Parallel Dysregulated Immune Response in Severe Forms of COVID-19 and Bacterial Sepsis via Single-Cell Transcriptome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Single-Cell Data
2.2. Subject Characteristics and Specimen Collection
2.3. Details of the Step-by-Step Method
2.4. scRNA Sequencing by Seq-Well
2.5. scRNA-seq Computational Pipelines and Analysis

2.6. Functional Analysis
3. Results
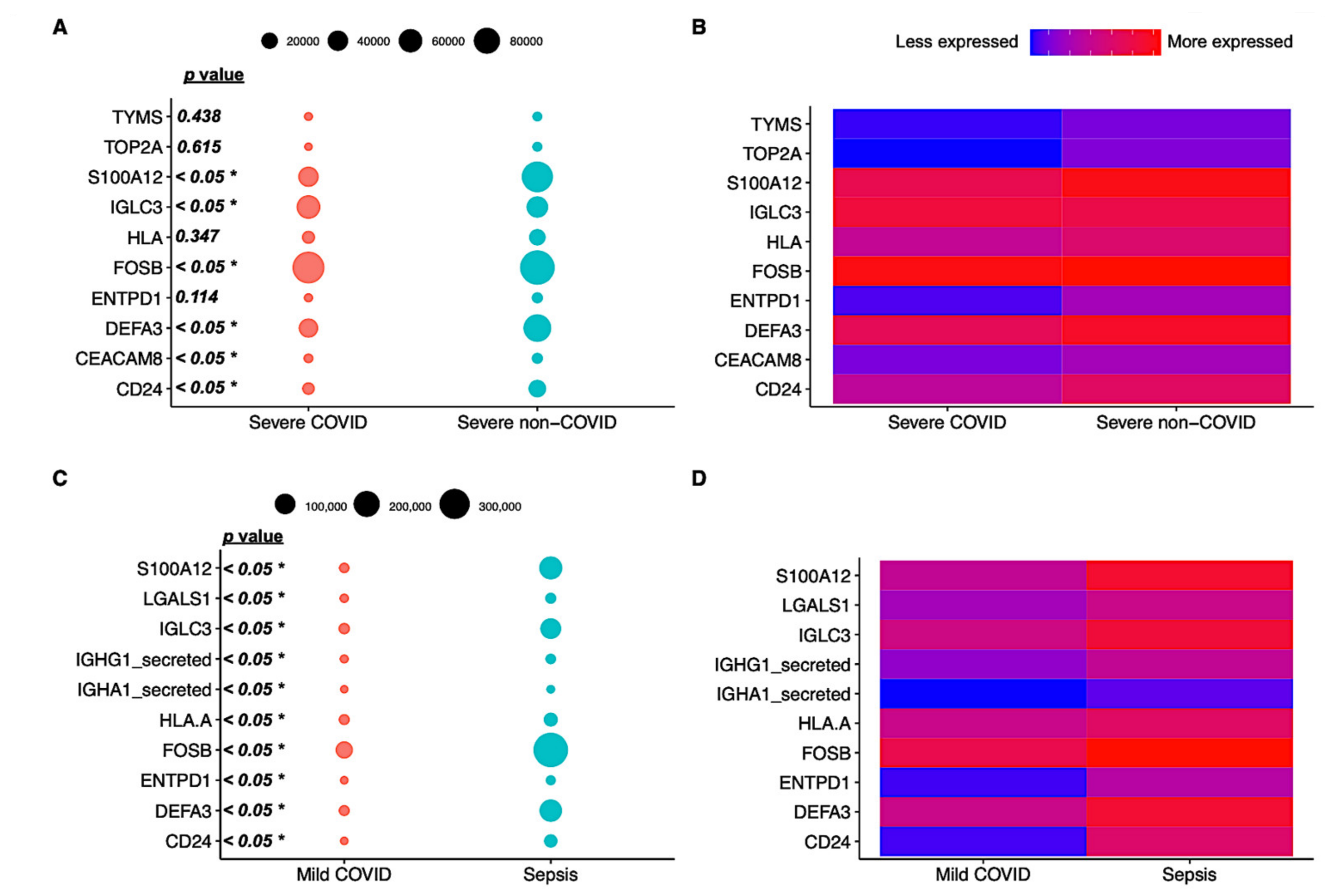
3.1. Differential Expression Profiles in Bacterial and Viral-Induced Sepsis
3.2. Mild and Severe Manifestations of Infections Have Varied Immune Cell Profiling
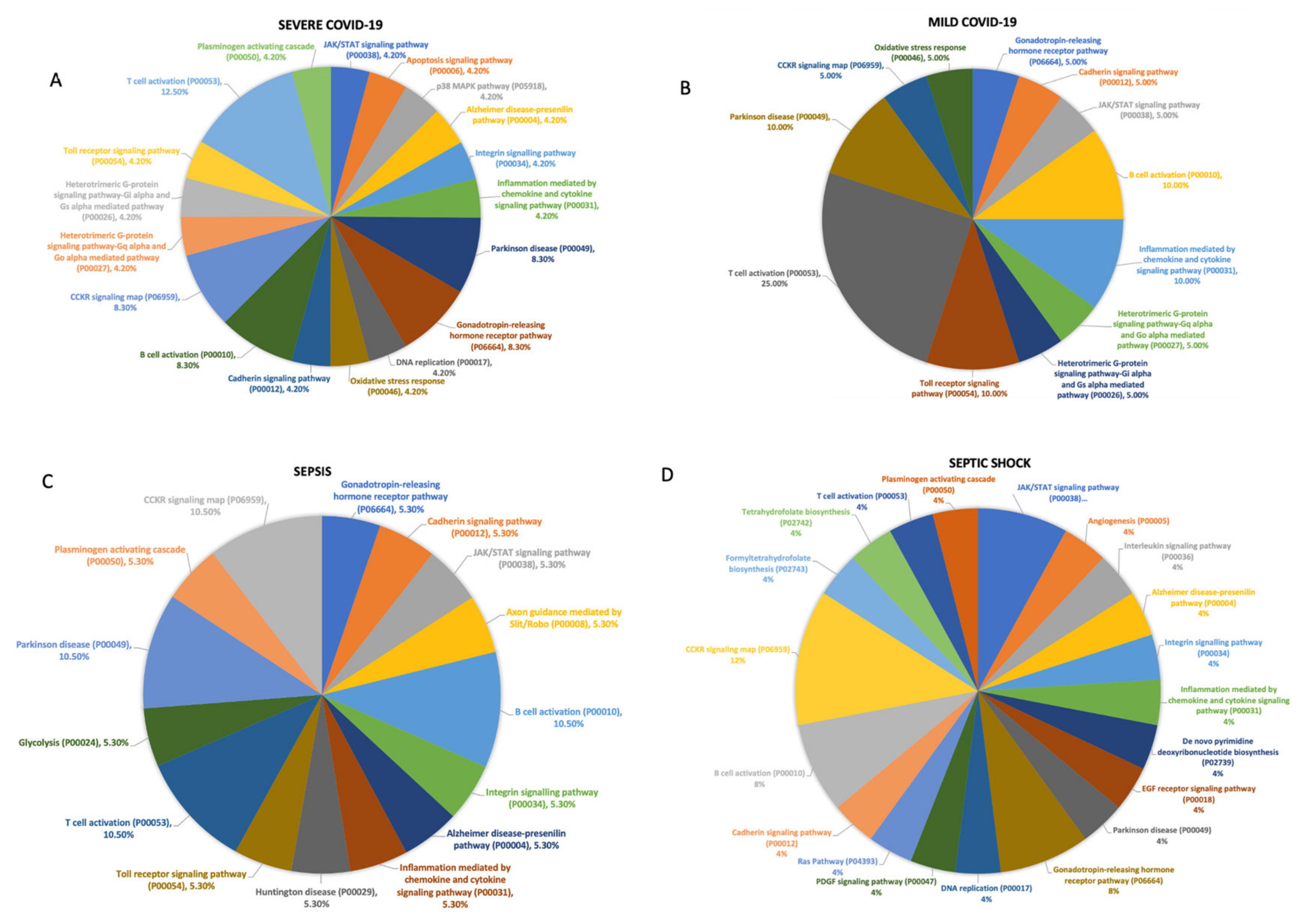
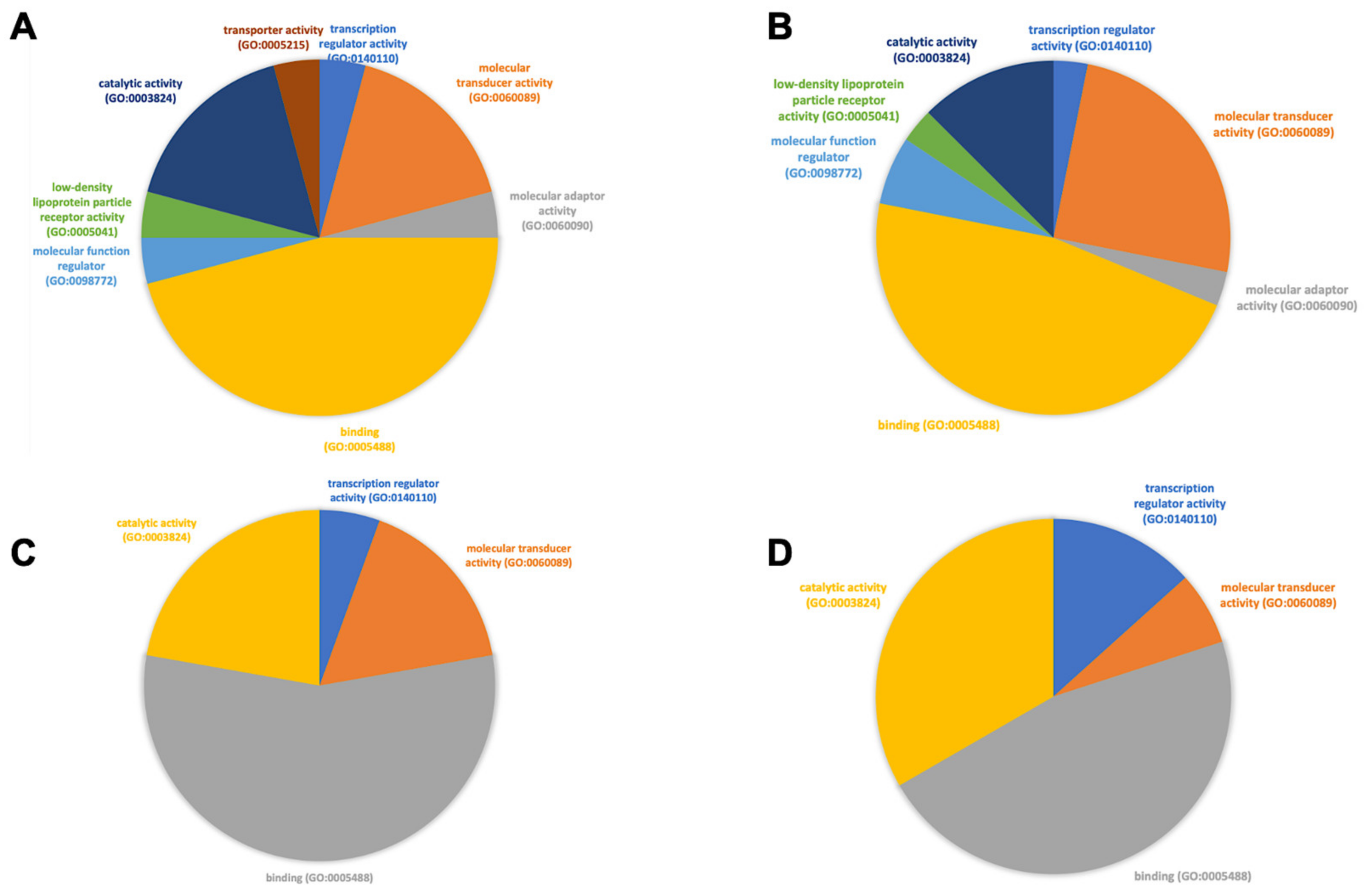
