---|
| AB5075 | AMA40 | |||
| MIC (mg/L) | MBC (mg/L) | MIC (mg/L) | MBC (mg/L) | |
| Untreated | 0.25 (S) | 0.25 (S) | 0.5 (S) | 32 (R) |
| HSA pre-Chelex® treatment | 8 (I) | 32 (R) | 2 (S) | 64 (R) |
| HSA Fe-Free (post-Chelex® treatment) | 0.125 (S) | 8 (I) | 1 (S) | 16 (R) |
| HSA Fe-Free + 100µM FeCl3 | 32 (R) | 256 (R) | 128 (R) | 128 (R) |
| HSA Fe-Free + 3.5% HSA | 8 (I) | 64 (R) | 4 (S) | 64 (R) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escalante, J.; Nishimura, B.; Tuttobene, M.R.; Subils, T.; Mezcord, V.; Actis, L.A.; Tolmasky, M.E.; Bonomo, R.A.; Ramirez, M.S. The Iron Content of Human Serum Albumin Modulates the Susceptibility of Acinetobacter baumannii to Cefiderocol. Biomedicines 2023, 11, 639. https://doi.org/10.3390/biomedicines11020639
Escalante J, Nishimura B, Tuttobene MR, Subils T, Mezcord V, Actis LA, Tolmasky ME, Bonomo RA, Ramirez MS. The Iron Content of Human Serum Albumin Modulates the Susceptibility of Acinetobacter baumannii to Cefiderocol. Biomedicines. 2023; 11(2):639. https://doi.org/10.3390/biomedicines11020639
Chicago/Turabian StyleEscalante, Jenny, Brent Nishimura, Marisel R. Tuttobene, Tomás Subils, Vyanka Mezcord, Luis A. Actis, Marcelo E. Tolmasky, Robert A. Bonomo, and María Soledad Ramirez. 2023. "The Iron Content of Human Serum Albumin Modulates the Susceptibility of Acinetobacter baumannii to Cefiderocol" Biomedicines 11, no. 2: 639. https://doi.org/10.3390/biomedicines11020639
APA StyleEscalante, J., Nishimura, B., Tuttobene, M. R., Subils, T., Mezcord, V., Actis, L. A., Tolmasky, M. E., Bonomo, R. A., & Ramirez, M. S. (2023). The Iron Content of Human Serum Albumin Modulates the Susceptibility of Acinetobacter baumannii to Cefiderocol. Biomedicines, 11(2), 639. https://doi.org/10.3390/biomedicines11020639