3.3. Functional Analysis of Genes Expressed in Different Cell Subtypes across Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torres, L.K.; Pickkers, P.; van der Poll, T. Sepsis-induced immunosuppression. Annu. Rev. Physiol. 2022, 84, 157–165. [Google Scholar] [CrossRef]
- Du, J.; Wei, L.; Li, G.; Hua, M.; Sun, Y.; Wang, D.; Han, K.; Yan, Y.; Song, C.; Song, R.; et al. Persistent high percentage of HLA-DR+CD38high CD8+ T cells associated with immune disorder and disease severity of COVID-19. Front. Immunol. 2021, 12, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.Q.; Li, Z.X.; Wang, L.X.; Li, Y.X.; Zheng, L.Y.; Dong, N.; Wu, Y.; Xia, Z.F.; Billiar, T.R.; Ren, C.; et al. Single-cell transcriptome profiling of the immune space-time landscape reveals dendritic cell regulatory program in polymicrobial sepsis. Theranostics 2022, 12, 4606. [Google Scholar] [CrossRef] [PubMed]
- Benlyamani, I.; Venet, F.; Coudereau, R.; Gossez, M.; Monneret, G. Monocyte HLA-DR measurement by flow cytometry in COVID-19 patients: An interim review. Cytom. Part A 2020, 97, 1217–1221. [Google Scholar] [CrossRef]
- Rhee, C.; Jones, T.M.; Hamad, Y.; Pande, A.; Varon, J.; O’Brien, C.; Anderson, D.J.; Warren, D.K.; Dantes, R.B.; Epstein, L.; et al. Prevalence, underlying causes, and preventability of sepsis-associated mortality in US acute care hospitals. JAMA Netw. Open 2019, 18, 4–11. [Google Scholar] [CrossRef]
- Alliance, G.S. Sepsis-A Global Health Crisis. 2019. Available online: https://www.global-sepsis-alliance.org/sepsis (accessed on 27 October 2022).
- Karakike, E.; Giamarellos-Bourboulis, E.J.; Kyprianou, M.; Fleischmann-Struzek, C.; Pletz, M.W.; Netea, M.G.; Reinhart, K.; Kyriazopoulou, E. Coronavirus disease 2019 as cause of viral sepsis: A systematic review and meta-analysis. Crit. Care Med. 2021, 49, 2042. [Google Scholar] [CrossRef] [PubMed]
- Ahern, D.J.; Ai, Z.; Ainsworth, M.; Allan, C.; Allcock, A.; Angus, B.; Ansari, M.A.; Arancibia-Cárcamo, C.V.; Aschenbrenner, D.; Attar, M.; et al. A Blood Atlas of COVID-19 Defines Hallmarks of Disease Severity and Specificity. Cell 2022, 185, 916–938.e58. [Google Scholar] [CrossRef]
- He, L.; Zhang, Q.; Zhang, Y.; Fan, Y.; Yuan, F.; Li, S. Single-cell analysis reveals cell communication triggered by macrophages associated with the reduction and exhaustion of CD8+ T cells in COVID-19. Cell Commun. Signal. 2021, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Ghaferi, A.A.; Birkmeyer, J.D.; Dimick, J.B. Complications, failure to rescue, and mortality with major inpatient surgery in medicare patients. Ann. Surg. 2009, 250, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Alhazzani, W.; Møller, M.H.; Arabi, Y.M.; Loeb, M.; Gong, M.N.; Fan, E.; Oczkowski, S.; Levy, M.M.; Derde, L.; Dzierba, A.; et al. Surviving Sepsis Campaign: Guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med. 2020, 48, 854–887. [Google Scholar] [CrossRef]
- Li, H.; Liu, L.; Zhang, D.; Xu, J.; Dai, H.; Tang, N.; Su, X.; Cao, B. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 2020, 395, 1517–1520. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.-Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Herminghaus, A.; Osuchowski, M.F. How sepsis parallels and differs from COVID-19. eBioMedicine 2022, 86, 104355. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020, 182, 1419–1440. [Google Scholar] [CrossRef] [PubMed]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Kaiser, K.M.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 2021, 54, 2650–2669. [Google Scholar] [CrossRef]
- Reyes, M.; Filbin, M.R.; Bhattacharyya, R.P.; Sonny, A.; Mehta, A.; Billman, K.; Kays, K.R.; Pinilla-Vera, M.; Benson, M.E.; Cosimi, L.A.; et al. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci. Transl. Med. 2021, 13, eabe9599. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021, 184, 1895–1913. [Google Scholar] [CrossRef] [PubMed]
- Feyaerts, D.; Hedou, J.; Gillard, J.; Chen, H.; Tsai, E.S.; Peterson, L.S.; Ando, K.; Manohar, M.; Do, E.; Dhondalay, G.K.R.; et al. Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19. Cell Rep. Med. 2021, 7, 100680. [Google Scholar] [CrossRef]
- COVID Coronavirus. Global Cases by the Center for Systems Science and Engineering (CSSE). Available online: https://coronavirus.jhu.edu/map.html (accessed on 29 September 2021).
- Riché, F.; Chousterman, B.G.; Valleur, P.; Mebazaa, A.; Launay, J.M.; Gayat, E. Protracted immune disorders at one year after ICU discharge in patients with septic shock. Crit. Care 2018, 22, 42. [Google Scholar] [CrossRef]
- Shankar-Hari, M.; Saha, R.; Wilson, J.; Prescott, H.C.; Harrison, D.; Rowan, K.; Rubenfeld, G.D.; Adhikari, N.K.J. Rate and risk factors for rehospitalisation in sepsis survivors: Systematic review and meta-analysis. Intensive Care Med. 2020, 46, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Calsavara, A.J.C.; Nobre, V.; Barichello, T.; Teixeira, A.L. Post-sepsis cognitive impairment and associated risk factors: A systematic review. Aust. Crit. Care 2016, 31, 242–253. [Google Scholar] [CrossRef]
- Lai, C.C.; Lee, M.T.; Lee, W.C.; Chao, C.C.; Hsu, T.C.; Lee, S.H.; Lee, C.C. Susceptible period for cardiovascular complications in patients recovering from sepsis. CMAJ 2018, 36, E1062–E1069. [Google Scholar] [CrossRef]
- Amornphimoltham, P.; Yuen, P.S.T.; Star, R.A.; Leelahavanichkul, A. Gut Leakage of Fungal-Derived Inflammatory Mediators: Part of a Gut-Liver-Kidney Axis in Bacterial Sepsis. Dig. Dis. Sci. 2019, 64, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Sepsis Alliance: Post-Sepsis Syndrome—PSS. Available online: http://www.sepsis.org/life-after-sepsis/post-sepsis-syndrome (accessed on 20 September 2022).
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Iba, T.; Arakawa, M.; Mochizuki, K.; Nishida, O.; Wada, H.; Levy, J.H. Usefulness of measuring changes in SOFA score for the prediction of 28-day mortality in patients with sepsis-associated disseminated intravascular coagulation. Clin. Appl. Thromb. Hemost. 2019, 25, 1076029618824044. [Google Scholar] [CrossRef] [PubMed]
- Ståhlberg, A.; Rusnakova, V.; Kubista, M. The added value of single-cell gene expression profiling. Brief. Funct. Genom. 2013, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.; Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 2022, 43, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, B.; Cosme, J.; Dupuis, C.; Coupez, E.; Adda, M.; Calvet, L.; Fabre, L.; Saint-Sardos, P.; Bereiziat, M.; Vidal, M.; et al. Severe COVID-19 is characterized by the co-occurrence of moderate cytokine inflammation and severe monocyte dysregulation. EBioMedicine 2021, 73, 103622. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, X.; Liu, Z.; Yao, T.; Zheng, D.; Gan, J.; Yu, S.; Li, L.; Chen, P.; Sun, J. Single-cell RNA sequencing reveals the sustained immune cell dysfunction in the pathogenesis of sepsis secondary to bacterial pneumonia. Genomics 2021, 113, 1219–1233. [Google Scholar] [CrossRef]
- Zhang, B.; Moorlag, S.J.; Dominguez-Andres, J.; Bulut, Ö.; Kilic, G.; Liu, Z.; van Crevel, R.; Xu, C.J.; Ab Joosten, L.; Netea, M.G.; et al. Single-cell RNA sequencing reveals induction of distinct trained-immunity programs in human monocytes. J. Clin. Investig. 2022, 132, e147719. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Vuerich, M.; Kalbasi, A.; Graham, J.J.; Csizmadia, E.; Manickas-Hill, Z.J.; Woolley, A.; David, C.; Miller, E.M.; Gorman, K.; et al. Limited TCR repertoire and ENTPD1 dysregulation mark late-stage COVID-19. iScience 2021, 24, 103205. [Google Scholar] [CrossRef]
- Leventogiannis, K.; Kyriazopoulou, E.; Antonakos, N.; Kotsaki, A.; Tsangaris, I.; Markopoulou, D.; Grondman, I.; Rovina, N.; Theodorou, V.; Antoniadou, E.; et al. Toward personalized immunotherapy in sepsis: The PROVIDE randomized clinical trial. Cell Rep. Med. 2022, 11, 100817. [Google Scholar] [CrossRef] [PubMed]
- Washburn, M.L.; Wang, Z.; Walton, A.H.; Goedegebuure, S.P.; Figueroa, D.J.; Van Horn, S.; Grossman, J.; Remlinger, K.; Madsen, H.; Brown, J.; et al. T cell–and monocyte-specific RNA-sequencing analysis in septic and nonseptic critically ill patients and in patients with cancer. J. Immunol. 2019, 203, 1897–1908. [Google Scholar] [CrossRef]
- Yao, C.; Bora, S.A.; Parimon, T.; Zaman, T.; Friedman, O.A.; Palatinus, J.A.; Surapaneni, N.S.; Matusov, Y.P.; Chiang, G.C.; Kassar, A.G.; et al. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021, 34, 108943. [Google Scholar] [CrossRef]
- Bos, L.D.; Sjoding, M.; Sinha, P.; Bhavani, S.V.; Lyons, P.G.; Bewley, A.F.; Botta, M.; Tsonas, A.M.; Neto, A.S.; Schultz, M.J.; et al. Longitudinal respiratory subphenotypes in patients with COVID-19-related acute respiratory distress syndrome: Results from three observational cohorts. Lancet Respir. Med. 2021, 12, 1377–1386. [Google Scholar] [CrossRef]
- Lee, S.; Song, J.; Park, D.W.; Seok, H.; Ahn, S.; Kim, J.; Park, J.; Cho, H.J.; Moon, S. Diagnostic and prognostic value of presepsin and procalcitonin in non-infectious organ failure, sepsis, and septic shock: A prospective observational study according to the Sepsis-3 definitions. BMC Infect. Dis. 2022, 22, 8. [Google Scholar] [CrossRef] [PubMed]
- Seymour, C.W.; Liu, V.X.; Iwashyna, T.J.; Brunkhorst, F.M.; Rea, T.D.; Scherag, A.; Rubenfeld, G.; Kahn, J.M.; Shankar-Hari, M.; Singer, M.; et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 762–774. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Crit. Care Med. 2021, 49, e1063–e1143. [Google Scholar] [CrossRef] [PubMed]
- Lacquaniti, A.; Campo, S.; Falliti, G.; Caruso, D.; Gargano, R.; Giunta, E.; Monardo, P. Free Light Chains, High Mobility Group Box 1, and Mortality in Hemodialysis Patients. J. Clin. Med. 2022, 23, 6904. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Lacquaniti, A.; Trombetta, D.; Smeriglio, A.; Monardo, P. Immune System Dysfunction and Inflammation in Hemodialysis Patients: Two Sides of the Same Coin. J. Clin. Med. 2022, 11, 3759. [Google Scholar] [CrossRef] [PubMed]
- Wattenberg, M.; Viégas, F.; Johnson, I. How to use t-SNE effectively. Distill 2016, 10, e2. [Google Scholar] [CrossRef]
- Riva, G.; Castellano, S.; Nasillo, V.; Ottomano, A.M.; Bergonzini, G.; Paolini, A.; Lusenti, B.; Milić, J.; De Biasi, S.; Gibellini, L.; et al. Monocyte Distribution Width (MDW) as novel inflammatory marker with prognostic significance in COVID-19 patients. Sci. Rep. 2021, 11, 12716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Xia, H.; Guo, J.; He, K.; Huang, C.; Luo, R.; Chen, Y.; Xu, K.; Gao, H.; et al. Identification of monocytes associated with severe COVID-19 in the PBMCs of severely infected patients through single-cell transcriptome sequencing. Engineering, 2021; in press. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, C.; Cai, P.; Shen, Z.; Sun, W.; Xu, H.; Fang, M.; Yao, X.; Zhu, L.; Gao, X.; et al. Single-cell analysis of COVID-19, sepsis, and HIV infection reveals hyperinflammatory and immunosuppressive signatures in monocytes. Cell Rep. 2021, 37, 109793. [Google Scholar] [CrossRef]
- Kaufmann, S.H.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35. [Google Scholar] [CrossRef]
- Carvelli, J.; Piperoglou, C.; Bourenne, J.; Farnarier, C.; Banzet, N.; Demerlé, C.; Gainnier, M.; Vély, F. Imbalance of Circulating Innate Lymphoid Cell Subpopulations in Patients with Septic Shock. Front. Immunol. 2019, 10, 2179. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.F.; Almansa, R.; Tedim, A.P.; De La Fuente, A.; Eiros, J.M.; Torres, A.; Kelvin, D.J. Mounting evidence of impaired viral control in severe COVID-19. Lancet Microbe 2021, 2, e228–e229. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.; Ortiz de Lejarazu, R.; Pumarola, T.; Rello, J.; Almansa, R.; Ramirez, P.; Martin-Loeches, I.; Varillas, D.; Gallegos, M.; Seron, C.; et al. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care 2009, 13, R201. [Google Scholar] [CrossRef]
- Mukund, K.; Nayak, P.; Ashokkumar, C.; Rao, S.; Almeda, J.; Betancourt-Garcia, M.M.; Sindhi, R.; Subramaniam, S. Immune Response in Severe and Non-Severe Coronavirus Disease 2019 (COVID-19) Infection: A Mechanistic Landscape. Front. Immunol. 2021, 12, 12. [Google Scholar] [CrossRef]
- Foell, D.; Wittkowski, H.; Kessel, C.; Lüken, A.; Weinhage, T.; Varga, G.; Vogl, T.; Wirth, T.; Viemann, D.; Björk, P.; et al. Proinflammatory S100A12 can activate human monocytes via toll-like receptor 4. Am. J. Respir. Crit. Care Med. 2013, 187, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Han, N.; Shen, Y. S100A12 promotes inflammation and cell apoptosis in sepsis-induced ARDS via activation of NLRP3 in fl ammasome signaling. Mol. Immunol. 2020, 122, 38–48. [Google Scholar] [CrossRef]
- Liu, D.; Huang, S.Y.; Sun, J.H.; Zhang, H.C.; Cai, Q.L.; Gao, C.; Li, L.; Cao, J.; Xu, F.; Zhou, Y.; et al. Sepsis-induced immunosuppression: Mechanisms, diagnosis and current treatment options. Mil. Med. Res. 2022, 9, 56. [Google Scholar] [CrossRef]
- Jin, B.; Liang, Y.; Liu, Y.; Zhang, L.X.; Xi, F.Y.; Wu, W.J.; Li, Y.; Liu, G.H. Notch signaling pathway regulates T cell dysfunction in septic patients. Int. Immunopharmacol. 2019, 76, 105907. [Google Scholar] [CrossRef]
- Brady, J.; Horie, S.; Laffey, J.G. Role of the adaptive immune response in sepsis. Intensive Care Med. Exp. 2020, 8, 20. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, D.; Li, X.; Griffith, L.; Chang, J.; An, P.; Guo, J.T. Interferon control of human coronavirus infection and viral evasion: Mechanistic insights and implications for antiviral drug and vaccine development. J. Mol. Biol. 2022, 434, 167438. [Google Scholar] [CrossRef]
- Kim, O.Y.; Monsel, A.; Bertrand, M.; Coriat, P.; Cavaillon, J.M.; Adib-Conquy, M. Differential down-regulation of HLA-DR on monocyte subpopulations during systemic inflammation. Crit. Care 2010, 14, R61. [Google Scholar] [CrossRef]
- De Waele, J.J.; Akova, M.; Antonelli, M.; Canton, R.; Carlet, J.; De Backer, D.; Dimopoulos, G.; Garnacho-Montero, J.; Kesecioglu, J.; Lipman, J.; et al. Antimicrobial resistance and antibiotic stewardship programs in the ICU: Insistence and persistence in the fight against resistance. A position statement from ESICM/ESCMID/WAAAR round table on multi-drug resistance. Intensive Care Med. 2018, 44, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Zahar, J.R.; Timsit, J.F. Risk stratification for selecting empiric antibiotherapy during and after COVID-19. Curr. Opin. Infect. Dis. 2022, 35, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Q.; Cai, S.; Peng, H.; Huyan, T.; Yang, H. The role of NK cells in fighting the virus infection and sepsis. Int. J. Med. Sci. 2021, 18, 3236–3248. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraman, M.; Muthu, S.; Bapat, A.; Jain, R.; Sushmitha, E.S.; Gulati, A.; Anudeep, T.C.; Dilip, S.J.; Jha, N.K.; Kumar, D.; et al. Bracing NK cell based therapy to relegate pulmonary inflammation in COVID-19. Heliyon 2021, 7, e07635. [Google Scholar] [CrossRef] [PubMed]
- Jensen, I.J.; McGonagill, P.W.; Butler, N.S.; Harty, J.T.; Griffith, T.S.; Badovinac, V.P. NK Cell-Derived IL-10 Supports Host Survival during Sepsis. J. Immunol. 2021, 206, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, L.; Luo, Z.; Xu, J.; Guan, W.; Xie, L. A Shift Toward Activated Receptors and Enhanced Effector Functions on NK Cells Contribute to Immune Imbalance During Sepsis. Res. Sq. 2022, 2–20. [Google Scholar]


| DIAGNOSIS | SEPSIS | SEPTIC SHOCK | SEVERE COVID-19 | MILD COVID-19 |
|---|---|---|---|---|
| Age | 57.25 | 65.00 | 65.25 | 76.33 |
| male Sex (%) | 75 | 75 | 50 | 66 |
| Weight (kg) | 86.25 | 100.00 | 82.35 | 76.27 |
| Height (cm) | 183.63 | 159.00 | 170.50 | 171.00 |
| Surgical Admission | 0.25 | 0.50 | 0.25 | 0.33 |
| TEMP (°C) | 37.88 | 39.70 | 37.30 | 37.47 |
| RR (bpm) | 29 | 36 | 30 | 22 |
| GCS | 3 | 3 | 7 | 15 |
| PaO2/FiO2 (worst) | 15 | 17 | 15 | 29 |
| LACTATE (mmol/L) | 2.3 | 2.86 | 2.43 | 2.77 |
| Noradrenaline dose day of sample, min-max (mean), mcg/kg/min | 0–0.667(0.149) | 0.02–0.353(0.115) | 0–0.169(0.031) | 0.00 |
| CRRT (%) | 25 | 25 | 50 | 0 |
| CREATININE (mmol/L) | 152.50 | 182.00 | 92.50 | 95.67 |
| egfr (ml/min/1.73 m2) | 19.67 | 33.00 | 59.50 | 52.00 |
| BILIRUBIN (µmol/L) | 21 | 18 | 12 | 17 |
| HB (g/dL) | 10.33 | 7.10 | 12.35 | 11.67 |
| PLT (109/L) | 305.00 | 717.00 | 346.75 | 332.00 |
| WCC (109/L) | 28.83 | 18.60 | 12.83 | 8.70 |
| APTT (seconds) | 42.73 | 30.20 | 54.50 | 31.67 |
| CRP (mg/L) | 229.70 | 341.89 | 74.35 | 96.80 |
| PCT (ng/mL) | 0.32 | 0.48 | 0.21 | |
| FERRITIN (µg/L) | 15,207.40 | 1469.00 | 763.93 | |
| DDIMER (mg/L FEU) | 1378.00 | 3386.75 | 780.00 | |
| NEUTS (109/L) | 25.43 | 14.90 | 10.85 | 5.17 |
| LYMPHOCYTES (109/L) | 0.38 | 0.80 | 0.98 | 0.77 |
| APACHE | 37.00 | 41.00 | 21.00 | 9.00 |
| SOFA | 9.75 | 12.75 | 7.00 | 1.33 |
| Hospital Length of Stay (days) | 124 | 149 | 17 | 31 |
| Survivors (%) | 75 | 75 | 75 | 66 |
| ICU LOS (days) | 24.25 | 38.5 | 24.5 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garduno, A.; Martinez, G.S.; Ostadgavahi, A.T.; Kelvin, D.; Cusack, R.; Martin-Loeches, I. Parallel Dysregulated Immune Response in Severe Forms of COVID-19 and Bacterial Sepsis via Single-Cell Transcriptome Sequencing. Biomedicines 2023, 11, 778. https://doi.org/10.3390/biomedicines11030778
Garduno A, Martinez GS, Ostadgavahi AT, Kelvin D, Cusack R, Martin-Loeches I. Parallel Dysregulated Immune Response in Severe Forms of COVID-19 and Bacterial Sepsis via Single-Cell Transcriptome Sequencing. Biomedicines. 2023; 11(3):778. https://doi.org/10.3390/biomedicines11030778
Chicago/Turabian StyleGarduno, Alexis, Gustavo Sganzerla Martinez, Ali Toloue Ostadgavahi, David Kelvin, Rachael Cusack, and Ignacio Martin-Loeches. 2023. "Parallel Dysregulated Immune Response in Severe Forms of COVID-19 and Bacterial Sepsis via Single-Cell Transcriptome Sequencing" Biomedicines 11, no. 3: 778. https://doi.org/10.3390/biomedicines11030778
APA StyleGarduno, A., Martinez, G. S., Ostadgavahi, A. T., Kelvin, D., Cusack, R., & Martin-Loeches, I. (2023). Parallel Dysregulated Immune Response in Severe Forms of COVID-19 and Bacterial Sepsis via Single-Cell Transcriptome Sequencing. Biomedicines, 11(3), 778. https://doi.org/10.3390/biomedicines11030778







